Abstract
The possibility of transplanting adult stem cells into damaged organs has opened new prospects for the treatment of several human pathologies. The purpose of this study was to develop a culture system for the expansion and production of human Periodontal Ligament Stem Cells (hPDLSCs) using a new xeno-free media formulation and ensuring the maintenance of the stem cells features comprising the multiple passage expansion, mesengenic lineage differentiation, cellular phenotype, and genomic stability, essential elements for conforming to translation to cell therapy. Somatic stem cells were isolated from the human periodontium using a minimally invasive periodontal access flap surgery in healthy donors. Expanded hPDLSCs in a xeno-free culture showed the morphological features of stem cells, expressed the markers associated with pluripotency, and a normal karyotype. Under appropriate culture conditions, hPDLSCs presented adipogenic and osteogenic potential; indeed, a very high accumulation of lipid droplets was evident in the cytoplasm of adipogenic-induced cells, and indisputable evidence of osteogenic differentiation, investigated by transmission electron microscopy, and analyzed for gene expression analysis has been shown. Based on these data, the novel xeno-free culture method might provide the basis for Good Manufacturing Procedure culture of autologous stem cells, readily accessible from human periodontium, and can be a resource to facilitate their use in human clinical studies for potential therapeutic regeneration.
Introduction
Human adult stem cells, identified in the stromal tissue-like bone marrow, spleen, and thymus, are postnatal stem cells that are able to self-renew and differentiate into multiple cell lineages such as bone, cartilage, tendon, skeleton muscle, and neuron and oral tissues.1
The oral area is a rich source of stem cells, and their characterization is important to develop new and effective strategies for dental applications and for the treatment of degenerative diseases of the skeleton.2
In the oral tissues, six different human dental stem cells have been described in literature until now: dental pulp stem cells (DPSCs),3 exfoliated deciduous teeth stem cells (SHED),4 periodontal ligament stem cells (PDLSCs),5,6 apical papilla stem cells,7 dental follicle stem cells (DFSCs),8 and gingiva stem cells.2,9
In particular, the periodontal ligament contains a population of multipotent postnatal stem cells that can be expanded ex vivo, providing an exceptional reservoir of autologous stem cells with biological characteristics similar to bone marrow-derived undifferentiated stem cells.5,10,11 It has been demonstrated that PDLSCs can differentiate into periodontal ligament (PDL) fibroblasts, osteoblasts, and cementoblasts,6 repair allogenic bone defects without immunological rejection, likely due to their low immunogenicity and immunosuppressive function,12 and in vivo PDLSCs were capable of offering optimal treatment for periodontitis.13 Considering that the periodontal disease plays a key role in a variety of systemic14–16 and oral diseases becomes urgent to find advanced therapeutic clinical interventions for periodontal regeneration using stem cells.17
Currently, in vitro expansion and culture of mesenchymal stem cells (MSCs) is founded on supplementing cell culture and differentiation media with fetal calf serum (FCS), which contains numerous growth factors inducing cell attachment to plastic surfaces, cell proliferation, and differentiation.17 Although these traditional formulations provide a high expansion of stem cells, their presence in the culture medium of FCS may trigger a xenogenic immune response, immunological reactions, and the potential transmission of prion diseases and zoonoses.18–20 Moreover, one of the central issues regarding limitations in using animal sera for cell therapy is that its components are highly variable and often unknown, and differences between lots are possible.21 Previous studies report that human platelet lysate and human plasma can replace FCS in terms of clinical-scale expansion22,23 and in vivo bone-forming capacity of human mesenchymal stromal cells.24 Human serum could be considered a suitable alternative, due to its possibility to promote osteogenic differentiation in DPSCs and to induce an efficient expansion of umbilical cord-derived stem cells,25 but this approach could be limited by the amount of autologous serum necessary to expand MSCs for clinical use and the variability of serum, especially for patients receiving previous chemotherapy.26
In any case, the elaboration of a culture medium, adaptable to the production of stem cells for the clinical application of cell therapy, remains a crucial matter, as a serum-free medium with no growth factors is unable to amplify these cells in vitro, and the type of serum used (FBS, human serum, and allogenic or autologous serum or plasma) is highly debated.27
The aim of the present study was to develop an efficient xeno-free culture system of human Periodontal Ligament Stem Cells (hPDLSCs) using a commercial medium that ensures the maintenance of stem cell features, including the multiple passage expansion, mesengenic lineage differentiation, cellular phenotype, and genomic stability, which are essential to be utilized in cell therapy.
Materials and Methods
Cell culture
Five human periodontal ligament biopsies were carried out from human premolar teeth, scheduled to be removed for orthodontic purposes, on healthy volunteers aged 20–35 years. All patients provided written consent for clinical research and for the processing of personal data. All periodontal ligament biopsies were de-identified.
Each patient was prepared for surgery by pretreatment for 1 week with professional dental hygiene. The periodontal ligament tissue was collected after tooth extraction. Explants were obtained from alveolar crest and horizontal fibers of the periodontal ligament by scraping the roots of noncarious molar teeth with a Gracey's curette.28
To assess how serum supplementation of the culture media affects the cell characteristics, hPDLSCs were isolated under two different culturing conditions: in medium containing 10% FBS (MSCGM BulletKit), standard conditions, or in TheraPEAK™MSCGM-CD™ BulletKit serum free, chemically defined (MSCGM-CD) medium, xeno-free conditions, for the growth of human MSCs (Lonza).
The specimens of periodontal tissue were cut into small pieces and after washing several times with PBS (LiStarFish), they were subsequently collected in MSCGM or in MSCGM-CD.
The medium was changed twice a week, and cells spontaneously migrating from the explant fragments after reaching about 80% of confluence were trypsinized (LiStar Fish), subsequently split, and subcultured until passage 10 (P10). Cells utilized for the experimental assays were at the second passage (P2).
Cell proliferation and viability assay
Ex vivo expanded hPDLSCs were seeded at 1×103 cells/well in triplicate using a 96-well flat-bottom plate and maintained in MSCGM or MSCGM-CD medium for 24, 48, 72 h and 1 week. After the incubation period, 15 μL/well of MTT were added to culture medium and cells were incubated for 3 h at 37°C. The supernatants were read at 650 nm wavelength using a microplate reader (Synergy HT; BioTek Instruments). Moreover, the doubling time of the trypan blue harvested cells, at 24, 48, 72 h and 1 week of culture, was calculated by using an algorithm available online (www.doubling-time.com).
Karyotyping of hPDLSCs
Metaphase chromosomes were prepared from hPDLSCs cultured with MSCGM-CD. When culture reached confluence, cells were treated with 0.05% trypsin (LiStar Fish) for 4 min at 37°C and 0.02% EDTA, replaced in amniodish, and incubated at 37°C for 24 h. For cytogenetic analysis, cultures were incubated for 40 min with Colcimide (100 ng/mL; Beit Haemek), washed with PBS, dissociated with Trypsin (LiStar Fish) for 4 min at 37°C, centrifuged at 100 g for 5 min, resuspended in 1 mL of growth medium into which 5 mL of warm KCl and sodium citrate hypotonic solution (Sigma Chemical Co) were added dropwise, incubated for 30 min at 37°C, followed by fixation at Room Temperature (RT) with 3:1 methanol/acetic acid for 5 min. GTG banding was performed by incubating the glass slides in a 0.05% trypsin solution at 37°C for 15 s, followed by rinsing the slides in phosphate-buffered saline buffer and staining in a 5% Giemsa stain for 8 min. The slides were rinsed with water and air dried.29 Thirty metaphases were examined for each sample.
Cytofluorimetric assay
Antibodies. Fluorescein isothiocyanate-conjugated anti-CD13 (CD13 FITC), phycoerythrin-conjugated anti-CD29 (CD29 PE), FITC-conjugated: anti-CD44 (CD44 FITC), anti-CD45 (CD45 FITC), anti-CD105 (CD105 FITC), and anti-CD166 (CD166 FITC) were obtained from Ancell; FITC-conjugated anti-CD14 (CD14 FITC) and PE-conjugated anti-CD133 (CD133 PE) were purchased from Milteny Biotec; PE-conjugated anti-CD73 (CD73 PE), FITC-conjugated anti-CD90 (CD90 FITC), allophycocyanin-conjugated anti-CD117 (CD117-APC), PE-conjugated anti-CD146 (CD146 PE), PE-conjugated anti-CD271 (CD271-PE), Alexa488-conjugated anti-Sox2 (Sox2 Alexa488), FITC-conjugated anti-SSEA-4 (SSEA-4 FITC), Alexa488-conjugated anti-HLA-ABC (HLA-ABC Alexa488), PE-conjugated anti-HLA-DR (HLA-DR PE), and PE-conjugated anti-OCT3/4 (OCT3/4 PE) were obtained from Becton Dickinson; FITC-conjugated anti-CD144 (CD144-FITC) was obtained from Acris Antibodies; PE-conjugated anti-CD34 (CD34-PE) was purchased from Beckman Coulter; and an appropriate secondary FITC-conjugated antibody was obtained from Jackson Immunoresearch Laboratories.Washing buffer (phosphate-buffered saline, PBS, 0.1% sodium azide, and 0.5% bovine serum albumine, BSA) was used for all washing steps (3 mL of washing buffer and centrifugation, 400 g 8 min at 4°C). Briefly, 5×105 cells/sample were incubated with 100 μL of 20 mM ethylenediaminetetraacetic acid (EDTA) at 37°C for 10 min and washed.
Staining of surface antigens and intracellular antigens was carried out according to Eleuterio et al.11 Quality control included a regular check-up with Rainbow Calibration Particles (BD Biosciences). Debris was excluded from the analysis by gating on morphological parameters; 20,000 nondebris events in the morphological gate were recorded for each sample. To assess nonspecific fluorescence, we used specific irrelevant controls. All antibodies were titrated under assay conditions, and optimal photomultiplier (PMT) gains were established for each channel. Data were analyzed using FlowJo™ software (TreeStar). Mean Fluorescence Intensity Ratio (MFI Ratio) was calculated by dividing the MFI of positive events by the MFI of negative events.30
Analysis of hPDLSC differentiation into mesengenic lineages
Adipogenic differentiation
Expanded cells at the second passage, cultured with MSCGM and MSCGM-CD, were seeded into dishes at a density of 2×104cells/cm2. At 100% confluence, three cycles of induction/maintainance stimulated adipogenic differentiation. Each cycle consisted of feeding the hPDLSCs with supplemented adipogenesis induction medium (Lonza) for 3 days, followed by 3 days in supplemented adipogenic maintenance medium (Lonza). After three complete cycles of induction/maintenance, the culture was incubated for 7 days in supplemented adipogenic maintenance medium, replacing the medium every 2–3 days. The cells were fixed in 10% formalin for 15 min and washed with dH2O. Subsequently, the cells were stained with Oil Red O (ORO) working solution (300 mg of Oil red O/100 mL of isopropanol) for 5 min and counterstained with hematoxylin. The differentiation of hPDLSCs into adipogenic lineage was evaluated by AdipoRed™ assay reagent hydrophilic Nile Red fluorescence (Lonza). After differentiation, the plates were rinsed with PBS and 140 μL/well of AdipoRed was added; after 10 min, the fluorescence with an excitation at 485 nm and an emission at 572 nm was measured with a fluorimeter (Synergy HT).
Osteogenic differentiation
For osteogenic induction, the primary cells at the second passage were seeded at 4×103 cells/cm2 in MSCGM-CD and in MSCGM culture medium and maintained in culture at 37°C, in a humidified 5% CO2 atmosphere. At subconfluence, cells were incubated with standard hMSC Osteogenic BulletKit (Lonza) or MSCGM-CD medium with the addition of osteogenic supplements, that is, 100 nM dexamethasone (Applichem GmbH), 10 nM β-glycerol-phosphate (Applichem), and 0.05 mM 2-phosphate-ascorbic acid (Sigma-Aldrich).31
Visualization of calcium deposition and extracellular matrix (ECM) mineralization was obtained by Alizarin Red S (ARS) staining assay performed after 21 days of culture. According to Gregory et al.,32 cells were washed with PBS, fixed in 10% (v/v) formaldehyde (Sigma-Aldrich) for 30 min, and washed twice with abundant dH2O before addition of 0.5% Alizarin red S in H2O, pH 4.0, for 1 h at RT.
After cell incubation under gentle shaking, cells were washed with dH2O four times for 5 min. For staining quantification, 800 μL 10% (v/v) acetic acid was added to each well. Cells were incubated for 30 min with shaking, then scraped from the plate, transferred into a 1.5-mL vial, and vortexed for 30 s. The obtained suspension, overlaid with 500 μL mineral oil (Sigma-Aldrich), was heated to 85°C for 10 min, then transferred to ice for 5 min, carefully avoiding opening of the tubes until fully cooled, and centrifuged at 20,000 g for 15 min. Five hundred microliters of the supernatant were placed into a new 1.5-mL vial, and 200 μL of 10% (v/v) ammonium hydroxide was added (pH 4.1–pH 4.5). One hundred fifty microliters of the supernatant obtained from differentiated and undifferentiated hPDLSCs were read in triplicate at 405 nm by a spectrophotometer (Synergy HT).
Light and electron microscopy analysis
Glass-adherent hPDLSCs at P2 and cells cultured at 1, 2, and 3 weeks on thermanox coverslips in the presence of osteoinductive medium, in xeno-free conditions, were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer pH 7.4 for 1 h. Some samples of glass-adherent cells were stained with toluidine blue and directly observed by light microscopy. After the fixation, the samples were postfixed with 1% osmium tetroxide, dehydrated in a graded series of ethanol, and embedded in Epon. Semithin sections were stained with toluidine blue and used for light microscopy analysis. All sections were observed with a Zeiss Axiophot apparatus, and images were captured using a Nikon digital camera Digital Sight. Thin sections were stained with uranyl acetate and lead citrate; some sections were also stained with tannic acid, and finally observed with a Zeiss EM 109 transmission electron microscope. Images were captured using a Nikon digital camera Dmx 1200F and ACT-1 software.
RNA extraction and TaqMan quantitative real-time PCR
Total RNA from 1, 2, and 3 weeks of ex vivo differentiated and un-differentiated hPDLSCs xeno-free cultures was extracted using the RNeasy Mini Kit (Quiagen). RNA quantity and quality was assessed by the Agilent 8453 Spectrophotometer (Agilent Technologies). Six microgram of RNA from each sample were reverse transcribed using the High-Capacity RNA-to-cDNA Kit following the manufacturer's instructions (Applied Biosystems). Retrotranscribed RNA was mixed with DNase-free water to reach a final volume of 1080 μL and then added to 1080 μL of TaqMan Universal PCR Master Mix No AmpErase UNG (Applied Biosystems). A 96-well TaqMan® Array Human Osteogenesis was loaded with 20 μL per well of the mix and run on an Abi 7900HT Sequencing Detection System (Applied Biosystems). The amplification conditions were 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Three independent experiments were run for each condition for a total of 18 plates.
Raw data were analyzed by DataAssist software (Applied Biosystems). Each analysis underwent a global normalization using GAPDH, 18s, and HPRT1 as selected controls. Only genes showing no outlier replicates and a maximum Ct value=35 were included in the analysis. A gene was considered differentially expressed if it showed a fold change >1.4 or <0.7 and a p-value<0.05; p-values were adjusted using Benjamini–Hochberg FDR. Gene biological functions and networks were inferred using Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems). IPA predicts functional networks based on known protein–protein and functional interactions and ranks them by a significance score.33
Statistical analysis
The Statistical Package for Social Science (SPSS, v.21.0, Inc.) was used for data analysis.
Parametrical methods were used after having verified the existence of the required assumptions. In particular, the normality of the distribution and the equality of variances were assessed by the Shapiro–Wilk and Levene's tests, respectively.
The factors under investigation were the time elapsed and the culture medium for Trypan blue test, MTT assay, and osteogenic differentiation; the culture medium and the addition or not of an induction supplement, on the other hand, defined the two factors for the adipogenic differentiation test. Data were expressed as means and standard deviation of the recorded dependent variables: the cell count (Trypan blue test) and the optical density (MTT assay, osteogenic differentiation, and adipogenic differentiation). The differences among the levels of the two factors under investigation were evaluated by performing four distinct two-way ANOVA tests, one for each experiment. Tukey tests were applied for pairwise comparisons. A value of p<0.05 was considered statistically significant in all tests.
Results
hPDLSC morphological analyses
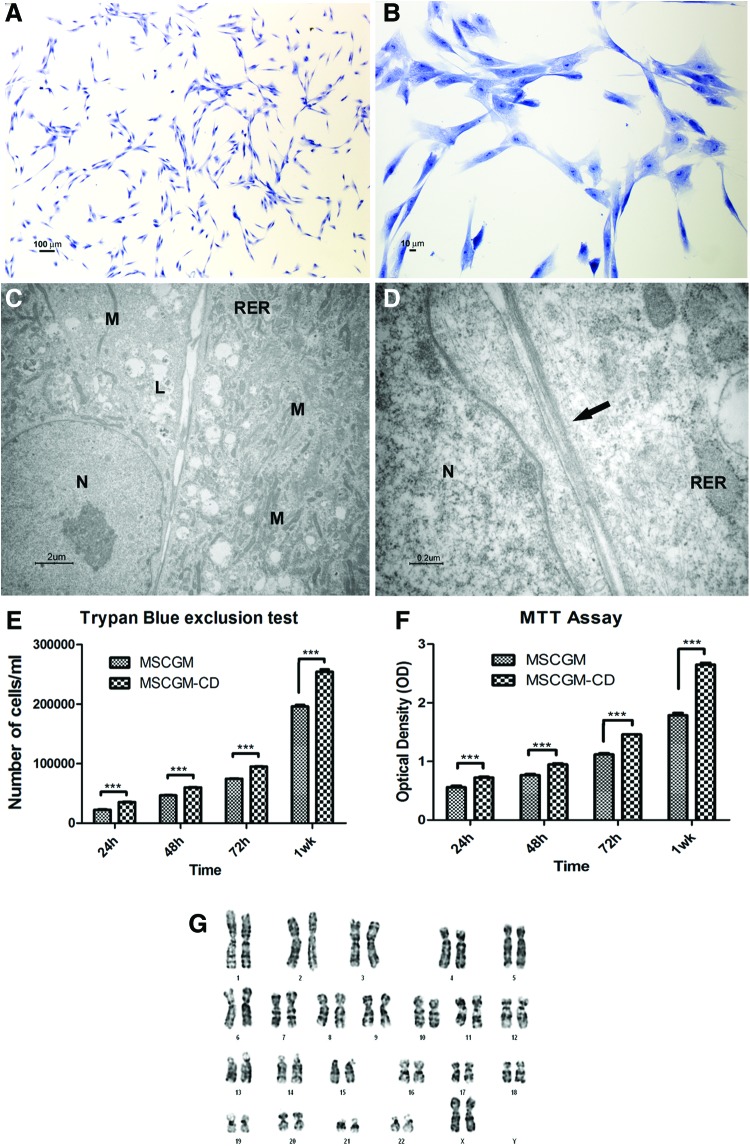
Toluidine blue-stained primary cultures of hPDLSCs in xeno-free condition were analyzed at the second passage. Cells adherent to glass coverslips showed a spindle-shaped appearance with long cytoplasmic processes; some polygonal cells were also present (Fig. 1A, B). At the ultrastructural level, the cells were flattened and showed a large cytoplasm containing numerous mitochondria, rough endoplasmic reticulum (RER) profiles, residual lysosomal bodies, and vacuoles; large euchromatic nuclei with one or two nucleoli were present (Fig. 1C). Adjacent cells showed junctional-like complexes (Fig. 1D).
FIG. 1.
Light and transmission electron microscopy of primary cultures of human periodontal ligament stem cells (hPDLSCs) at the second passage in xeno-free medium. (A, B) Light microscopy of semithin section at different magnification. The cells show mainly a spindle-shaped appearance with long cytoplasmatic processes; some polygonal cells are present. Bars respectively: 100 μm, 10 μm. (C, D) Transmission electron microscopy of thin sections. (C) Cells with euchromatic nuclei (N) show a large cytoplasm containing numerous mitochondria (M), rough endoplasmic reticulum (RER) profiles, and residual lysosomal bodies (L). Bar: 2 μm. (D) Contact zones between cells (arrow) are shown at high magnification. Bar: 0.2 μm. (E) Trypan blue exclusion test shows the proliferation rate and viability of hPDLSCs maintained in culture with MSCGM (standard conditions) and MSCGM-CD (xeno-free medium). An increase in cell growth, time dependent, is evident in xeno-free culture. The y-axis shows cell number, and x-axis shows culture time. The data represent the mean of three separate experiments with five different cell lines. (F) Proliferation rate is assessed by MTT assay in five hPDLSCs grown with xeno-free medium and MSCGM. The proliferation rate is measured as the absorbance detected at 650 nm OD. Results are expressed as mean and standard deviation of three independent experiments, in which five different samples are used (***P<0.001). (G) A representative karyotyping of hPDLSCs primary culture appears completely normal. Color images available online at www.liebertpub.com/tec
Cell growth
An MTT assay was performed at indicated time points (24, 48, 72 h and 1 week) in both standard and xeno-free conditions (Fig. 1F) to measure the viability and proliferation rate of each stem cell population. The assay displayed an increasing trend in the proliferation rate for all the examined time points. The previous data were confirmed through the analysis of Trypan Blue exclusion test staining of hPDLSCs at the second passage (Fig. 1E) showing the logarithmic growth during the culture. Each test was repeated thrice for each of the primary culture cells. In particular, cell growth was higher in hPDLSCs under xeno-free conditions.
Karyotyping analysis
Each of the cell line obtained from different patients, cultured in xeno-free conditions, was subject to cytogenetic analysis of karyotypes to ensure its genomic stability. The results indicated that five hPDLSC samples analyzed had a normal karyotype; in fact, no chromosomal translocation, delection, or extra-chromosome number was observed (Fig. 1G).
Flow cytometry immunophenotype
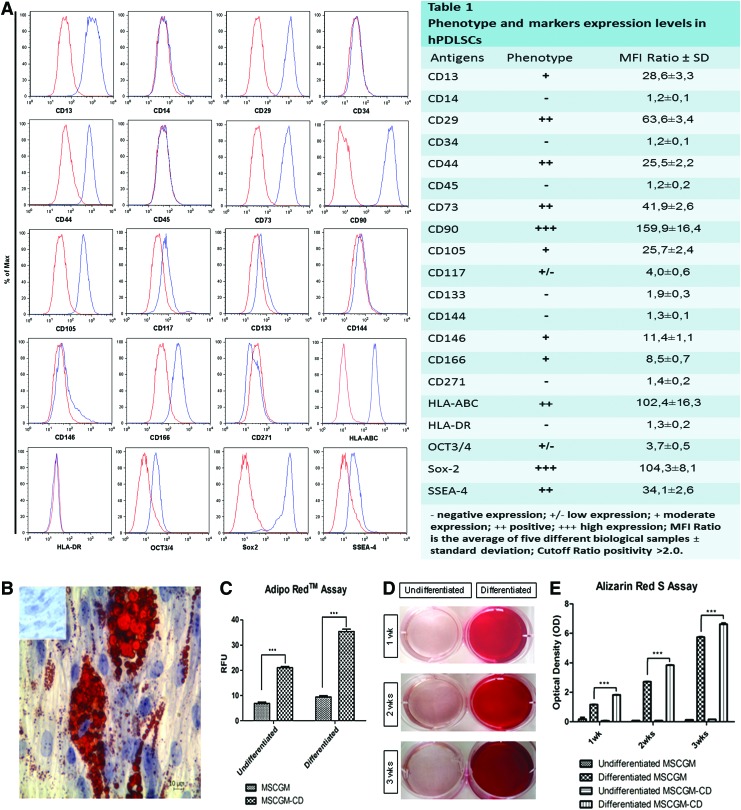
Using flow cytometry, we extensively characterized the hPDLSC expression profile of the principal mesenchymal markers under normal and xeno-free culture condition. The use of the xeno-free culture medium does not change immunophenotype, and even the expression levels for each marker hPDLSC under xeno-free condition did not display surface expression of any hematopoietic marker (CD14, CD34, and CD45). On the contrary, they expressed a variety of mesenchymal markers (CD13, CD29, CD73, CD90, CD105, and CD117), several surface adhesion molecules (CD44, CD146, and CD166), and the stemness markers Sox-2, Oct ¾, and SSEA-4; while CD133 and CD271 as well as the surface endothelial marker CD144 were not expressed. Consistent with a stem cell profile, these cells stained positive for HLA-ABC and negative for HLA-DR (Fig. 2A, Table 1).
FIG. 2.
Cytofluorimetric analysis of xeno-free hPDLSCs culture. (A) The blue histogram shows the distribution of the antigen expression, whereas the red histogram represents the respective isotype control. (A) is representative of five separate experiments performed using cells at the second passage. The value are expressed as mean fluorescence ratio (MFI) obtained by dividing the MFI of positive events by the MFI of negative events. (A, Table 1) The values are the mean and standard deviation of five separate experiments. (B) The adipogenic potential of hPDLSCs, grown in MSCGM and MSCGM-CD media, is analyzed through the quantification of intracellular lipid droplets at light microscopy using Oil Red O staining after adipogenic induction. Control hPDLSCs are visualized in inset. (C) Quantitative comparison of lipid vacuole formation in control and induced cells is evaluated by AdipoRed™ assay. (D) Mineralization of hPDLSCs, grown for 1, 2 and 3 weeks in osteogenic medium, evaluated by cell staining with Alizarin red S (ARS) and quantification of staining via extraction with ammonium hydroxide (E) at a different commitment time. The amount of released dye is measured by a microplate reader at 405 nm. The values, expressed as units of optical density (OD), are the mean and standard deviation of five independent experiments, in which different cell samples were used (***P<0.001). Color images available online at www.liebertpub.com/tec
Multipotentiality of hPDLSCs under xeno-free conditions
Adipogenesis-induced cells at light microscopy showed the intracellular lipid droplets single or grouped stained with Oil Red O (Fig. 2B). These data were supported by the AdipoRed test, consisting of a solution of the Hydrophilic Stain Nile Red reagent that enables the quantification of intracellular lipid droplets (Fig. 2C).
The osteogenic differentiation process induced in primary culture after 1, 2, and 3 weeks was evaluated by ARS staining (Fig. 2D) and colorimetric detection (Fig. 2E). A progressive significantly increased extracellular deposition was evident in cells grown under osteogenic condition in comparison with control cells. Adipogenesis and osteogenesis differentiation were more evident in xeno-free cells culture media.
Toluidine blue-stained semithin sections and thin sections of samples at 1, 2, and 3 weeks of culture in the osteogenic medium were analyzed by light and electron microscopy (Figs. 3–5).
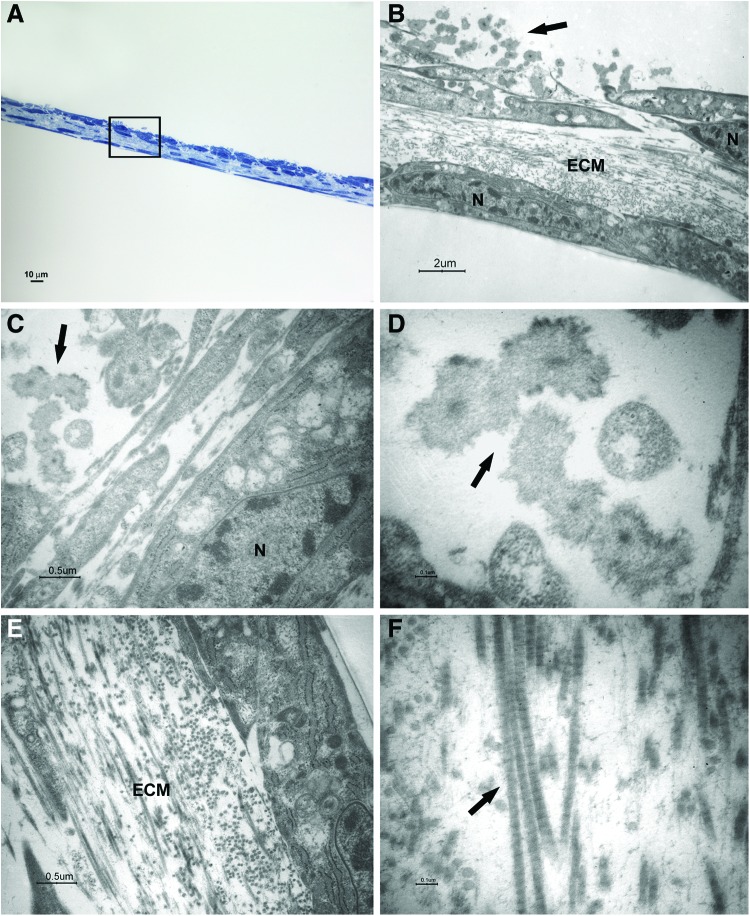
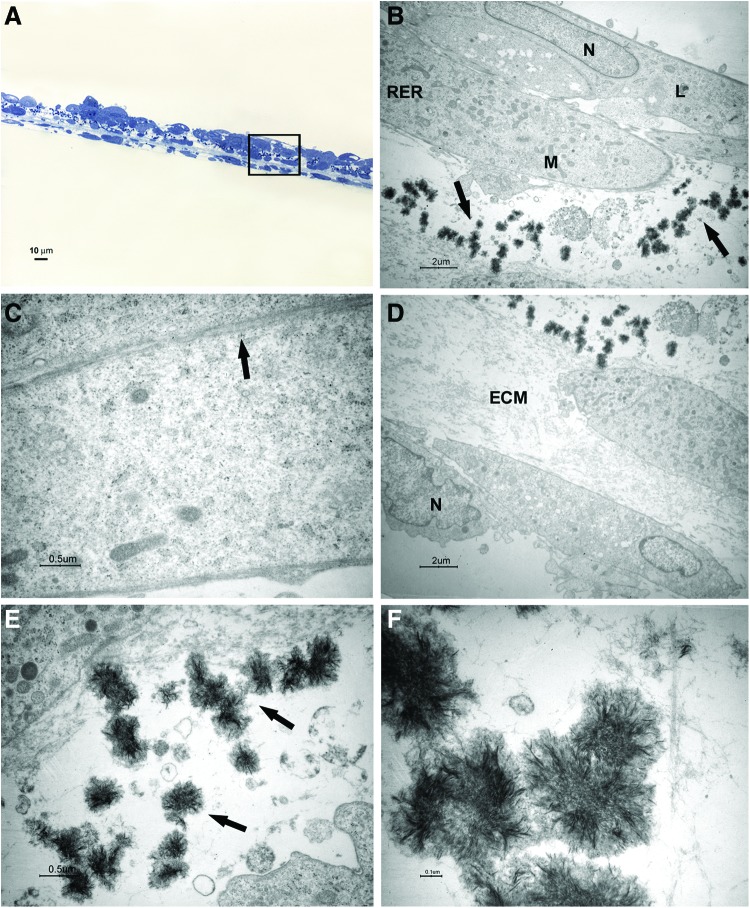
FIG. 3.
Light and transmission electron microscopy of cells at 1 week of culture in the osteogenic xeno-free medium. Sections cut perpendicularly to the surface of seeding. (A) Light microscopy of semithin section. Bar: 10 μm. (B–F) Transmission electron microscopy of thin sections also stained with tannic acid. (B) A magnification of an area similar to that marked by square in (A). The cells show heterochromatic nuclei (N). Mineralizing nodules (arrow) at an early stage of formation were close to the free surface. Bar: 2 μm. (C, D) Higher magnification of mineralizing nodules (arrow). Bars respectively: 0.5 μm, 0.1 μm. (E, F) Higher magnification of extracellular matrix (ECM) and fibrils (arrow). Bars respectively: 0.5 μm, 0.1 μm. Color images available online at www.liebertpub.com/tec
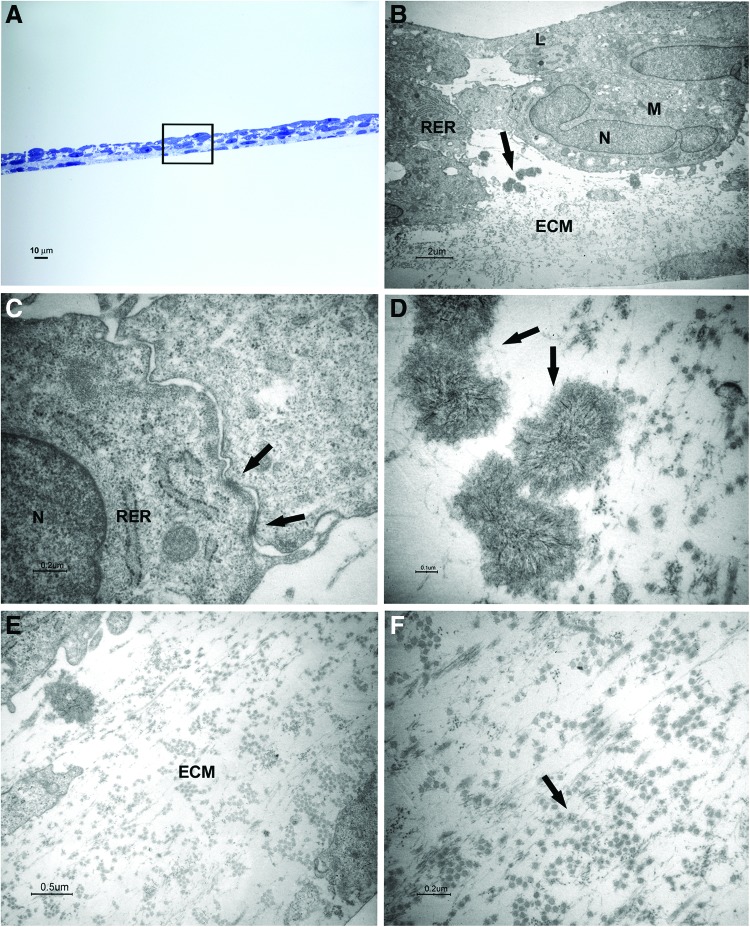
FIG. 4.
Light and transmission electron microscopy of cells at 2 weeks of culture in the osteogenic xeno-free medium. Sections cut perpendicularly to the surface of seeding. (A) Light microscopy of semithin section. Bar: 10 μm. (B–F) Transmission electron microscopy of thin sections also stained with tannic acid. (B) A magnification of an area similar to that marked by square in (A). The osteoblast-like cells with a cuboidal shape show euchromatic nuclei (N), well-represented RER profiles, numerous mitochondria (M), and some lysosomal bodies (L). Mineralizing nodules are pointed out by arrow. Bar: 2 μm. (C) Desmosome-like junctions are shown (arrows). Bar: 0.2 μm. (D) Higher magnification of mineralizing nodules (arrows). Bar: 0.1 μm. (E, F) Higher magnification of ECM and fibrils (arrow). Bars respectively: 0.5 μm, 0.2 μm. Color images available online at www.liebertpub.com/tec
FIG. 5.
Light and transmission electron microscopy of cells at 3 weeks of culture in the osteogenic xeno-free medium. Sections cut perpendicularly to the surface of seeding. (A) Light microscopy of semithin section. Bar: 10 μm. (B–F) Transmission electron microscopy of thin sections. (B, D) Taken together, the two photos represent a magnification of an area similar to that marked by a square in (A). Mineralizing nodules (arrows) appear numerous. Bar: 2 μm. (C) Contact zones between cells are shown. Bar: 0.5 μm. (E,F) Higher magnification of mineralizing nodules (arrows in E). Bars respectively: 0.5 μm, 0.1 μm. Color images available online at www.liebertpub.com/tec
After 1 week of culture, elongated cells were able to organize in two to three layers; a consistent ECM was evident between a superficial cell layer and the deeper one (Fig. 3A, B, E). Cross striated fibrils could be highlighted at high magnification (Fig. 3F). The cells showed nuclei with condensed chromatin, suggesting that a proliferative activity was taking place. Some structures recognized as mineralizing nodules at an early stage of formation were present close to the cell surface (Fig. 3B–D).
After 2 weeks of culture, cuboidal shape cells were organized in three to five layers; mineralizing nodules with the typical needle structure were present between cells and ECM (Fig. 4A, B, D, E). At the ultrastructural level, the cells presented euchromatic nuclei, numerous mitochondria, some lysosomal bodies, and well-represented RER profiles, indicating an active protein synthesis (Fig. 4B). Some desmosome-like junctions were observed between cells (Fig. 4C).
After 3 weeks of culture, the number of cuboidal cells increased and the mineralizing nodules also increased (Fig. 5A, B, D–F). Some membrane contacts were present between superficial cells (Fig. 5C).
Osteogenic gene modulation
The osteogenic differentiation data of hPDLSCs cultured under xeno-free conditions were assessed by a qRT-PCR of an osteogenesis-related assays panel. The plate contains 92 assays, enabling the analysis of pathways related to the osteogenic determination and differentiation, including transcription and growth factors, genes involved in the physiological processes of bone and tooth formation, mineralization, and maintenance, including collagens and matrix metalloproteinases.
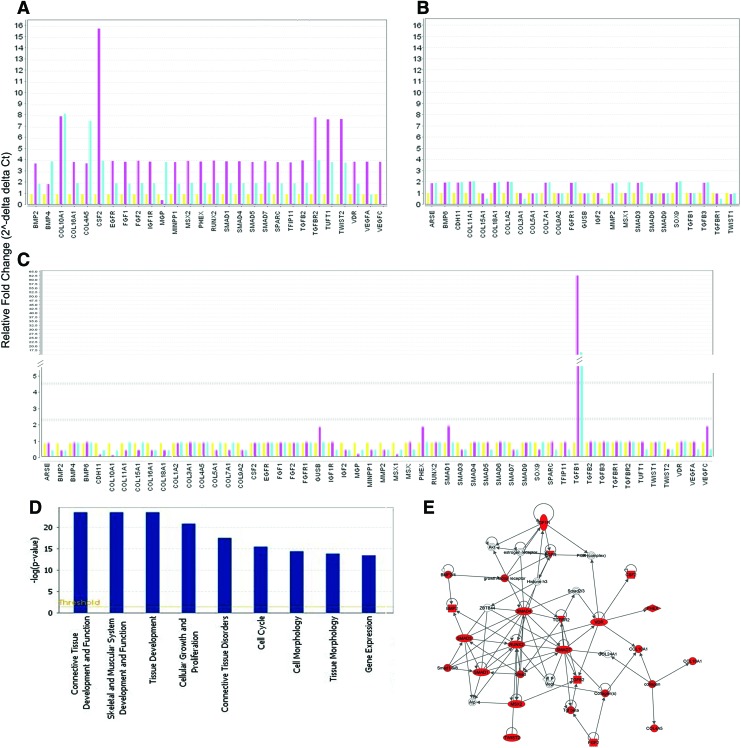
The analysis of hPDLSCs differentiated at 2 and 3 weeks versus 1 week unraveled a significant altered expression of 52 genes (Fig. 6A, B). Twenty-three genes showed an early overexpression at 2 weeks and at 3 weeks of treatment, while 15 genes were up-expressed at 3 weeks (Supplementary Tables S1–S4; Supplementary Data are available online at www.liebertpub.com/tec). Moreover, gene expression analysis revealed the early down-expression of 12 transcripts at 2 weeks and three out of these genes were also downregulated at 3 weeks (Supplementary Tables S1–S4). The significant dataset of genes (52 transcripts) in the undifferentiated 2- and 3-week hPDLSCs versus undifferentiated 1-week hPDLSCs revealed a mainly unmodified expression profile, except the TGFβ1 gene, which is highly upregulated at 2 and 3 weeks (Fig. 6C).
FIG. 6.
Genes significantly regulated by the osteogenic differentiation under xeno-free conditions and IPA functional analysis of osteogenic-related genes expression. Bar charts show the relative fold changes of the 52 genes significantly regulated by osteogenic differentiation. (A) Genes with higher expression modulation during differentiation. (B) Genes with lower expression modulation during differentiation. To every gene is shown the expression at first, second and third week. (C) The 52 genes, modulated during differentiation, are mainly non modulated in undifferentiated cells. To every gene is shown the expression at first, second and third week. (D) IPA-inferred biological functions analysis shows the key functions regulated by the 52 genes modulated during osteogenic differentiation. (E) IPA cluster analysis of the 23 early upregulated genes (second week of osteogenic differentiation vs. first week) shows the central role played by RUNX2 during the osteogenic differentiation. IPA, Ingenuity Pathways Analysis (software). Color images available online at www.liebertpub.com/tec
IPA functional analysis of the 52 genes dataset indicated that the main biological functions associated are connective tissue development, functions and disorders, skeletal and muscular system development and function, cellular growth and proliferation, cell cycle and morphology, and gene expression (Fig. 6D).
IPA network analysis of the 23 early up-expressed genes generated 1 network with a score=53 (Fig. 6E).
Discussion
It has recently been described that dental tissue-originated SCs stand out as a broad potential stem cell source for bone regeneration in oral and maxillofacial surgery, craniofacial anomalies, and orthopedics.34
The marked ability of allogenic PDLSCs to repair periodontal defects by forming bone, PDL, and cementum-like tissue in vivo suggested that stem cells from oral tissue could be good candidates for periodontal regeneration in rodent fenestration defect.35
hPDLSCs exhibited a higher number of growing cells compared with Bone Marrow Stem Cells.11 The multipotent capacity and the immunomodulatory properties, both in vitro and in vivo of PDLSCs, retained these cells as a more accessible cell source than bone marrow-derived MSCs for cell-based therapy of immune and inflammation-related diseases.36 In particular, the autologous periodontal ligament progenitors, mixed with bone-grafting material, when implanted into deep periodontal defects may be effective in regenerative dentistry.37
Our previous studies provided evidence that the human periodontal ligament showed an accessible niche of stem cells which could differentiate into osteoblasts and adipocytes, secrete IL-7 and SDF-1a,10,38,39 and express stemness markers such as nanog, Oct-4, SSEA-1, and SSEA-4.40 hPDLSCs showed differentially regulated proteins that were potentially related to growth, regulation, and genesis of neuronal cells, suggesting that stem cells derived from periodontal tissue possessed the potential ability of neuronal differentiation in agreement with its neural crest origin.11
Clinical applications of cell therapy represent a complex challenge for the necessity of operating under strict Good Manufacturing Procedure and for the absolute need to demonstrate not just the safety of the proposed approach but also, first and foremost, its feasibility, without compromising the safety of the patients, and an acceptable degree of reproducibility for clinical application.41,42 Although many efforts have shown that human placenta, bone marrow, and skin43 can be isolated and expanded in serum-free medium, actually, no published work has shown the possibility to expand hPDLSCs under serum-free conditions.
Then, the purpose of this study was to develop an innovative xeno-free culture system for the expansion of hPDLSCs for tissue engineering. hPDLSCs can be identified as a cell population showing the essential features for human clinical use; in fact, they are autologous and easily obtainable by scraping of the alveolar crest and of the horizontal fibers of the periodontal ligament. The current results demonstrated that they could be expanded ex vivo using a new xeno-free media formulation without enzymatic digestion. The multipotent ability of xeno-free hPDLSCs and their high ex vivo-expansive potential demonstrated that a biopsy of periodontal tissue was sufficient to obtain an elevated number of stem cells with genomic stability which were capable of differentiating into adipogenic and osteogenic precursors. Furthermore, the xeno-free culture maintained the characteristic immunophenotype11 as well as the cells grown in FBS-containing medium. The proliferation rate of hPDLSCs xeno-free expansion was significantly increased with regard to hPDLSCs cultured with MSCGM; moreover, the cells retained their functionality, indicating that the medium was suitable for periodontium stem cell growth. To our knowledge, the current study showed for the first time the morphological characteristics and gene modulation of xeno-free hPDLSCs cultured in osteogenic inductive medium. Some differentiation steps of osteogenic cells and, in particular, the secretion of ECM resembling the secretion pattern during in vivo osteogenesis44,45 were documented.
These results were consistent with quantitative Real-Time PCR expression of 92 osteogenesis-associated genes that revealed a significant modulation of 52 transcripts during the xeno-free hPDLSCs osteogenic differentiation; while in the undifferentiated hPDLSCs, the same genes were mainly not modulated. Fifty-two transcripts were upregulated at the third week, and many of these genes (23 transcripts) were already upregulated at the second week, showing an early osteogenic differentiation. Early upregulated genes were grouped into one network that focused on the RUNX2 gene, a master transcription factor essential for osteoblastic differentiation and skeletal morphogenesis, which has been shown to interact through its C-terminal segment with Smads, and this interaction was required for both in vivo endochondral and intramembranous bone formation.46,47 The network also showed other powerful markers, directly related to RUNX2, promoting MSCs differentiation into osteoblasts in vitro and inducing bone formation in vivo (SMAD1,-4,-5,-7, MSX2, TWIST2, CSF2, VDR, TGFβ2, and BMP2).48–51 In particular, BMPs and RUNX2 cooperatively interacted and stimulated osteoblast gene expression, and direct evidence showed that RUNX2 was essential for the execution and completion of BMP2 signaling for osteoblast differentiation.52 The differentiation process was endorsed by the 12 genes that were early down-expressed (COL15A1, COL3A1, COL5A1, COL9A2, GUSB, IGF2, SMAD9, TWIST1, TGFβ R1, and TGFβ1). TGFβ1 was instead highly upregulated in undifferentiated cells, probably due to its role in the transition of mesenchymal cells.53 TGFβ superfamily, multifunctional cytokine regulated various cellular activities, induced the proliferation of PDL cells and the expression of ECM components in PDL cells.54 It has been hypothesized that TGFβ1 might be important for the induction of the fiber synthesis necessary to regenerate PDL tissue. Furthermore, the synthesis of TGFβ1 in PDL cells depended on their differentiation stage and could contribute to the differentiation of immature PDL cells into functional PDL fibroblasts.55
However, TGFβ1 also blocked osteogenesis by various mechanisms depending on its concentration, cell density, and differentiation stage of the cells; in particular, it inhibited osteoblast differentiation via suppression of IGF1 expression and subsequent downregulation of the PI3K/Akt pathway. In fact, during osteogenic commitment, it appeared to stimulate the proliferation and development of early osteoblasts, but inhibited their maturation and mineralization, and, consequently, the differentiation process was associated with a decrease of TGFβ.56 This was in agreement with the downregulation of TGFβ during osteogenesis commitment of hPDLSCs.
The TWIST family of basic helix-loop-helix transcription factors, in particular TWIST1, is known as a mediator of mesodermal tissue development. Overexpression of TWIST1 is associated with a decrease in the gene expression of osteoblast-associated markers, bone morphogenic protein-2, bone sialoprotein, osteopontin, alkaline phosphatase, and osteocalcin; a decreased capacity for osteo/chondrogenic differentiation; and an enhanced capacity to undergo adipogenesis.57 Our results demonstrated that hPDLSCs in basal conditions express TWIST1 which appears downregulated during osteogenic differentiation.
Therefore, the expression of specific osteogenic markers suggested that xeno-free hPDLSCs underwent an effective osteogenesis process beginning at the second week of differentiation induction as evidenced in previous works.58 Then, hPDLSCs can be manipulated ex vivo under xeno-free conditions without compromising their functional properties and offering various potential advantages when compared with a standard culture system; these results are according to current literature showing that the modified serum-free media support the expansion of oral stem cells and the maintenance of their multipotency.21
Conclusion
In synthesis, our findings may be of high interest for the understanding of xeno-free standardized protocols for hPDLSCs preparations, in order not to interfere with their self-renewal and differentiation processes and for an effective large clinical-scale production of a vast number of functionally competent stem cells for the passage from basic to translational research in transplantation, immune-therapy, and regenerative medicine.
Supplementary Material
Acknowledgments
This work has been supported by OT ex 60% University of Chieti fund, and partly by PRIN 20102ZLNJ5 “Stem cells and 3D scaffolds: a novel construct in bone regeneration” financed by the Ministry of Education, University, and Research (M.I.U.R.), Rome, Italy.
The authors thank Prof. Paolo Guanciali (University of Chieti) for karyotyping analysis.
Disclosure Statement
No competing financial interests exist.
References
- 1.Huang G.T., Gronthos S., and Shi S.Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res 88,792, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egusa H., Sonoyama W., Nishimura M., Atsuta I., and Akiyama K.Stem cells in dentistry—Part II: clinical applications. J Prosthodont Res 56,229, 2012 [DOI] [PubMed] [Google Scholar]
- 3.Gronthos S., Mankani M., Brahim J., Robey P.G., and Shi S.Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A 97,13625, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miura M., Gronthos S., Zhao M., Lu B., Fisher L.W., Robey P.G., et al. . SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A 100,5807, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo B.M., Miura M., Gronthos S., Bartold P.M., Batouli S., Brahim J., et al. . Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 364,149, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Ivanovski S., Gronthos S., Shi S., and Bartold P.M.Stem cells in the periodontal ligament. Oral Dis 12,358, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Sonoyama W., Liu Y., Fang D., Yamaza T., Seo B.M., Zhang C., et al. . Mesenchymal stem cell-mediated functional tooth regeneration in swine. PloS One 1,e79, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morsczeck C., Gotz W., Schierholz J., Zeilhofer F., Kuhn U., Mohl C., et al. . Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol 24,155, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Fournier B.P., Larjava H., and Hakkinen L.Gingiva as a source of stem cells with therapeutic potential. Stem Cells Dev 20,611, 2014 [DOI] [PubMed] [Google Scholar]
- 10.Trubiani O., Di Primio R., Traini T., Pizzicannella J., Scarano A., Piattelli A., et al. . Morphological and cytofluorimetric analysis of adult mesenchymal stem cells expanded ex vivo from periodontal ligament. Int J Immunopathol Pharmacol 18,213, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Eleuterio E., Trubiani O., Sulpizio M., Di Giuseppe F., Pierdomenico L., Marchisio M., et al. . Proteome of human stem cells from periodontal ligament and dental pulp. PloS One 8,e71101, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding G., Liu Y., Wang W., Wei F., Liu D., Fan Z., et al. . Allogeneic periodontal ligament stem cell therapy for periodontitis in swine. Stem Cells 28,1829, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Zheng Y., Ding G., Fang D.J., Zhang C.M., Bartold P.M., et al. . Periodontal ligament stem cell-mediated treatment for periodontitis in miniature swine. Stem Cells 26,1065, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiran M., Arpak N., Unsal E., and Erdogan M.F.The effect of improved periodontal health on metabolic control in type 2 diabetes mellitus. J Clin Periodontol 32,266, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Mealey B.L., and Rose L.F.Diabetes mellitus and inflammatory periodontal diseases. Curr Opin Endocrinol Diabetes Obes 15,135, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Manau C., Echeverria A., Agueda A., Guerrero A., and Echeverria J.J.Periodontal disease definition may determine the association between periodontitis and pregnancy outcomes. J Clin Periodontol 35,385, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Washio K., Iwata T., Mizutani M., Ando T., Yamato M., Okano T., et al. . Assessment of cell sheets derived from human periodontal ligament cells: a pre-clinical study. Cell Tissue Res 341,397, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Mannello F., and Tonti G.A.Concise review: no breakthroughs for human mesenchymal and embryonic stem cell culture: conditioned medium, feeder layer, or feeder-free; medium with fetal calf serum, human serum, or enriched plasma; serum-free, serum replacement nonconditioned medium, or ad hoc formula? All that glitters is not gold! Stem Cells 25,1603, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Dimarakis I., and Levicar N.Cell culture medium composition and translational adult bone marrow-derived stem cell research. Stem Cells 24,1407, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Shahdadfar A., Fronsdal K., Haug T., Reinholt F.P., and Brinchmann J.E.In vitro expansion of human mesenchymal stem cells: choice of serum is a determinant of cell proliferation, differentiation, gene expression, and transcriptome stability. Stem Cells 23,1357, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Tarle S.A., Shi S., and Kaigler D.Development of a serum-free system to expand dental-derived stem cells: PDLSCs and SHEDs. J Cell Physiol 226,66, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Schallmoser K., Bartmann C., Rohde E., Reinisch A., Kashofer K., Stadelmeyer E., et al. . Human platelet lysate can replace fetal bovine serum for clinical-scale expansion of functional mesenchymal stromal cells. Transfusion 47,1436, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Bieback K., Hecker A., Kocaomer A., Lannert H., Schallmoser K., Strunk D., et al. . Human alternatives to fetal bovine serum for the expansion of mesenchymal stromal cells from bone marrow. Stem Cells 27,2331, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Prins H.J., Rozemuller H., Vonk-Griffioen S., Verweij V.G., Dhert W.J., Slaper-Cortenbach I.C., et al. . Bone-forming capacity of mesenchymal stromal cells when cultured in the presence of human platelet lysate as substitute for fetal bovine serum. Tissue Eng Part A 15,3741, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Hatlapatka T., Moretti P., Lavrentieva A., Hass R., Marquardt N., Jacobs R., et al. . Optimization of culture conditions for the expansion of umbilical cord-derived mesenchymal stem or stromal cell-like cells using xeno-free culture conditions. Tissue Eng Part C Methods 17,485, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Kocaoemer A., Kern S., Kluter H., and Bieback K.Human AB serum and thrombin-activated platelet-rich plasma are suitable alternatives to fetal calf serum for the expansion of mesenchymal stem cells from adipose tissue. Stem Cells 25,1270, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Tonti G.A., and Mannello F.From bone marrow to therapeutic applications: different behaviour and genetic/epigenetic stability during mesenchymal stem cell expansion in autologous and foetal bovine sera? Int J Dev Biol 52,1023, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Carranza F.A.The Thoot-Supporting structures. In: Newman F, ed. Clinical Periodontology. Philadelphia: W.B. Saunders, 1996, pp. 31–50 [Google Scholar]
- 29.Sanchez O., Escobar J.I., and Yunis J.J.A simple G-banding technique. Lancet 2,269, 1973 [DOI] [PubMed] [Google Scholar]
- 30.Miscia S., Ciccocioppo F., Lanuti P., Velluto L., Bascelli A., Pierdomenico L., et al. . Abeta(1–42) stimulated T cells express P-PKC-delta and P-PKC-zeta in Alzheimer disease. Neurobiol Aging 30,394, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., et al. . Multilineage potential of adult human mesenchymal stem cells. Science 284,143, 1999 [DOI] [PubMed] [Google Scholar]
- 32.Gregory C.A., Gunn W.G., Peister A., and Prockop D.J.An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem 329,77, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Gatta V., D'Aurora M., Lanuti P., Pierdomenico L., Sperduti S., Palka G., et al. . Gene expression modifications in Wharton's Jelly mesenchymal stem cells promoted by prolonged in vitro culturing. BMC Genomics 14,635, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hakki S.S., Bozkurt B., Hakki E.E., Kayis S.A., Turac G., Yilmaz I., et al. . Bone morphogenetic protein-2, -6, and -7 differently regulate osteogenic differentiation of human periodontal ligament stem cells. J Biomed Mater Res Part B Appl Biomater 102,119, 2013 [DOI] [PubMed] [Google Scholar]
- 35.Han J., Menicanin D., Marino V., Ge S., Mrozik K., Gronthos S., et al. . Assessment of the regenerative potential of allogeneic periodontal ligament stem cells in a rodent periodontal defect model. J Periodontal Res 49,333, 2013 [DOI] [PubMed] [Google Scholar]
- 36.Li Z., Jiang C.M., An S., Cheng Q., Huang Y.F., Wang Y.T., et al. . Immunomodulatory properties of dental tissue-derived mesenchymal stem cells. Oral Dis 20,25, 2014 [DOI] [PubMed] [Google Scholar]
- 37.Feng F., Akiyama K., Liu Y., Yamaza T., Wang T.M., Chen J.H., et al. . Utility of PDL progenitors for in vivo tissue regeneration: a report of 3 cases. Oral Dis 16,20, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trubiani O., Isgro A., Zini N., Antonucci I., Aiuti F., Di Primio R., et al. . Functional interleukin-7/interleukin-7Ralpha, and SDF-1alpha/CXCR4 are expressed by human periodontal ligament derived mesenchymal stem cells. J Cell Physiol 214,706, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Trubiani O., Orsini G., Zini N., Di Iorio D., Piccirilli M., Piattelli A., et al. . Regenerative potential of human periodontal ligament derived stem cells on three-dimensional biomaterials: a morphological report. J Biomed Mater Res Part A 87,986, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Trubiani O., Zalzal S.F., Paganelli R., Marchisio M., Giancola R., Pizzicannella J., et al. . Expression profile of the embryonic markers nanog, OCT-4, SSEA-1, SSEA-4, and frizzled-9 receptor in human periodontal ligament mesenchymal stem cells. J Cell Physiol 225,123, 2010 [DOI] [PubMed] [Google Scholar]
- 41.Banfi A., Muraglia A., Dozin B., Mastrogiacomo M., Cancedda R., and Quarto R.Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: implications for their use in cell therapy. Exp Hematol 28,707, 2000 [DOI] [PubMed] [Google Scholar]
- 42.Salvade A., Belotti D., Donzelli E., D'Amico G., Gaipa G., Renoldi G., et al. . GMP-grade preparation of biomimetic scaffolds with osteo-differentiated autologous mesenchymal stromal cells for the treatment of alveolar bone resorption in periodontal disease. Cytotherapy 9,427, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Chase L.G., Lakshmipathy U., Solchaga L.A., Rao M.S., and Vemuri M.C.A novel serum-free medium for the expansion of human mesenchymal stem cells. Stem Cell Res Ther 1,8, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto T., Nagaoka N., Hirata A., Nakamura H., Inoue M., Kawai M., et al. . Ultrastructural and immunohistochemical studies of medullary bone calcification, with special reference to sulphated glycosaminoglycans. J Electron Microsc 54,29, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Struys T., Moreels M., Martens W., Donders R., Wolfs E., and Lambrichts I.Ultrastructural and immunocytochemical analysis of multilineage differentiated human dental pulp- and umbilical cord-derived mesenchymal stem cells. Cells Tissues Organs 193,366, 2011 [DOI] [PubMed] [Google Scholar]
- 46.Greenblatt M.B., Shim J.H., and Glimcher L.H.Mitogen-activated protein kinase pathways in osteoblasts. Ann Rev Cell Dev Biol 29,63, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Agas D., Sabbieti M.G., Marchetti L., Xiao L., and Hurley M.M.FGF-2 enhances Runx-2/Smads nuclear localization in BMP-2 canonical signaling in osteoblasts. J Cell Physiol 228,2149, 2013 [DOI] [PubMed] [Google Scholar]
- 48.Chen G., Deng C., and Li Y.P.TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci 8,272, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishimura R., Hata K., Matsubara T., Wakabayashi M., and Yoneda T.Regulation of bone and cartilage development by network between BMP signalling and transcription factors. J Biochem 151,247, 2012 [DOI] [PubMed] [Google Scholar]
- 50.Lieben L., and Carmeliet G.Vitamin D signaling in osteocytes: effects on bone and mineral homeostasis. Bone 54,237, 2013 [DOI] [PubMed] [Google Scholar]
- 51.Hisada K., Hata K., Ichida F., Matsubara T., Orimo H., Nakano T., et al. . Retinoic acid regulates commitment of undifferentiated mesenchymal stem cells into osteoblasts and adipocytes. J Bone Miner Metab 31,53, 2013 [DOI] [PubMed] [Google Scholar]
- 52.Javed A., Afzal F., Bae J.S., Gutierrez S., Zaidi K., Pratap J., et al. . Specific residues of RUNX2 are obligatory for formation of BMP2-induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs 189,133, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tirino V., Camerlingo R., Bifulco K., Irollo E., Montella R., Paino F., et al. . TGF-beta1 exposure induces epithelial to mesenchymal transition both in CSCs and non-CSCs of the A549 cell line, leading to an increase of migration ability in the CD133+ A549 cell fraction. Cell Death Dis 4,e620, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujita T., Shiba H., Van Dyke T.E., and Kurihara H.Differential effects of growth factors and cytokines on the synthesis of SPARC, DNA, fibronectin and alkaline phosphatase activity in human periodontal ligament cells. Cell Biol Int 28,281, 2004 [DOI] [PubMed] [Google Scholar]
- 55.Fujii S., Maeda H., Tomokiyo A., Monnouchi S., Hori K., Wada N., et al. . Effects of TGF-beta1 on the proliferation and differentiation of human periodontal ligament cells and a human periodontal ligament stem/progenitor cell line. Cell Tissue Res 342,233, 2010 [DOI] [PubMed] [Google Scholar]
- 56.Koli K., Ryynanen M.J., and Keski-Oja J.Latent TGF-beta binding proteins (LTBPs)-1 and -3 coordinate proliferation and osteogenic differentiation of human mesenchymal stem cells. Bone 43,679, 2008 [DOI] [PubMed] [Google Scholar]
- 57.Isenmann S., Arthur A., Zannettino A.C., Turner J.L., Shi S., Glackin C.A., et al. . TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells 27,2457, 2009 [DOI] [PubMed] [Google Scholar]
- 58.Proksch S., Steinberg T., Stampf S., Schwarz U., Hellwig E., and Tomakidi P.Crosstalk on cell behavior in interactive cocultures of hMSCs with various oral cell types. Tissue Eng Part A 18,2601, 2012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.