Abstract
The NAv1.5 sodium channel α subunit is the predominant α-subunit expressed in the heart and is associated with cardiac arrhythmias. We tested five previously identified SCN5A variants (rs7374138, rs7637849, rs7637849, rs7629265, and rs11129796) for an association with PR interval and QRS duration in two unique study populations: the Third National Health and Nutrition Examination Survey (NHANES III, n= 552) accessed by the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) and a combined dataset (n= 455) from two biobanks linked to electronic medical records from Vanderbilt University (BioVU) and Northwestern University (NUgene) as part of the electronic Medical Records & Genomics (eMERGE) network. A meta-analysis including all three study populations (n~4,000) suggests that eight SCN5A associations were significant for both QRS duration and PR interval (p<5.0E-3) with little evidence for heterogeneity across the study populations. These results suggest that published SCN5A associations replicate across different study designs in a meta-analysis and represent an important first step in utility of multiple study designs for genetic studies and the identification/characterization of genetic variants associated with ECG traits in African-descent populations.
Keywords: electrocardiographic traits, African Americans, genetic association study, electronic medical records, eMERGE, epidemiology, NHANES
1 Introduction
A necessary step to establish a robust genotype-phenotype relationship in the literature is statistical replication. There are multiple challenges with replication studies, one of which is the phenotypic heterogeneity that exists between the original study and the replication study population(s). Although the use of quantitative traits or intermediate phenotypes for both the discovery and replication study can alleviate some of the expected phenotypic heterogeneity observed for complex diseases, between study differences may not be eliminated. Therefore, it is ideal to include multiple independent studies to confirm a robust genotype-phenotype association.
The quantitative traits examined here are derived from electrocardiograms (ECGs). The ECG is a useful tool in assessing electrical conduction in the heart, and perturbations in the ECG are routinely used to diagnose cardiac arrhythmias, myocardial infarction, pericarditis, and other cardiac abnormalities by measuring and recording the electrical activity of the heart [1]. The ECG begins with the P wave that occurs during atrial depolarization, and represents the electrical impulse from the sinoatrial (SA) node towards the atrioventricular (AV) node that then spreads to the left and right atrium. The PR interval is the time the electrical impulse takes to go to the sinus node to the AV node then to the ventricles (lower chambers of the heart). It is measured from the P wave to the start of the QRS complex. The QRS marks the start of depolarization of the ventricles, and the ST segment represents the ventricle once they have fully depolarized. After depolarization, the left and right ventricles repolarize, represented as the T wave on the ECG. The QT interval represents the time it takes the ventricles to depolarize and repolarize and is measured in the ECG from the start of the QRS complex to the end of the T wave. In this study, we evaluate the associations with PR interval (representing atrioventricular conduction) and QRS duration (representing intraventricular conduction).
Slower cardiac conduction is thought to contribute to cardiac arrhythmias[2]. The PR interval is influenced primarily by body mass index, increased age, and height. QRS duration is influenced sex and somewhat by body mass index and height[3]. Clinical factors, such as hypertension, cardiac disease, and medications, in addition to genetic factors, can also cause abnormal electrical activity in the heart. Heritability studies suggest that >35% of the variation in ECG traits can be explained by genetics [4–6]. Several genetic association studies of ECG traits have been focused on genes that encode for proteins in voltage-gated ion channels [6–12]. The NAv1.5 sodium channel α subunit is the predominant α-subunit expressed in myocytes and is encoded by the SCN5A gene, located on chromosome 3 [11,13]. Genetic association studies have identified variants in the SCN5A gene that are associated with long QT syndrome and Brugada syndrome [14–17]. Common variants in SCN5A that are associated with longer QRS are also associated with atrial fibrillation[18]. These associations have been reported to explain ~2% of the variation of ECG traits [19].
There have been several candidate gene studies performed in African Americans for SCN5A and ECG traits [19–21]. Although there are few genome-wide association studies in African descent populations for ECG traits, there have been at least three GWAS or fine-mapping studies on various ECG traits in African Americans [22–24]. These studies typically include African Americans ascertained from epidemiological longitudinal studies focused on cardiovascular diseases (CVD). In the present study we sought to replicate previously reported associations in SCN5A in two study populations of African Americans: the Third National Health and Nutrition Survey (NHANES III, n= 552) and a combined dataset (n= 455) from two biobanks linked to electronic medical records from Vanderbilt University (BioVU) and Northwestern University (NUgene) as part of the Electronic Medical Records and Genomics (eMERGE) network.
2 Methods
2.1 Study populations and ECG measurements
African Americans from two study populations were used for the present study (Table 1): the Third National Health and Nutrition Examination Survey (NHANES III) and participants from two biobanks, the Vanderbilt Genome-Electronic Records (VGER) and the Northwestern biobank (NUgene) as part of the eMERGE network [31]. All ECG traits followed a normal distribution and participants with QRS duration >120 m/sec were excluded from all analyses [32].
Table 1.
Population characteristics.
| Trait | Original Study (n= 3,054 )* |
NHANES III (n= 552) |
eMERGE (n = 455) |
|||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
|
Age (yrs) |
56.5 | ±11.73 | 53.9 | ±11.61 | 46 | ±15 |
|
Sex (% female) |
62% | n/a | 67% | n/a | 77% | n/a |
|
Body mass index (BMI) |
not reported | 29 | ±6 | 34 | ±10 | |
|
Type 2 diabetes (%) |
19% | n/a | 8% | n/a | 20% | n/a |
|
PR interval (msec) |
171.6 | ±33.02 | 164.6 | ±25.56 | 159 | ±21 |
|
QRS duration (msec) |
92.3 | ±10.12 | 95.5 | ±10.90 | 82 | ±8 |
Original Study data represent data from the Jackson Heart Study abstracted from Jeff et al 2011[19]. Mean and standard deviation was calculated for ECG traits, age, and sex for African Americans from the epidemiologic (NHANES III) and clinic-based (VGER/NUgene) study populations. Analysis of variance statistical test was performed to determine significant differences across study populations (bolded italicized data denote p<0.001 for all tests).
NHANES III was conducted from 1988–1994 as a complex survey that over-sampled minorities, the young, and the elderly [33]. Biospecimens for DNA extraction were collected in phase 2 of NHANES III (1991–1994). All NHANES participants were interviewed for demographic, socioeconomic, dietary, and health-related data. Additionally, all NHANES study participants undergo a detailed medical examination at a central location known as the Mobile Examination Center (MEC). Electrocardiograms (ECGs) were recorded on adult (40 years of age or greater) men and women in the mobile examination center (MEC) using a standard 12-lead resting ECG [33]. ECGs were recorded using the Marquette MAC 12 (Marquette Medical Systems, Inc, Milwaukee, Wisconsin) (U.S. DHHS, 1996). NHANES III 12-lead ECG data were recorded with eight independent components of the 12 standard leads simultaneously. ECG data were also sampled at 250 samples per second per channel; giving the availability of multiple simultaneous ECG leads for analysis. This study was limited to self-identified non-Hispanic blacks (referred to here as African Americans) in NHANES III with normal ECG measurements. All procedures were approved by the CDC Ethics Review Board and written informed consent was obtained from all participants. Because no identifying information was accessed by the investigators, Vanderbilt University’s Institutional Review Board determined that this study met the criteria of “non-human subjects.”
VGER and NUgene are study sites of the National Human Genome Research Institute’s electronic MEdical Records and GEnomics (eMERGE) Network [31]. The Vanderbilt study site (VGER) accesses BioVU, which is a collection of DNA samples extracted from discarded blood samples collected for routine clinical care linked to de-identified electronic medical records (EMRs) [34]. The Northwestern biobank, NUgene, combines DNA samples from consented participants with an enrollment questionnaire and longitudinal data from the EMR [31]. Study individuals from both sites were identified using a previously validated algorithm that used ECGs, laboratory data, medication exposures, and natural language processing of clinical notes [35]. Study participants included in this study had a normal ECG without evidence of cardiac disease (or abnormal ECG) before or within one month following the ECG, without concurrent use of medications that interfere with QRS duration, and who did not have abnormal electrolyte values at the time of the ECG. All ECGs had normal Bazett’s corrected QT intervals (<450ms), heart rates (between 50–100 bpm), and QRS duration (65–120 ms). All participants were African American indicated by either observer reported (VGER) or self-reported (NUgene) ancestry. Both biobanks were approved by Institutional Review Boards at their respective sites.
2.2 Genotyping and statistical analysis
DNA was extracted from crude cell lysates from lymphoblastoid cell lines established for NHANES III participants aged 12 over [36]. We chose five SNPs that were significant at p 1.0E-4 from the original study [19] for genotyping in NHANES III. All genotyping was performed in the Center for Human Genetics Research DNA Resources Core using either Sequenom’s iPLEX Gold assay on the MassARRAY platform (San Diego, CA) or Illumina’s BeadXpress. All genotype data reported here passed CDC quality control (QC) metrics and are available for secondary analysis through CDC. All statistical analyses in NHANES III were performed using the Statistical Analysis Software (SAS v.9.2; SAS Institute, Cary, NC) either locally or via the Analytic Data Research by Email (ANDRE) portal of the CDC Research Data Center (RDC) in Hyattsville, MD.
Genotyping for VGER and NUgene was performed by the Center for Inherited Disease Research (CIDR) and the Broad Institute. All individuals that met the inclusion criteria (n = 501) were genotyped for >1.1 million SNPs using the Illumina 1M BeadChip at the Broad Institute. Data were cleaned by the eMERGE QC pipeline [37]. There were 46 individuals that did not meet the QC thresholds and were removed from further analysis.
Using standard linear regression, assuming an additive model, we tested each SNP for an association with PR interval and QRS duration. We did not test SCN5A variants with heart rate and QT interval since these SNPs were not associated with these traits in the original analysis [19]. All tests were limited to African Americans and adjusted for age and sex. We declared significance at p<0.05 uncorrected for multiple testing. Using a fixed-effects inverse-variance weighted approach, we performed a meta-analysis using the effect sizes, standard errors, and p-values from each study populations using METAL [38]. Pairwise FST was calculated between the original study and each study population using the Platform for the Analysis, Translation, and Organization of large-scale data (PLATO) [39] (Table 2).
Table 2.
Comparison of minor allele frequencies across studies.
| SNP | MA | Original Study* (n= 3,054) |
NHANES III (n =552) |
eMERGE (n= 455) |
||
|---|---|---|---|---|---|---|
| MAF | MAF | FST | MAF | FST | ||
| rs7374138 | G | 0.23 | 0.15 | <0.0001 | 0.23 | <0.0001 |
| rs7637849 | A | 0.19 | 0.20 | 0.014 | 0.20 | <0.0001 |
| rs11129796 | T | 0.15 | 0.08 | 0.019 | 0.11 | 0.005 |
| rs7629265 | T | 0.08 | 0.09 | <0.0001 | 0.08 | <0.0001 |
| rs6768664 | G | 0.36 | 0.36 | <0.0001 | 0.38 | <0.0001 |
Original Study data represent data from the Jackson Heart Study abstracted from Jeff et al 2011[19]. We calculated three-way FST for all SCN5A SNPs to test for differences between study populations.
Abbreviations: minor allele (MA) and minor allele frequency (MAF).
3 Results
We abstracted data from the previously published Jackson Heart Study [19] referred to here as the “original study” for comparison with our population-based (NHANES III) and clinical (eMERGE) collections. We compared the three study populations and observed differences across study populations for age, sex, and ECG measurements (Table 1). On average, eMERGE (46 years) and NHANES III (54 years) participants were younger compared with the original study (57 years). Both eMERGE and NHANES III had more female participants (77% and 67%, respectively) compared with the original study (62%; Table 1). Additionally, the measurements QRS duration and PR interval in eMERGE were shorter compared to the other studies, which is a reflection of the more stringent selection criteria within eMERGE to select subjects without any prior heart disease or abnormalities on their ECG.
To further characterize similarities and differences between the original study and the other two study populations, we first calculated the minor allele frequency and compared these estimates across study populations. Though not statistically significant, NHANES III had a lower minor allele frequency for SCN5A rs7374138 (0.15) and rs11129796 (0.08) compared with the original study and eMERGE (Table 2). To further characterize study population differences at these loci, we calculated FST using the Weir and Cockerham algorithm [25] between the original study and each study site separately for each SNP (Table 2). The fixation index FST is a measure of population differentiation, and an F statistic >0.15 is indicative as a significant difference between populations. As might be expected, there were no significant differences between studies for any of the SNPs tested at this stringent threshold for population differentiation (FST<0.019, Table 2).
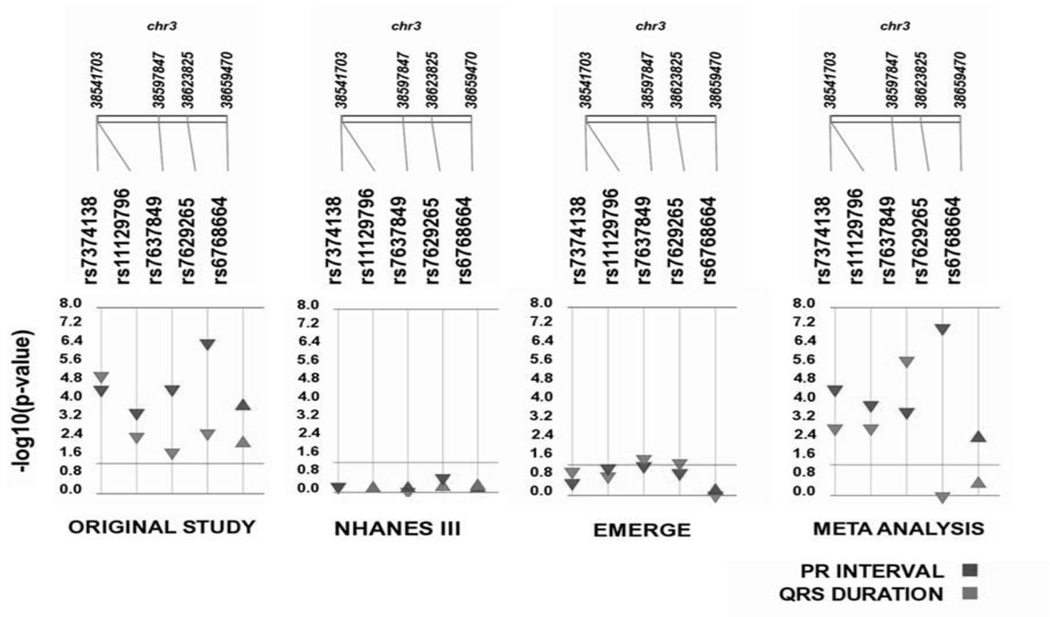
We performed single SNP tests of association for SCN5A SNPs identified in the original study with PR interval and QRS duration in eMERGE and NHANES III [19]. There were no significant associations (p<0.05) observed between SCN5A SNPs for any ECG traits in African Americans from NHANES III (Table 3, Figure). Three SNPs (rs7374138, rs7629265, and rs6768664) have a consistent direction of effect compared to the original study, despite not being statistically significant for PR interval. Likewise for QRS duration three SNPs (rs7637849, rs7374138, and rs6768664) have a consistent direction of effect compared to the original study in NHANES III.
Table 3.
Association results across three independent African American study populations and meta-analysis results across all study populations.
| SNP | CA | Original study (n = 3,054) |
NHANES III (n = 552) |
eMERGE (n = 455) |
Meta Analysis (n~4,000) |
|
|---|---|---|---|---|---|---|
| β (p) |
β (p) |
β (p) |
β (p) |
Q P |
||
| PR Interval | ||||||
| rs7374138 | G | −4.00 (2.4E-5) |
−1.65 (0.40) |
−1.94 (0.21) |
−3.17 (2.2E-5) |
0.37 |
| rs11129796 | T | −3.50 (2.4E-4) |
0.12 (0.96) |
−3.89 (0.05) |
−3.14 (1.0E-4) |
0.36 |
| rs7637849 | A | −4.20 (2.3E-5) |
0.11 (0.96) |
−3.11 (0.04) |
−3.11 (2.0E-4) |
0.17 |
| rs7629265 | T | −7.80 (2.4E-7) |
−4.13 (0.17) |
−4.00 (0.08) |
−6.32 (5.3E-8) |
0.30 |
| rs6768664 | G | 3.00 (2.4E-4) |
0.16 (0.92) |
0.16 (0.90) |
1.80 (5.0E-3) |
0.13 |
| QRS duration | ||||||
| rs7374138 | G | −1.30 (6.2E-6) |
−0.70 (0.41) |
−1.15 (0.07) |
−1.22 (1.0E-3) |
0.79 |
| rs11129796 | T | −1.10 (2.5E-3) |
0.09 (0.94) |
−1.40 (0.11) |
−1.03 (1.0E-3) |
0.52 |
| rs7637849 | A | −0.76 (1.2E-2) |
−0.39 (0.66) |
−1.55 (0.02) |
−0.85 (1.3E-6) |
0.48 |
| rs7629265 | T | −1.50 (1.8E-3) |
1.07 (0.82) |
−2.08 (0.03) |
−0.15 (0.77) |
0.01 |
| rs6768664 | G | 0.67 (9.5E-3) |
1.93 (0.73) |
−0.20 (0.71) |
0.21 (0.45) |
0.11 |
For each test of association, SNP rs number, coded allele (CA), beta, and p-value are given. For the meta-analysis, the Q p-value is also given.
Original Study data represent data from the Jackson Heart Study abstracted from Jeff et al 2011[19].
Figure. Association results for SCN5A SNPs by study site.
We performed single SNP tests of association for five previously identified SCN5A SNPs with PR interval (red) and QRS duration (blue). The direction of effect is indicated by the direction of the arrow and the −log p-value is plotted in the y-axis. SNPs are sorted based on location on chromosome 3. SNPs that met our liberal significance threshold p<0.05 are above the line.
To better understand the impact SCN5A variation has on these ECG traits, we performed a meta-analysis using the effect estimates from all three study populations (Table 3). All SCN5A SNPs were significant (p<0.05) and the direction of effect was consistent with the original study for PR interval. For QRS duration, all SCN5A SNPs have a consistent direction of effect; however, rs7629265 and rs6768664 did not meet our liberal significance threshold in NHANES III samples (Table 3). In NHANES III, having two copies of the risk allele “T” for rs7629265 increased QRS duration, whereas in both the original study and in eMERGE; having two copies of the “T” allele decreased the QRS duration.
In contrast, having two copies of the risk allele “G” at rs6768664 is associated with increased QRS duration in both the original study and NHANES III but with decreased QRS duration in eMERGE samples.
4 Discussion
In the present study we replicate and or generalize previously identified SCN5A associations in African Americans ascertained from clinical and population-based collections. Associated variants in the clinical-based study population, eMERGE, had a consistent direction of effect with previous studies in African Americans for four of the five SNPs between the two traits. However, in African Americans from the US population-based cohort, NHANES III, several associations had opposing direction of effects, and none of the SNPs tested reached our liberal significance threshold (p=0.05) for any ECG trait.
Here we tested five SNPs: rs7374138, rs11129796, rs7637849, rs7629265, and rs6768664, all located in various SCN5A introns. Consistent with published data, three of the five tested SNPs are specific to African-descent populations and are monomorphic or rare in European and Asian-descent populations [19,20,22,24,26]. While there is not a direct biological correlation with the role intronic SNPs have on protein function, intronic regions are known to play an important role in splicing, which could possibly affect protein function. Previous studies report that in African-descent populations one SNP, rs7629265, is in high linkage disequilibrium (LD, r2 = 0.87) with coding non-synonymous variant, rs7626962 (S1103Y), and has been consistently associated with long QT syndrome [26]. This SNP is associated with PR interval in the combined analysis and is not associated with QRS duration, which is also consistent with the literature [19] (Table 3).
We did not test all SCN5A SNPs reported in the literature; as a consequence, there are several important associations we did not test. In spite of this, there are several trends we observed that are consistent with previous studies which suggest testing more SNPs in these regions will likely yield the same results [19,22,24]. Another limitation to this study is sample size. Compared to the previous reports for SCN5A, both eMERGE and NHANES III were limited in sample size: n= 455 and n=520, respectively. However, despite this limitation we were able to observe a consistent direction of effect compared to original reports for most SCN5A SNPs in African Americans from eMERGE.
In addition to small sample size, differences in study population may explain the lack of replication in NHANES III African Americans. Indeed, there are several similarities and differences across study populations. We sought to replicate associations originally detected in African Americans from the Jackson Heart Study (JHS). The Jackson Heart Study is a longitudinal study collected with the primary objective to identify and explain the disparity of cardiovascular diseases in African Americans [27–29]. As a result, the JHS has more samples with in-depth phenotype information for cardiovascular diseases and related traits compared to the other studies and can possibly include individuals with CVD. Another difference between the NHANES III population compared to the other study populations could be explained by geography. Unlike NHANES III, which is representative of the US population, the JHS is limited to African Americans from the southeastern United States. Similarly, most samples from eMERGE are limited to African Americans that visit the clinic or hospital in the Nashville metropolitan areas, also in the southeastern United States. The prevalence of CVD and related environmental risk factors are disproportionately higher in the southeastern US, which likely makes up majority of the eMERGE samples tested here and all of the JHS samples[30]. Lack of replication in NHANES III might be explained by this difference. Another possibility could be the time in which the study was conducted. Both eMERGE and JHS samples were collected fairly recently (in the past 10 years) compared to NHANES III, which was collected over 20 years ago. There are several environmental factors that have changed since then, such as diet, that might be interacting with these SNPs and thus have an impact on our study and explain our failure to replicate in NHANES III. Other non-genetic risk factors such as age, sex, type 2 diabetes mellitus (T2DM) status, and BMI are associated with ECG traits [3]. Indeed, we did observe significant differences across study populations for age, sex and BMI (Table 1). Study individuals from eMERGE were more likely to be female and were significantly younger compared to the original study and NHANES III. Also, eMERGE study individuals had a higher mean BMI and higher proportion of T2DM cases compared with NHANES III. It is possible that associations between SCN5A and ECG traits are modified by environmental factors (such as poor diet and BMI). Additional analyses are needed to statistically assess the impact of gene-environment interactions on this complex trait.
Perhaps the main reason for lack of replication in NHANES III is phenotype definition. It is important to note that despite small sample sizes in the replication cohorts, SCN5A associations have a much more consistent direction of effect in eMERGE than do the NHANES III samples compared to the original study (Table 3). One of the main goals of the eMERGE consortium was to successfully replicate GWAS associations in clinical populations, thus accurately identifying phenotypes from electronic medical record (EMR) data was critical. As previously mentioned, eMERGE study participants are limited to ECG measurements in normal range without any evidence of pre-existing heart conditions, laboratory values, or medications that may alter their ECG results. The algorithm was validated by blinded physician review with a positive predictive value of 97% at both eMERGE sites participating in this study[18]. This stringent phenotype definition was not used in either the original study or NHANES III, and both had longer PR interval and QRS duration compared to eMERGE (Table 1). The broad phenotype definition might have resulted in a loss of power in NHANES III and thus explain why SCN5A variants did not replicate. While the original study also had a broad phenotype definition, the loss of power is not as significant given the large sample size, which is almost seven times larger than NHANES III.
Despite these study population differences, most tests of association were significant at p<0.05 after meta-analysis for both PR interval and QRS duration. And, tests of heterogeneity suggested little detectable differences across the studies in this meta-analysis. Overall, our data validate SCN5A associations with QRS duration and PR interval in African Americans by meta-analysis. Most importantly, this work high-lights the challenge of conducting and interpreting genetic association studies in multiple study populations with differing ascertainment strategies. This latter finding is of particular important in this era of meta-analysis of genetic association studies where study design and phenotypic precision are traded in favor of larger sample sizes for greater power.
Acknowledgements
This work was supported by NIH U01HG004798 and its ARRA supplements (EAGLE) as well as U01HG004609 (Northwestern University as part of eMERGE); U01HG04603 (Vanderbilt University as part of eMERGE, also serving as the Administrative Coordinating Center). Portions of the dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Contributor Information
Janina M. Jeff, Email: janina.jeff@mssm.edu.
Kristin Brown-Gentry, Email: kristin.gentry@healthspring.com.
Robert Goodloe, Email: robert.goodloe@gmail.com.
Marylyn D. Ritchie, Email: marylyn.ritchie@psu.edu.
Joshua C. Denny, Email: josh.denny@vanderbilt.edu.
Abel N. Kho, Email: a-kho@northwestern.edu.
Loren L. Armstrong, Email: loren-armstrong@northwestern.edu.
Bob McClellan, Jr, Email: bob.mcclellan@vanderbilt.edu.
Ping Mayo, Email: pxm304@case.edu.
Melissa Allen, Email: mjallentn@hotmail.com.
Hailing Jin, Email: hailing.jin@vanderbilt.edu.
Niloufar B. Gillani, Email: nila.gillani@vanderbilt.edu.
Nathalie Schnetz-Boutaud, Email: Nathalie.boutaud@vanderbilt.edu.
Holli H. Dilks, Email: holli.h.dilks@vanderbilt.edu.
Melissa A. Basford, Email: melissa.basford@vanderbilt.edu.
Jennifer A. Pacheco, Email: japacheco@northwestern.edu.
Gail P. Jarvik, Email: gjarvik@medicine.washington.edu.
Rex L. Chisholm, Email: r-chisholm@northwestern.edu.
Dan M. Roden, Email: dan.roden@vanderbilt.edu.
M. Geoffrey Hayes, Email: ghayes@northwestern.edu.
Dana C. Crawford, Email: dana.c.crawford@vanderbilt.edu.
References
- 1.Chizner MA. Clinical Cardiology Made Ridiculously Simple. MedMaster; Miami: 2004. [Google Scholar]
- 2.Capone RJ, Pawitan Y, el-Sherif N, Geraci TS, Handshaw K, Morganroth J, Schlant RC, Waldo AL. Events in the cardiac arrhythmia suppression trial: baseline predictors of mortality in placebo-treated patients. J Am Coll Cardiol. 1991;18:1434–1438. doi: 10.1016/0735-1097(91)90671-u. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez AH, Schildcrout JS, Blakemore DL, Masys DR, Pulley JM, Basford MA, Roden DM, Denny JC. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm. 2011;8:271–277. doi: 10.1016/j.hrthm.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akylbekova EL, Crow RS, Johnson WD, Buxbaum SG, Njemanze S, Fox E, Sarpong DF, Taylor HA, Newton-Cheh C. Clinical correlates and heritability of QT interval duration in blacks: the Jackson Heart Study. Circ Arrhythm Electrophysiol. 2009;2:427–432. doi: 10.1161/CIRCEP.109.858894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedlander Y, Siscovick DS, Weinmann S, Austin MA, Psaty BM, Lemaitre RN, Arbogast P, Raghunathan TE, Cobb LA. Family history as a risk factor for primary cardiac arrest. Circulation. 1998;97:155–160. doi: 10.1161/01.cir.97.2.155. [DOI] [PubMed] [Google Scholar]
- 6.George AL., Jr Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George AL, Jr, Varkony TA, Drabkin HA, Han J, Knops JF, Finley WH, Brown GB, Ward DC, Haas M. Assignment of the human heart tetrodotoxin-resistant voltage-gated Na+ channel alpha-subunit gene (SCN5A) to band 3p21. Cytogenet Cell Genet. 1995;68:67–70. doi: 10.1159/000133892. [DOI] [PubMed] [Google Scholar]
- 8.Gouas L, Nicaud V, Berthet M, Forhan A, Tiret L, Balkau B, Guicheney P D.E.S.I.R. Study Group. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. Eur J Hum Genet. 2005;13:1213–1222. doi: 10.1038/sj.ejhg.5201489. [DOI] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O'Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai LP, Deng CL, Moss AJ, Kass RS, Liang CS. Polymorphism of the gene encoding a human minimal potassium ion channel (minK) Gene. 1994;151:339–340. doi: 10.1016/0378-1119(94)90685-8. [DOI] [PubMed] [Google Scholar]
- 11.Gellens ME, George AL, Jr, Chen LQ, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci USA. 1992;89:554–558. doi: 10.1073/pnas.89.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 13.George AL, Jr, Iyer GS, Kleinfield R, Kallen RG, Barchi RL. Genomic organization of the human skeletal muscle sodium channel gene. Genomics. 1993;15:598–606. doi: 10.1006/geno.1993.1113. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Shen J, Li Z, Timothy K, Vincent GM, Priori SG, Schwartz PJ, Keating MT. Cardiac sodium channel mutations in patients with long QT syndrome, an inherited cardiac arrhythmia. Hum Mol Genet. 1994;4:1603–1607. doi: 10.1093/hmg/4.9.1603. [DOI] [PubMed] [Google Scholar]
- 15.Bezzina C, Veldkamp MW, van den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, van Der Hout AH, Mannens MM, Wilde AA. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 16.Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, Wilde AA, Escande D, Mannens MM, Le Marec H. Cardiac conduction defects associate with mutations in SCN5A. Nat Genetics. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, Ramirez AH, Mosley JD, Pulley JM, Basford MA, Bradford Y, Rasmussen LV, Pathak J, Chute CG, Kullo IJ, McCarty CA, Chisholm RL, Kho AN, Carlson CS, Larson EB, Jarvik GP, Sotoodehnia N, Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) QRS Group. Manolio TA, Li R, Masys DR, Haines JL, Roden DM. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–1385. doi: 10.1161/CIRCULATIONAHA.112.000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeff JM, Brown-Gentry K, Buxbaum SG, Sarpong DF, Taylor HA, George AL, Roden DM, Crawford DC. SCN5A Variation is Associated with Electrocardiographic Traits in the Jackson Heart Study. Circ Cardiovasc Genet. 2011;4:139–144. doi: 10.1161/CIRCGENETICS.110.958124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burke A, Creighton W, Mont E, Li L, Hogan S, Kutys R, Fowler D, Virmani R. Role of SCN5A Y1102 polymorphism in sudden cardiac death in blacks. Circulation. 2005;112:798–802. doi: 10.1161/CIRCULATIONAHA.104.482760. [DOI] [PubMed] [Google Scholar]
- 21.Plant LD, Bowers PN, Liu Q, Morgan T, Zhang T, State MW, Chen W, Kittles RA, Goldstein SA. A common cardiac sodium channel variant associated with sudden infant death in African Americans, SCN5A S1103Y. J Clin Invest. 2006;116:430–435. doi: 10.1172/JCI25618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JG, Magnani JW, Palmer C, Meng YA, Soliman EZ, Musani SK, Kerr KF, Schnabel RB, Lubitz SA, Sotoodehnia N, Redline S, Pfeufer A, Muller M, Evans DS, Nalls MA, Liu Y, Newman AB, Zonderman AB, Evans MK, Deo R, Ellinor PT, Paltoo DN, Newton-Cheh C, Beonjamin EJ, Mehra R, Alonso A, Heckbert SR, Fox ER. Candidate-gene Association Resource (CARe) Consortium: Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7:e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler AM, Yin X, Evans DS, Nalls MA, Smith EN, Tanaka T, Li G, Buxbaum SG, Whitsel EA, Alonso A, Arking DE, Benjamin EJ, Berenson GS, Bis JC, Chen W, Deo R, Ellinor PT, Heckbert SR, Heiss G, Hsueh WC, Keating BJ, Kerr KF, Li Y, Limacher MC, Liu Y, Lubitz SA, Marciante KD, Mehra R, Meng YA, Newman AB, Newton-Cheh C, North KE, Palmer CD, Psaty BM, Quibrera PM, Redline S, Reiner AP, Rotter JI, Schnabel RB, Schork NJ, Singleton AB, Smith JG, Soliman EZ, Srinivasan SR, Zhang ZM, Zonderman AB, Ferrucci L, Murray SS, Evans MK, Sotoodehnia N, Magnani JW, Avery CL. Novel Loci Associated with PR Interval in a Genome-Wide Association Study of Ten African American Cohorts. Circ Cardiovasc Genet. 2012;5:639–646. doi: 10.1161/CIRCGENETICS.112.963991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avery CL, Sethupathy P, Buyske S, He Q, Lin DY, Arking DE, Carty CL, Duggan D, Fesinmeyer MD, Hindorff LA, Jeff JM, Klein L, Patton KK, Peters U, Shohet RV, Sotoodehnia N, Yong AM, Kooperberg C, Haiman CA, Mohlke KL, Whitsel EA, North KE. Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans. PLoS Genet. 2012;8:e1002870. doi: 10.1371/journal.pgen.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weir BS, Cockerham CC. Estimating F- Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 26.Splawski I, Timothy KW, Tateyama M, Clancy CE, Malhotra A, Beggs AH, Cappuccio FP, Sgnella GA, Kass RS, Keating MT. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 27.Sempos CT, Bild DE, Manolio TA. Overview of the Jackson Heart Study: a study of cardiovascular diseases in African American men and women. Am J Med Sci. 1999;317:142–146. doi: 10.1097/00000441-199903000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Wilson JG, Rotimi CN, Ekunwe L, Royal CD, Crump ME, Wyatt SB, Steffes MW, Adeyemo A, Zhou J, Taylor HA, Jr., Jaquish C. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis. 2005;15:S6–S37. [PubMed] [Google Scholar]
- 29.Wyatt SB, Diekelmann N, Henderson F, Andrew ME, Billingsley G, Felder SH, Fugua S, Jackson PB. A community-driven model of research participation: the Jackson Heart Study Participant Recruitment and Retention Study. Ethn Dis. 2003;13:438–455. [PubMed] [Google Scholar]
- 30.Crook ED, Taylor H. Traditional and nontraditional risk factors for cardiovascular and renal disease in African Americans (Part 2): a project of the Jackson Heart Study investigators. Am J Med Sci. 2003;325:305–306. doi: 10.1097/00000441-200306000-00001. [DOI] [PubMed] [Google Scholar]
- 31.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. eMERGE Team: The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeff JM, Ritchie MD, Denny JC, Kho AN, Ramirez AH, Crosslin D, Armstrong L, Basford MA, Wolf WA, Pacheco JA, Chisholm RL, roden DM, Hayes MG, Crawford DC. Generalization of Variants Identified by Genome-Wide Association Studies for Electrocardiographic Traits in African Americans. Ann Hum Genet. 2013;77:321–332. doi: 10.1111/ahg.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Center for Health Statistics. Vital Health Stat 1. 1994. Centers for Disease Control and Prevention. Plan and operation of the Third National Health and Nutrition Examination Survey, 1988–1994. [PubMed] [Google Scholar]
- 34.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, Basford MA, Masys DR, Haines JL, Roden DM. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinberg KK, Sanderlin KC, Ou CY, Hannon WH, McQuillan GM, Sampson EJ. DNA banking in epidemiologic studies. Epidemiol Rev. 1997;19:156–162. doi: 10.1093/oxfordjournals.epirev.a017938. [DOI] [PubMed] [Google Scholar]
- 37.Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes MG, Jarvik GP, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto ME, McCarty CA, McDavid AN, Mirel DB, Olson LM, Paschall JE, Pugh EW, Rasmussen LV, Rasmussen-Torvik LJ, Turner SD, Wilke RA, Ritchie MD. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35:887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]