Abstract
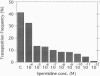
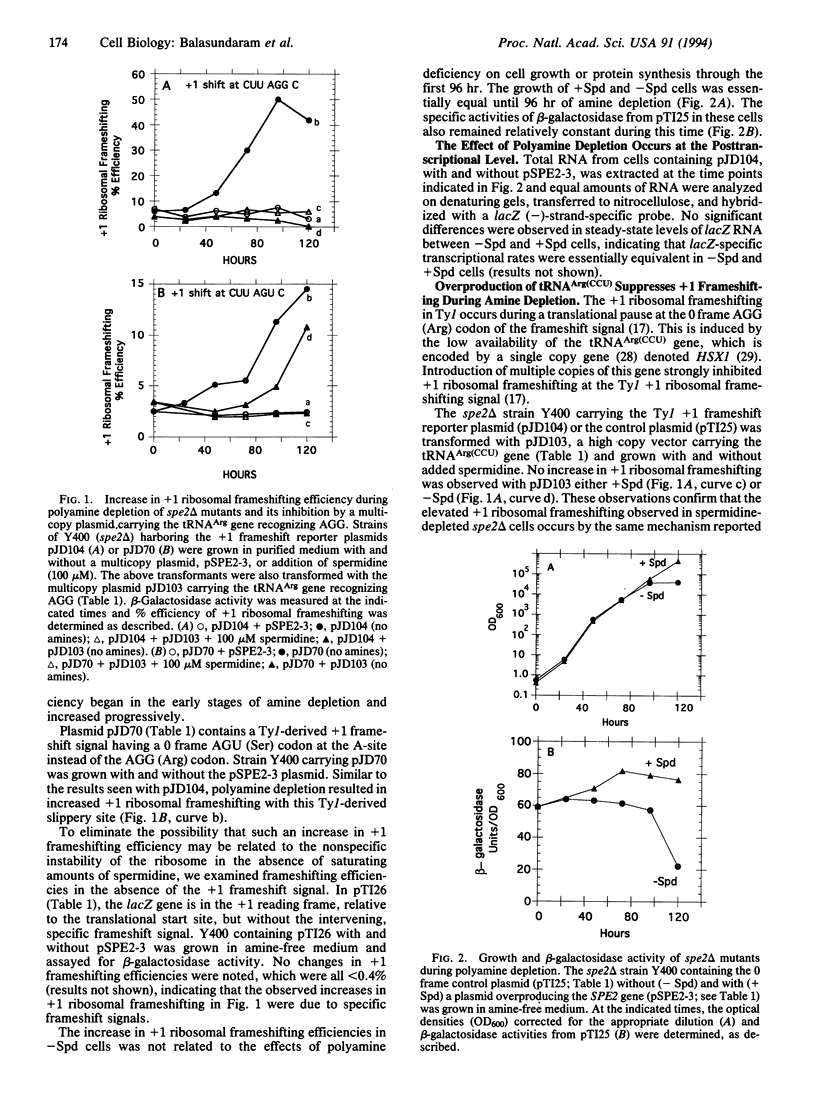
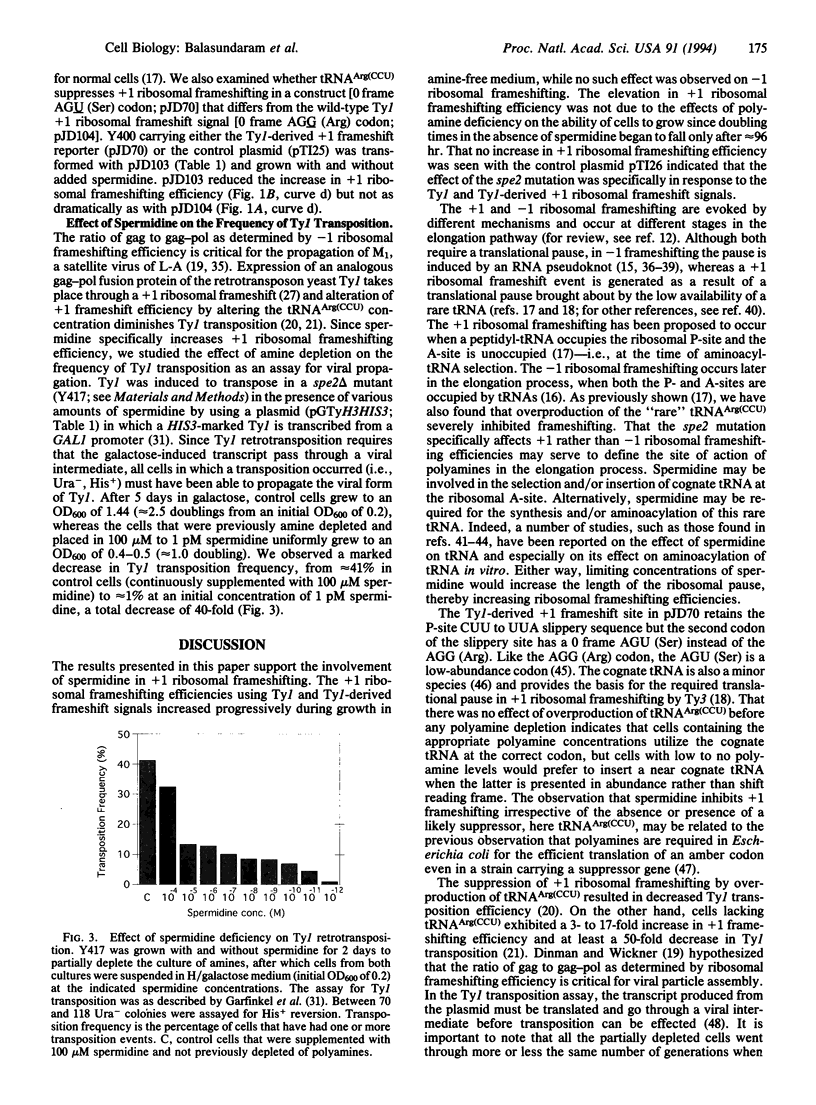
Polyamines have been implicated in nucleic acid-related functions and in protein biosynthesis. RNA sequences that specifically direct ribosomes to shift reading frame in the -1 and +1 directions may be used to probe the mechanisms controlling translational fidelity. We examined the effects of spermidine on translational fidelity by an in vivo assay in which changes in beta-galactosidase activity are dependent on yeast retrovirus Ty +1 and yeast double-stranded RNA virus L-A -1 ribosomal frameshifting signals. In spe2 delta mutants of Saccharomyces cerevisiae, which cannot make spermidine as a result of a deletion in the SPE2 gene, there is a marked elevation in +1 but no change in -1 ribosomal frameshifting. The increase in +1 ribosomal frameshifting efficiency is accompanied by a striking decrease in Ty1 retrotransposition.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balasundaram D., Tabor C. W., Tabor H. Oxygen toxicity in a polyamine-depleted spe2 delta mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4693–4697. doi: 10.1073/pnas.90.10.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D., Tabor C. W., Tabor H. Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5872–5876. doi: 10.1073/pnas.88.13.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D., Tyagi A. K. Polyamine--DNA nexus: structural ramifications and biological implications. Mol Cell Biochem. 1991 Feb 2;100(2):129–140. doi: 10.1007/BF00234162. [DOI] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991 Aug 20;220(4):889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992 Jan 2;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Clare J. J., Belcourt M., Farabaugh P. J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M. S., Tabor C. W., Tabor H. Isolation and characterization of Saccharomyces cerevisiae mutants deficient in S-adenosylmethionine decarboxylase, spermidine, and spermine. J Bacteriol. 1978 Apr;134(1):208–213. doi: 10.1128/jb.134.1.208-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Wickner R. B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992 Jun;66(6):3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Zhao H., Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993 Jul 16;74(1):93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafner J., Robertis E. M., Philippsen P. Delta sequences in the 5' non-coding region of yeast tRNA genes. EMBO J. 1983;2(4):583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Mastrangelo M. F., Sanders N. J., Shafer B. K., Strathern J. N. Transposon tagging using Ty elements in yeast. Genetics. 1988 Sep;120(1):95–108. doi: 10.1093/genetics/120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Matsuzaki K., Takeda Y. Aminoacyl transfer RNA formation. I. Absence of pyrophosphate-ATP exchange in aminoacyl-tRNA formation stimulated by polyamines. Biochim Biophys Acta. 1971 Nov 29;254(1):91–103. [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K., Taneja S. K., Liu T. Y., Tabor C. W., Tabor H. Spermidine biosynthesis in Saccharomyces cerevisiae. Biosynthesis and processing of a proenzyme form of S-adenosylmethionine decarboxylase. J Biol Chem. 1990 Dec 25;265(36):22321–22328. [PubMed] [Google Scholar]
- Kawakami K., Pande S., Faiola B., Moore D. P., Boeke J. D., Farabaugh P. J., Strathern J. N., Nakamura Y., Garfinkel D. J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993 Oct;135(2):309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Shafer B. K., Garfinkel D. J., Strathern J. N., Nakamura Y. Ty element-induced temperature-sensitive mutations of Saccharomyces cerevisiae. Genetics. 1992 Aug;131(4):821–832. doi: 10.1093/genetics/131.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D., Gallant J. On the directional specificity of ribosome frameshifting at a "hungry" codon. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5469–5473. doi: 10.1073/pnas.90.12.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövgren T. N., Petersson A., Loftfield R. B. The mechanism of aminoacylation of transfer ribonucleic acid. The role of magnesium and spermine in the synthesis of isoleucyl-tRNA. J Biol Chem. 1978 Oct 10;253(19):6702–6710. [PubMed] [Google Scholar]
- Pochon F., Cohen S. S. 4-Thiouridine and the conformation of E. coli tRNA induced by spermidine. Biochem Biophys Res Commun. 1972 May 26;47(4):720–726. doi: 10.1016/0006-291x(72)90551-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991 Oct;7(7):657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- TABOR H., TABOR C. W. SPERMIDINE, SPERMINE, AND RELATED AMINES. Pharmacol Rev. 1964 Sep;16:245–300. [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu Rev Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985 Mar;49(1):81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Tabor H., Tabor C. W. Polyamine requirement for efficient translation of amber codons in vivo. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7087–7091. doi: 10.1073/pnas.79.23.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. C., Riles L. E., Wickner R. B. Localized mutagenesis and evidence for post-transcriptional regulation of MAK3. A putative N-acetyltransferase required for double-stranded RNA virus propagation in Saccharomyces cerevisiae. J Biol Chem. 1992 Oct 5;267(28):20270–20276. [PubMed] [Google Scholar]
- Tu C., Tzeng T. H., Bruenn J. A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Ribosomal frameshifting from -2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Wickner R. B. Double-stranded RNA virus replication and packaging. J Biol Chem. 1993 Feb 25;268(6):3797–3800. [PubMed] [Google Scholar]
- Wickner R. B. Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae. Annu Rev Microbiol. 1992;46:347–375. doi: 10.1146/annurev.mi.46.100192.002023. [DOI] [PubMed] [Google Scholar]
- Xu H., Boeke J. D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]