Abstract
Resistance to amikacin (AMK) and kanamycin (KAN) in clinical Mycobacterium tuberculosis strains is largely determined by specific mutations in the rrs gene and eis gene promoter. We developed a rapid, multiplexed sloppy molecular beacon (SMB) assay to identify these mutations and then evaluated assay performance on 603 clinical M. tuberculosis DNA samples collected in South Korea. Assay performance was compared to gold-standard phenotypic drug susceptibility tests, including Lowenstein-Jensen (LJ) absolute concentration, mycobacterial growth indicator tubes (MGIT), and TREK Sensititre MycoTB MIC plate (MycoTB) methods. Target amplicons were also tested for mutations by Sanger sequencing. The SMB assay correctly detected 115/116 mutant and mixed sequences and 487/487 wild-type sequences (sensitivity and specificity of 99.1 and 100%, respectively). Using the LJ method as the reference, sensitivity and specificity for AMK resistance were 92.2% and 100%, respectively, and sensitivity and specificity for KAN resistance were 87.7% and 95.6%, respectively. Mutations in the rrs gene were unequivocally associated with high-level cross-resistance to AMK and KAN in all three conventional drug susceptibility testing methods. However, eis promoter mutations were associated with KAN resistance using the MGIT or MycoTB methods but not the LJ method. No testing method associated eis promoter mutations with AMK resistance. Among the discordant samples with AMK and/or KAN resistance but wild-type sequence at the target genes, we discovered four new mutations in the whiB7 5′ untranslated region (UTR) in 6/22 samples. All six samples were resistant only to KAN, suggesting the possible role of these whiB7 5′ UTR mutations in KAN resistance.
INTRODUCTION
Tuberculosis (TB) was declared a global public emergency nearly 20 years ago (1). Although the rate of new cases of TB has been decreasing worldwide, the millennium developmental goal target of a 50% disease reduction by 2015 is unlikely to be achieved (1). An increase in the incidence of multidrug-resistant (MDR) and extensively drug resistant (XDR) TB is a serious threat to these reduction goals (1). Patients with drug-resistant TB are best identified as rapidly as possible so that appropriate infection control and treatments can be quickly initiated (2).
Conventional phenotypic methods can take weeks to months to fully define the drug resistance pattern of Mycobacterium tuberculosis isolates due to the very slow growth of this bacterium (3–5). Molecular tests offer the promise of more rapid drug resistance detection. A number of tests are available to detect resistance to many of the first-line anti-TB drugs (6–8). However, there are fewer rapid tests available to test for resistance to the injectable second-line drugs amikacin (AMK), kanamycin (KAN), and capreomycin (CAP). DNA sequencing studies suggest that most cases of resistance to injectable drugs can be identified by detecting mutations in positions 1401, 1402, and 1484 in the M. tuberculosis 16S rRNA (rrs) gene and between nucleotides −10 to −37 of the M. tuberculosis eis gene promoter region (9–13) although sensitivity and specificity for detecting resistance have varied in different studies. The reverse line blot hybridization assay, GenoType MTBDRsl test (Hain Lifescience, Germany), which targets mutations in the rrs gene, is currently undergoing validation in trials, as reported in several studies (14). However, the open hybridization system format of line probe assays, which require rigorous physical separation of different work areas to prevent amplicon cross-contamination (15, 16), and the absence of probes targeting the eis promoter mutations are likely to limit the sensitivity and specificity of the assay (17, 18). Several other molecular techniques which detect resistance to the injectable drugs have been described recently. These include in-house reverse line blot hybridization assays, multiplex allele-specific PCR, mass spectrometry of PCR products using iPLEX Gold assay (MassArray System; Sequenom, Inc., San Diego, CA, USA), oligonucleotide microarray, and melting curves using dually labeled probes (19–24). However, most of these techniques require open handling of PCR amplicons or have not been systematically validated in clinical settings.
We have recently described two separate sloppy molecular beacon (SMB) assays that rapidly and reliably identify the M. tuberculosis mutations that are largely responsible for rifampin and fluoroquinolone (FQ) resistance (25, 26). These assays have the advantage of being in real-time PCR format so that they are easy to use and not subject to amplicon cross-contamination. Moreover, mutation detection has been shown to be robust and amenable to high-throughput testing. Here, we present a further extension of our SMB TB drug resistance detection assay system, adding assays that enable detection of resistance to AMK and KAN. We also explore the causes of discordance between our assay and phenotypic susceptibility testing methods. Our results show that some of the most commonly used phenotypic methods can miss M. tuberculosis isolates with resistance-conferring mutations if these mutations only moderately increase MICs of KAN. Novel mutations in whiB7 that are associated with low-level KAN resistance were also discovered.
MATERIALS AND METHODS
DNA samples.
M. tuberculosis test samples consisted of DNA isolated from 603 sequential M. tuberculosis isolates cultured from 503 patients enrolled in a natural history study of MDR tuberculosis (registered at ClinicalTrials.gov under no. NCT00341601) in the National Masan Hospital in Changwon, Republic of Korea. Two cohorts were tested. Cohort A consisted of treatment-naive newly suspected TB cases (158 samples), and cohort B consisted of retreatment TB cases (445 samples). Fresh sputum samples were collected from each patient at the onset of treatment and cultured for M. tuberculosis. In a subset of patients, repeat sputum samples were collected at the 1st, 4th, and 6th month of treatment and also cultured for M. tuberculosis. Nontuberculosis mycobacteria (NTM) and Gram-positive and Gram-negative bacteria test samples were taken from the New Jersey Medical School (NJMS) DNA repository as described previously (26).
Phenotypic drug susceptibility testing.
Phenotypic drug susceptibility testing was performed on all 603 isolates by the absolute concentration method on LJ medium to determine the susceptibility to AMK and KAN using a critical concentration of 40 μg/ml (the standard concentration used during 2012 when the isolates were tested) for both the antibiotics (27) at the International Tuberculosis Research Center (ITRC), South Korea. MICs of AMK and KAN for 173/603 samples were also evaluated using TREK Sensititre MycoTB MIC plates (MycoTB; TREK Diagnostic Systems, Cleveland, OH, USA) as described previously (28). For 560/603 samples, resistance to KAN was also evaluated using a mycobacterial growth indicator tube (MGIT) system (Becton Dickinson, Franklin Lakes, NJ, USA) at a critical concentration of 2.5 μg/ml. For the samples with phenotypic susceptibility test results that were discordant with Sanger sequencing results of the target genes, the phenotypic susceptibility tests were repeated to confirm the initial findings. In cases where MGIT and LJ susceptibility test results showed discordance, both the assays were repeated to confirm or rectify the initial findings.
DNA preparation and sequencing.
DNA for both SMB assay testing and Sanger sequencing was prepared from cultured isolates by boiling one loopful of culture in 200 μl of Instagene Matrix resin (Bio-Rad Laboratories, Hercules, CA, USA) in the presence of 0.1% Triton X-100 for 10 to 15 min. The supernatant was recovered after centrifugation and quantified using a NanoDrop microvolume spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For Sanger sequencing, two different fragments of the rrs gene (nucleotides 420 to 980 and 1293 to 1537) and a part of the upstream eis coding region plus the entire eis promoter were amplified using 0.5 μM each forward and reverse primer, 1× PCR buffer, 250 mM deoxynucleoside triphosphates (dNTPs), 2.5 mM MgCl2, and 0.03 U/μl of AmpliTaq Gold DNA polymerase enzyme (Applied Biosystems, Foster City, CA, USA) according to the following parameters: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 10s, 58-60°C for 30s and 72°C for 10 to 30s depending on the amplicon size. The eis promoter region and the rrs gene fragments were amplified as described previously (13, 29). For a subset of samples, a 538-bp fragment from the whiB7 gene including 412 bp of the 5′ untranslated region (UTR) and 126 bp from the open reading frame (ORF) was amplified and sequenced using primers whiB7F (5′-AAACGCGCAGGTCAGAAAAT-3′) and whiB7R (5′-CAGTGTCTTGGCTACCTCGA-3′). Additionally, a 275-bp fragment from the whiB7 gene, which included almost the entire whiB7 ORF, was also amplified using the primers whiB7-ingene-F (5′-GTCGGTACTGACAGTCCCC-3′) and whiB7-ingene-R (5′-ATGCAACAGCATCCTTGCG-3′). The PCR products were subjected to bidirectional sequencing using gene-specific forward and reverse primers in a 3130XL genetic analyzer (Applied Bio-systems, Foster City, CA, USA) using a BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
Assay molecular beacons and primers.
The SMB assays targeted M. tuberculosis mutations in codons 1401 and 1402 of the rrs gene and mutations along the promoter region of the eis gene. A 113-bp fragment (nucleotides 1335 to 1451) was amplified from the rrs gene using the primers AMG-F (5′-GCTAGTAATCGCAGATCAGCAACGCTGC-3′) and AMG-R (5′-CCTCCCGAGGGTTAGGCCACT-3′), and a 98-bp fragment encompassing the promoter region and the initial five codons of the eis gene (nucleotides −81 to 17) was amplified using the primers eis-F (5′-CACAGGGTCACAGTCACAGAATC-3′) and eis-R (5′-GCATCGCGTGATCCTTTGCCAGAC-3′). The rrs primers were designed to be specific to Mycobacterium genus, and the eis primers were designed to be specific to the M. tuberculosis complex. One SMB probe, rrs-1400 (5′-6-carboxyfluorescein-CACGACCGCCCGTCACGTCATGAAAGTCGGTCGTG--BHQ1-3′), and two SMB probes, eis-1 (5′-Cy5-CAGGCGGTCGTAATATTCACGTGCACCTGGCCGCCGCCTG-BHQ2-3′) and eis-2 (5′-TexasRed-CTCGCGGCATATGCCACAGTCGGATTCTCTGACGCGAG-BHQ2-3′) (where underlined sequences represent the stem portion of the SMB and BHQ represents black hole quencher), were targeted against the rrs gene and the eis promoter region, respectively. The rrs probe was designed to be complementary to the antisense strand, and the eis probes were designed to be complementary to the sense strand. The SMBs were designed using the in silico DNA folding program at http://mfold.rna.albany.edu/?q_mfold/dna-folding-form, and the probe-target hybrid folding program at http://mfold.rna.albany.edu/?q_DINAMelt/Two-state-melting was used to predict the possible probe-target hybrid structures and melting temperatures (Tms). The probes were designed to generate a maximum Tm difference between wild-type and mutant sequences in their respective target regions to enable unambiguous mutation identification. Primers were obtained from Sigma-Aldrich (St. Louis, MO, USA), and SMB probes were from Biosearch Technologies (Novato, CA, USA).
Assay procedure.
All the samples were independently coded and randomly distributed to ensure that assay validation was performed in a blinded manner. The assay was tested at both ITRC in Masan, South Korea, and the New Jersey Medical School (NJMS), Rutgers, Newark, NJ. Once testing of the entire 603 sample set was completed, the samples were decoded, and the PCR results were compared to the corresponding sequencing and phenotypic drug susceptibility testing results. The results obtained at each site were also compared. Assay results were not reported to the treating physicians and were not used to guide any treatment decisions. PCR was performed in 384-well plates using a Roche LightCycler 480 II real-time PCR system (Roche Diagnostics Co., Indianapolis, IN, USA) in 20-μl reaction volumes containing 100 nM forward primer and 1 μM reverse primer for the rrs gene and 1 μM forward primer and 50 nM reverse primer for the eis promoter region, 1 ng/μl of rrs-1400 and eis-1 probes and 0.8 ng/μl of eis-2 probe, 4 mM MgCl2, 250 mM deoxynucleoside triphosphates (dNTPs), 1× PCR buffer, 8% glycerol, 0.06 U/μl of Platinum TfiExo(−) DNA polymerase (Life Technologies, Grand Island, NY, USA), and 2 to 5 ng of sample DNA or an equivalent volume of water. PCR was carried out with the following steps: activation of the enzyme for 2 min at 95°C, followed by 50 cycles of denaturation at 95°C for 10s and combined annealing and extension at 67°C for 30s. Following PCR cycling, post-PCR Tm analysis was performed by denaturation at 95°C for 2 min, followed by cooling down to 45°C and then gradual heating to 85°C, with continuous monitoring of fluorescence during the process at a rate of 1 datum acquisition per degree centigrade. Tm values were identified at the end of the reaction using the Tm calling software (LightCycler 480 software). However, each Tm value was also verified by a trained observer before the final identification of the Tm value was made. Samples showing distinct double peaks for any probes corresponding to wild-type and mutant Tms were considered to be indicative of heteroresistance. A no-template control using sterile water instead of DNA as the template was used as the DNA-negative control, and a DNA-positive control using 1 ng of genomic DNA from M. tuberculosis H37Rv as the template was also included in each assay plate.
Human subject approvals.
This study was approved by the National Masan Hospital, NIAID, and Rutgers (formerly University of Medicine and Dentistry of New Jersey [UMDNJ]) institutional review boards (IRBs), and all subjects gave written informed consent (Rutgers IRB protocol number 0120090104).
RESULTS
Identification of Tm values associated with wild-type and mutant sequences.
Our SMB-based assay detected resistance to AMK and KAN by looking for mutations in the M. tuberculosis rrs gene and eis promoter that have known associations with resistance. The assay consisted of a PCR step followed by a Tm analysis in the presence of SMB probes complementary to portions of the rrs and eis target amplicons. We first evaluated the capability of the assay to identify the target mutations on artificial oligonucleotides and sequenced DNA templates from selected wild-type and mutant M. tuberculosis strains (data not shown). Wild-type sequences were identified by the presence of Tm values within 1°C of the known mean values for wild-type targets. Mutant sequences were identified by a shift in Tm values of at least 5 standard deviations away from the mean wild-type Tm values. The ability of the assay to detect the most prevalent mutations associated with AMK and KAN resistance was then evaluated on the clinical DNA samples. Tests were performed on a panel of 603 clinical samples, consisting of 487 samples with wild-type sequences and 116 samples with mutations in the assay targets. Five of these samples had mixtures of both wild-type and mutant DNA detected on Sanger sequencing. The SMB assay correctly identified 115/116 (99%) mutant or mixed (heterogeneous samples containing both mutant and wild-type DNA) samples as mutant or mixed and 487/487 (100%) pure wild-type samples as wild type. A single mixed sample (as indicated by Sanger sequencing) was identified as a wild-type sample by our assay. The Tm values produced by each SMB probe in the setting of wild-type or mutant targets were highly reproducible. For wild-type targets, probes rrs-1400, eis-1, and eis-2 showed mean Tm values of 70.1 ± 0.15°C, 63.9 ± 0.19°C, and 69 ± 0.23°C, respectively (Table 1). For mutant Tm targets, A1401G and C1402T, the mutations resulted in 3.9°C (± 0.17°C) and 5.6°C (± 0.21°C) decreases, respectively, in Tm values in probe rrs-1400 (Table 1). Similarly, the eis-1 and the eis-2 probes robustly detected a range of mutations in the eis promoter region as mutant by developing a decrease in Tm values of 4.3°C to 6.5°C compared to the expected wild-type Tm values (Table 1). The PCR assays performed in the two different laboratories at Rutgers and ITRC were in complete agreement for all the samples detected as wild type and mutant as well as mixtures.
TABLE 1.
Melting temperature (Tm) values of the rrs and eis probes tested against clinical DNA with wild-type and mutant sequences
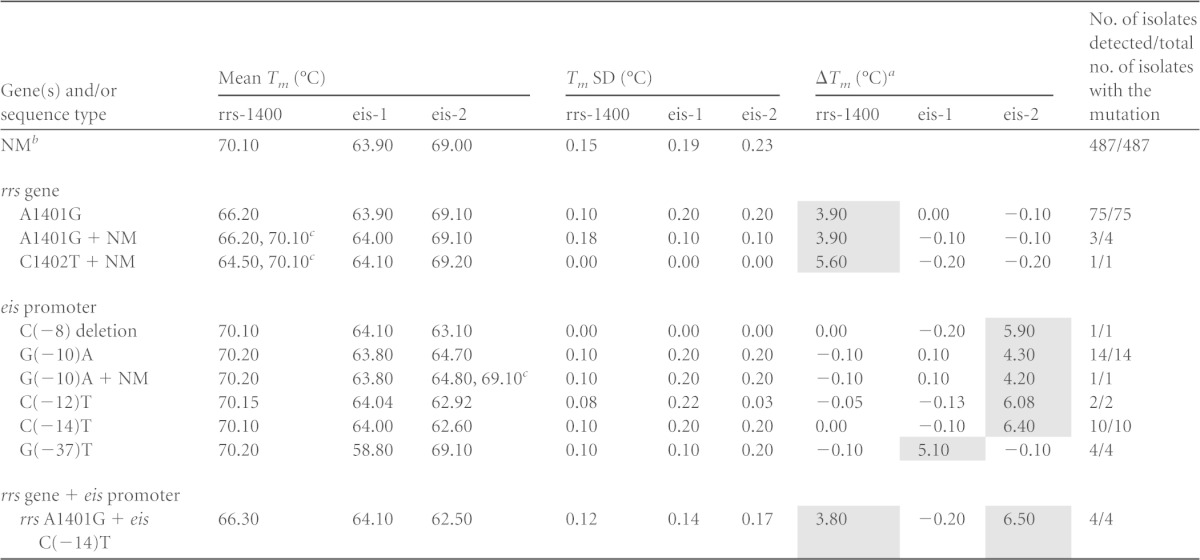
ΔTm, difference between the value of the mutant sequence and the wild-type sequence for each probe. The shaded areas represent the ΔTm indicating the mutations.
NM, no mutation (wild type). Heterogeneous samples contained both mutant and wild-type (+ NM) sequence.
Samples with mixed DNA. The two different Tms correspond to mutant and wild-type Tms, respectively.
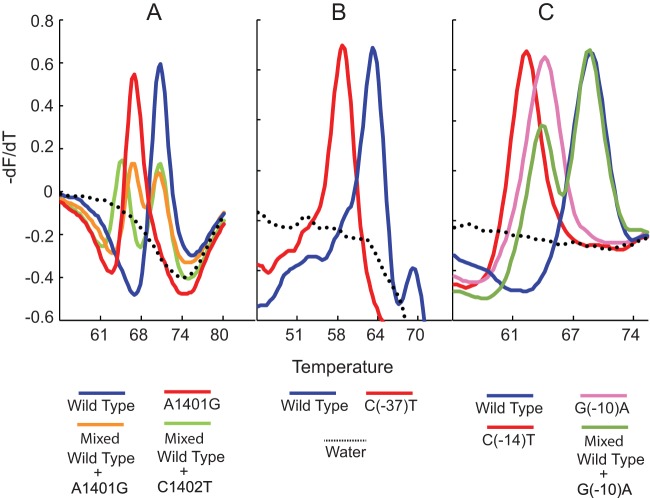
Our assay results enabled us to clearly segregate the 603 samples into wild-type and mutant Tm cluster types based on their individual three-point Tm patterns (Fig. 1). The assay correctly identified mutations in all 75 samples that contained only the A1401G mutation (Table 1 and Fig. 2A). Three of the four samples containing mixtures of A1401G and wild-type sequence were also detected as mixed wild type/mutant based on the presence of a double Tm peak. A single sample that contained a mixture of the C1402T mutation and wild-type DNA was also identified by the presence of double Tm peaks from the sample with a mutant Tm specific for the C1402T mutation (Table 1 and Fig. 2A). The 32 samples with eis promoter region mutations included five different polymorphisms (at positions −8, −10, −12, −14, and −37). All of these mutations were successfully detected by one of the eis SMBs (Table 1 and Fig. 2B and C). Four samples that had mutations in both the rrs gene and the eis promoter region were also correctly detected as double mutants (Table 1). Sequencing of the rrs gene did not identify any samples with a codon 1484 mutation, regardless of its drug susceptibility pattern.
FIG 1.
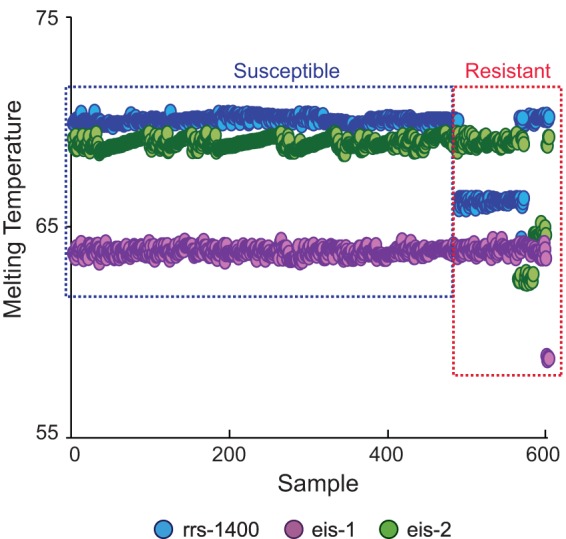
Detection of AMK and/or KAN resistance in 603 clinical DNA samples using an SMB probe-generated three-point Tm profile. Each of the three assay SMBs were tested against all M. tuberculosis DNA samples in a multiplex PCR. The results for each sample are shown as a three-point Tm plot on the x axis, with the Tm value of each SMB indicated on the y axis. Isolates are sorted from left to right as phenotypically susceptible and then as resistant. Distinct Tm shifts from at least one of the three probes can be seen in each resistant isolate.
FIG 2.
First-derivative melt peak profiles of each SMB probe. The melt peak profiles of wild-type, mutant, and mixed DNA samples are shown for the rrs-1400 SMB probe (A), the eis-1 SMB probe (B), and the eis-2 SMB probe (C). Each melt curve represents an individual strain.
Identification of amikacin resistance.
We next evaluated the performance of our molecular assay relative to phenotypic drug susceptibly test results. The apparent performance of a genotypic drug susceptibility test can vary depending on the mutations selected for inclusion in the test and the phenotypic assay that is used as a gold standard (4, 30, 31). Considering the LJ-based drug susceptibility testing method as the gold standard (performed for all of the 603 study samples), the SMB assay classified 82/90 of the AMK-resistant samples as resistant using the rrs Tm characteristic of the A1401G mutation (sensitivity of 91.1%; 95% confidence interval [CI], 82.8% to 96.8%). Using the wild-type Tm, the assay classified 512/513 of the AMK-susceptible samples as susceptible. A single isolate among the 513 AMK-susceptible isolates was identified as a mixture of wild-type and C1402T mutant DNA by our SMB assay due to the presence of a clear double peak generated by the rrs SMB probe, corresponding to the wild-type Tm and a specific C1402T mutant Tm (Fig. 2A). This was also confirmed by Sanger sequencing. Since previous studies have shown that the C1402T mutation does not code for AMK resistance (32), the specific Tm corresponding to the C1402T mutation can be considered an indicator of AMK susceptibility. This consideration resulted in our assay correctly detecting all 513/513 AMK-susceptible samples, resulting in a specificity of 100% (95% CI, 99 to 100%). Including Tm values characteristic for mutations in the eis promoter region in the analysis did not increase the sensitivity for detecting AMK resistance but decreased the specificity from 100% to 93.8% (95% CI, 91.2 to 95.6%). These results are consistent with previous reports which suggest that the eis promoter mutations are not associated with AMK resistance as defined by the LJ drug susceptibility testing (9, 33).
Identification of kanamycin resistance.
The performance of our assay to detect KAN resistance was also evaluated using LJ-based drug susceptibly testing as the gold standard for all 603 samples. Using Tm values generated by our rrs SMB typical for either the A1401G or the C1402T mutation to define resistance, the assay detected 82/106 samples as KAN resistant (sensitivity, 77.4%; 95% CI 68.0 to 84.7%). Conversely, using an rrs SMB Tm characteristic for a wild-type target to define susceptibility, the assay identified 496/497 KAN-susceptible samples as susceptible (Table 2) (specificity, 99.8%; 95% CI, 98.7 to 100%). Adding Tm values of the two eis SMBs characteristic for mutations in the eis promoter region to our definition of resistance increased the sensitivity of the assay for detecting KAN resistance from 77.4% to 87.7% (95% CI, 79.5 to 93%) as 11 additional KAN-resistant samples were classified as resistant. However, specificity decreased from 99.8% to 95.6% (95% CI, 93.3 to 97.1%) as 21 KAN-susceptible samples with eis promoter mutations were now “falsely” detected as KAN resistant (Table 2).
TABLE 2.
Susceptibility of the clinical strains to AMK and KAN by the LJ and MGIT methods and their relation to the mutations present in the rrs gene and the eis promoter region
| Test method and drug susceptibilitya | No. of isolates detected by genotype |
Total no. of isolates | |||
|---|---|---|---|---|---|
| rrs A1401G | rrs C1402T | eis promoter mutation | Wild-type rrs gene and eis promoter | ||
| LJ | |||||
| AMK resistant | 82 | 0 | 1 | 7 | 90 |
| AMK susceptible | 0 | 1 | 31 | 481 | 513 |
| KAN resistant | 82 | 0 | 11 | 13 | 106 |
| KAN susceptible | 0 | 1 | 21 | 475 | 497 |
| MGIT | |||||
| KAN resistant | 63 | 0 | 30 | 20 | 113 |
| KAN susceptible | 1 | 0 | 1 | 445 | 447 |
LJ and MGIT susceptibility as determined by the LJ proportions and the MGIT methods respectively.
We then performed a similar analysis using MGIT-based drug susceptibility test results as the gold standard for the 560 of the samples for which a MGIT result was available. This subset included all the samples harboring only eis promoter mutations. Comparison of our assay results to the MGIT-based gold standard helped to clarify the eis mutants with discordant KAN resistance in LJ medium. Using MGIT as the gold standard and only the rrs SMB Tm values characteristic for the A1401G or C1402T mutation to define KAN resistance, only 63/113 KAN-resistant samples were identified as resistant by the SMB assay (sensitivity, 55.8%; 95% CI, 46.1 to 65%). Conversely, using the rrs SMB Tm characteristic for the wild-type target to define susceptibility, the assay identified 445/447 samples as KAN susceptible (specificity, 99.8%; 95% CI, 98.5 to 100%) (Table 2). Unlike the case with LJ-based susceptibility testing, including the Tm values characteristic for eis promoter mutations in this case increased sensitivity for resistance testing from 55.8% to 82.3%, with the specificity of the assay for KAN resistance still remaining very high at 99.5% (95% CI, 98.2 to 100%). Thus, based on an MGIT-based susceptibility test, the eis assay allowed the detection of 29 additional KAN-resistant samples without affecting specificity (Table 2).
Relationship between mutations detected by the assay and MIC.
The discordance between resistance as defined by our assay and resistance as defined by our two phenotypic susceptibility test methods principally involved isolates with eis promoter mutations. Previous studies have shown that the eis promoter mutations give rise to relatively low levels of KAN resistance, while rrs gene mutations result in high-level resistance to AMK, KAN, and CAP (9, 10, 12, 33). An additional finding in our results was the discordance that we observed between LJ- and MGIT-based susceptibility test results. We performed MIC testing to more carefully explore the relationship between rrs and eis promoter mutations and their differential susceptibility patterns in the LJ and the MGIT systems. Samples that were either susceptible to both AMK and KAN (and wild type at both the target regions) or chosen to be representative of the most common mutation types in the two target genes [the A1401G mutation in rrs and the eis promoter region mutations G(−10)A, C(−14)T, and G(−37)T] were tested by the MycoTB method to determine the MICs. Additional isolates known to be wild type in both of our assay targets were also tested as controls. We observed that the AMK MICs of the isolates that had only eis promoter mutations (without rrs mutations) ranged between 0.25 μg/ml and 2 μg/ml, with the majority of samples showing MICs of 0.5 μg/ml to 1 μg/ml (Fig. 3). Only one eis promoter mutant had an AMK MIC of 4 μg/ml. Control isolates with no eis promoter or rrs mutations had AMK MICs between 0.25 μg/ml and 0.5 μg/ml. Thus, the AMK MICs of the isolates with either wild-type or mutant eis promoter sequences overlapped substantially. In contrast, the KAN MICs for most of the same eis promoter mutants ranged from 5 μg/ml to 20 μg/ml, with one isolate showing an MIC of 40 μg/ml (Fig. 3). Only one eis promoter mutant had a low KAN MIC (2.5 μg/ml). The isolates with wild-type eis promoter sequences showed KAN MICs between 0.6 μg/ml to 2.5 μg/ml, which is 2- to 30-fold less than the mean MIC of the eis promoter mutants (Fig. 3). Thus, in contrast to the situation with AMK, the KAN MICs of the wild-type isolates overlapped very little with the KAN MICs of the eis promoter mutants. These results strongly suggest that eis promoter mutants should be considered to have low- to moderate-level KAN resistance even if resistance is not detected on LJ-based or even MGIT-based susceptibility tests.
FIG 3.

MIC values of rrs and eis mutant and wild-type strains. The average AMK and KAN MIC values for the rrs and the eis mutants and the wild-type strains are shown. Error bars represent the means ± 1 standard deviation of the MIC values. eis-P, eis gene promoter.
Assay specificity against bacteria other than M. tuberculosis.
The analytical specificity of the assay was tested against a panel of 18 species of nontuberculous mycobacteria (NTM) obtained from the ATCC repository (Manassas, VA, USA), 121 clinical NTM strains representing 26 species, and 18 species of Gram-positive and Gram-negative bacteria. The rrs region targeted in our assay is highly conserved among different NTM species. Thus, the rrs assay generated a Tm of 70°C (which is identical to the Tm generated in the presence of wild-type M. tuberculosis DNA) for all the NTM strains tested, as expected based on sequence homology for Mycobacterium xenopi, which did not generate any Tm. The NTM species which generate Tm values identical to aminoglycoside-susceptible M. tuberculosis strains would not be expected to cause a false resistance test result. When M. tuberculosis DNA from rrs mutant AMK- and KAN-resistant strains was mixed with a 10- to 20-fold excess of NTM DNA, a distinct double Tm peak was produced by the assay, corresponding to a mutant Tm value from the M. tuberculosis target and a wild-type Tm value from the NTM sequence (data not shown), indicating that resistance-associated rrs mutations can be detected in M. tuberculosis by our assay even in the presence of a large background of NTM DNA. No visible melt curve was generated by the eis probes in the presence of any NTM species tested even when 107 genome equivalents of DNA was added to the PCR assay. None of the Gram-positive or Gram-negative bacteria produced Tm values to any of the rrs or eis SMBs; thus, they did not cause any false resistance calls to be made by the assay.
Additional genetic causes of AMK and KAN resistance.
Our study included 22 samples that were resistant to AMK and/or KAN but had wild-type rrs gene and eis promoter sequences. Recent investigations have suggested that mutations in the 5′ untranslated region (UTR) of the whiB7 gene may cause aminoglycoside resistance in M. tuberculosis (34). To determine whether whiB7 mutations could be responsible for some of our phenotypically resistant but assay-susceptible isolates, we sequenced all 22 samples in a 412-bp region upstream of the whiB7 gene start site plus a portion of the whiB7 open reading frame. As a control set, we also sequenced 30 randomly picked pan-susceptible isolates. Of the 22 discordant isolates, 6 isolates from three patients showed mutations in the whiB7 5′ UTR region. One sample had a cytosine deletion at position +138 in the 5′ UTR, two samples from one patient contained an A-to-G mutation at position +237, and the remaining three samples from a single patient showed an A-to-C mutation at position +273 (Table 3), considering the transcription start site as +1 (34). Three samples from a single patient failed to generate any amplification from the 5′ UTR after repeated PCR attempts despite functioning positive PCR controls. This suggested the presence of a large deletion in the 5′ UTR since we could easily amplify a 275-bp fragment from within the whiB7 ORF for all the three samples. All the samples with whiB7 mutations were resistant only to KAN, which is consistent with the presumed whiB7 mechanism of action by upregulation of the eis gene (34). The KAN MICs for these isolates were also low at 5 μg/ml, a value similar to that observed for eis promoter mutants. None of the 30 control samples that were susceptible to aminoglycosides had any mutations in the 5′ UTR of the whiB7 gene. Further studies are necessary to confirm the relationship of these mutations and the deletion in the 5′ UTR of the whiB7 gene with aminoglycoside resistance. However, the absence of such mutations in the susceptible strains implies that they might have some role to play in aminoglycoside resistance, and future assays could target these mutations to improve sensitivity for detecting low-level KAN resistance.
TABLE 3.
Susceptibility of the clinical strains to AMK and KAN and mutations in the 5′ untranslated region of the whiB7 gene
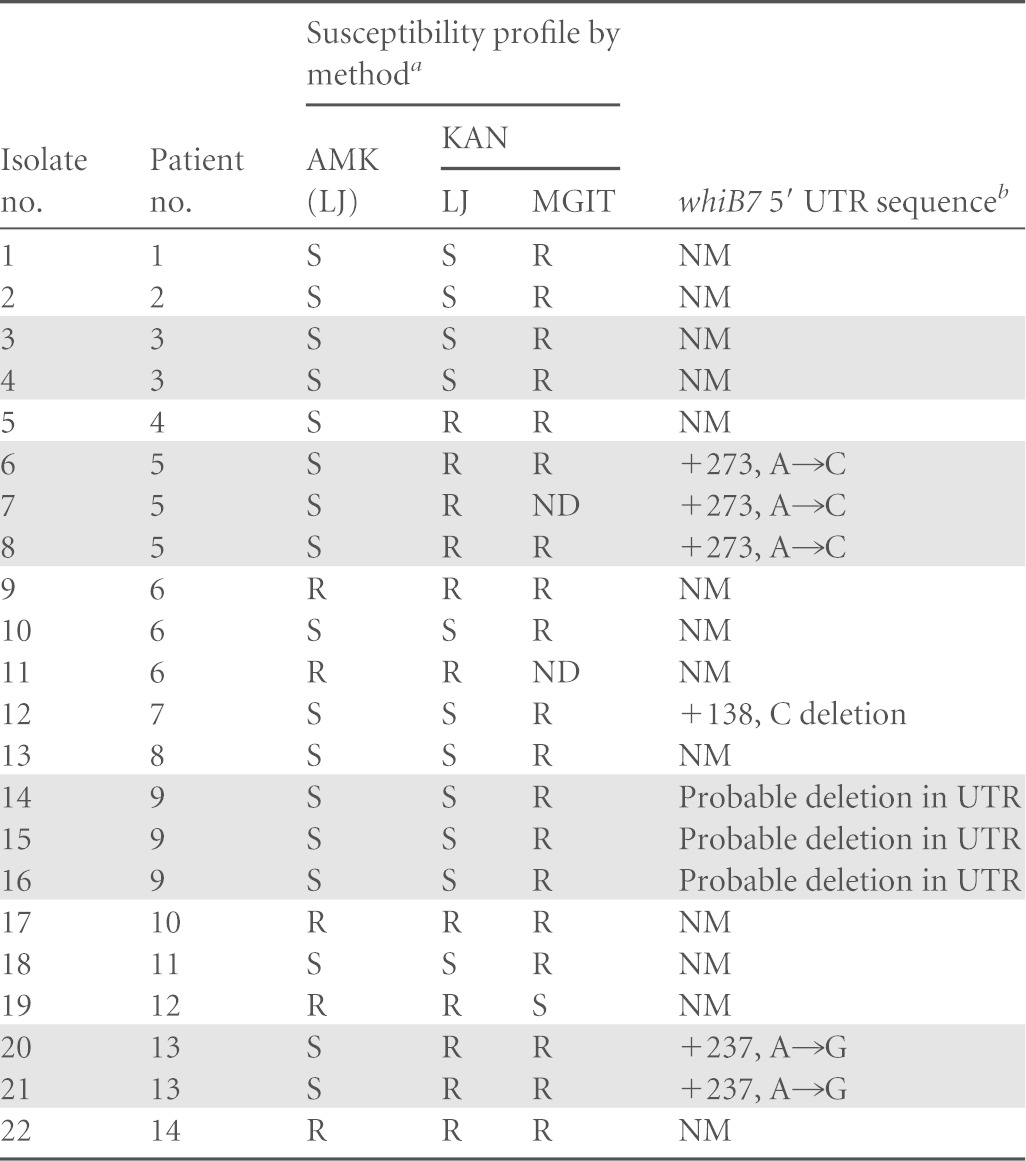
LJ and MGIT susceptibility as determined by the LJ proportions and the MGIT methods respectively. ND, not determined; R; resistant, S; susceptible.
NM; no mutation. Shaded rows represent multiple samples from an individual patient.
DISCUSSION
Our multiplexed SMB PCR and melt assay accurately identified mutations in the rrs gene and eis promoter associated with resistance to AMK and/or KAN. The assay did not produce false resistance calls when tested against NTM and Gram-positive and Gram-negative bacteria. Most cases of heteroresistance, when present, were also detected by the assay. Unlike the MTBDRsl platform, our assay can be performed in a closed real-time PCR system and can easily be adapted to high-throughput testing as all assay steps are performed in 96-well plates. The SMB assay also avoids potential problems associated with alternative methods for mutation detection. High-resolution melt curve analysis requires the ability to detect subtle variations in melt curves (22). Other post-PCR melt-based molecular assays must detect mutations by decoding complex fluorescence contours (35). In contrast, the SMB assay produces clear and easily distinguishable Tm peaks and definitive Tm shifts to identify the mutations of interest. Individual Tm values can also be used to cluster samples that have the same genotype.
We tested our assay on a panel of 603 clinical samples representing both new cases of TB as well as unresolved retreatment cases and evaluated the relationship of the targeted mutations with the susceptibility pattern of the clinical isolates. We observed that 100% of the isolates with rrs A1401G mutations had a strong correlation with high-level resistance to both AMK and KAN. However, eis promoter mutations resulted in only moderate- to low-level KAN resistance and no resistance to AMK, which is consistent with previous studies (9, 33). Our study also showed that the LJ absolute concentration method for susceptibility testing does not adequately detect moderate- to low-level KAN resistance. In fact, nearly two-thirds of the samples with eis promoter mutations were detected as KAN susceptible in the LJ medium. However, all but one sample with eis promoter mutations were detected as KAN resistant by the MGIT method. Two such isolates contained an eis C(−12)T mutation. These mutants were also resistant to KAN when tested by MycoTB, which showed a KAN MIC of 5 μg/ml. Previous studies have suggested that clinical isolates with C(−12)T mutations do not correlate (33) or correlate poorly (9, 17) with KAN resistance. These studies possibly missed the relation between this mutation and low-level KAN resistance due to the testing method used to establish phenotypic susceptibility. These results suggest that the MGIT or MycoTB method should be preferred for testing phenotypic resistance to KAN. They also highlight the power of genotypic resistance tests, such as ours, to detect mutations which cause low-level resistance and may be missed by phenotypic tests alone (30, 31, 36).
Our study set included one sample that was a mixture of rrs wild-type and rrs C1402T mutants. This sample was susceptible to both AMK and KAN in LJ medium. Isolates with C1402T mutations have been reported to be susceptible to AMK but resistant to KAN (32). In this particular case, repeated susceptibility tests using LJ medium showed susceptibility to KAN, presumably because of the heteroresistant nature of the sample. Here, the molecular assay served as a better predictor of potentially emerging resistance than the phenotypic assay as the SMB assay clearly detected the presence of both the wild-type and the mutant DNA types.
The incidence of rrs codon 1484 mutations in clinical strains with AMK or KAN resistance has been very low (12), making its clinical significance debatable. A separate version of our assay which targeted rrs codon 1484 did not detect any mutations in any of the 603 isolates in our study or in an additional 259 isolates from the New Jersey-New York area, which included 33 AMK- and KAN-resistant isolates. The lack of any rrs codon 1484 mutations in this enlarged study set was confirmed by Sanger sequencing (data not shown). In light of the very low prevalence of mutations in rrs codon 1484, this codon is unlikely to provide much value in predicting aminoglycoside resistance. Thus, we recommend that molecular assays for aminoglycoside resistance target only the 1401 and 1402 codons in the rrs gene.
We found 22 AMK- or KAN-resistant samples that had wild-type sequences in the rrs gene and the eis promoter region. A recent study has shown a possible association between mutations in the 5′ UTR of the whiB7 gene and KAN resistance by identifying a 5′ UTR whiB7 mutation in a single clinical strain with unexplained KAN resistance (34). To our knowledge, our study is the second to report that mutations in this region are associated with KAN resistance. We also describe several novel 5′ UTR whiB7 mutations, as well as a deletion, that appear to be associated with KAN resistance. No suitable universal biomarkers have been identified which can account for KAN and AMK resistance in the remaining 15 to 20% of clinical strains with both wild-type rrs and eis promoter region. Samples containing wild-type rrs gene and eis promoter region DNA mixed with a trace amount of mutant targets from a KAN- or AMK-resistant subpopulation could also account for the remaining discordances between phenotypic resistance tests and our SMB assay. However, extensive investigation of heteroresistance was beyond the scope of this study. Some recent studies have suggested that PPE60 and Rv3168 genes might be involved in unexplained KAN resistance (37, 38) although this remains to be verified in clinical settings. Further studies are necessary to confirm the relationship of these mutations to KAN resistance.
In summary, we have developed a sensitive and specific assay for detection of AMK and KAN resistance in M. tuberculosis and validated it in clinical isolates with a high prevalence of MDR and XDR TB. We show that rrs A1401G mutations encode high-level cross-resistance to both AMK and KAN and that eis promoter mutations encode moderate- to low-level KAN resistance, which is consistent with previous functional genomics studies (33). Comparing the performance of our assay with that of three different phenotypic susceptibility testing methods in solid and liquid media revealed that low- to moderate-level KAN resistance caused by eis promoter mutations is largely missed by the LJ-based susceptibility tests. These results strongly argue for the value of genotypic tests to detect aminoglycoside resistance, and the results demonstrate the specific utility of our SMB-based assay.
ACKNOWLEDGMENTS
This work was supported in part by U.S. National Institutes of Health (NIH) grants AI082174 and AI080653, the Intramural Research Program of the NIAID, NIH, and the South Korean Ministry of Health, Welfare and Family Affairs.
D.A. acknowledges that he is among a group of inventors who earn royalties from molecular beacon usage.
REFERENCES
- 1.World Health Organization. 2013. Global tuberculosis report 2013. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf. [Google Scholar]
- 2.Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT, Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L, Whitelaw A, Sagadevan K, Alexander H, Albert H, Cobelens F, Cox H, Alland D, Perkins MD. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heifets L, Linder T, Sanchez T, Spencer D, Brennan J. 2000. Two liquid medium systems, mycobacteria growth indicator tube and MB redox tube, for Mycobacterium tuberculosis isolation from sputum specimens. J Clin Microbiol 38:1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SJ. 2005. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J 25:564–569. doi: 10.1183/09031936.05.00111304. [DOI] [PubMed] [Google Scholar]
- 5.Kent PT, Kubica GP. 1985. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 6.Balasingham SV, Davidsen T, Szpinda I, Frye SA, Tonjum T. 2009. Molecular diagnostics in tuberculosis: basis and implications for therapy. Mol Diagn Ther 13:137–151. doi: 10.1007/BF03256322. [DOI] [PubMed] [Google Scholar]
- 7.Kalokhe AS, Shafiq M, Lee JC, Ray SM, Wang YF, Metchock B, Anderson AM, Nguyen ML. 2013. Multidrug-resistant tuberculosis drug susceptibility and molecular diagnostic testing. Am J Med Sci 345:143–148. doi: 10.1097/MAJ.0b013e31825d32c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Grady J, Maeurer M, Mwaba P, Kapata N, Bates M, Hoelscher M, Zumla A. 2011. New and improved diagnostics for detection of drug-resistant pulmonary tuberculosis. Curr Opin Pulm Med 17:134–141. doi: 10.1097/MCP.0b013e3283452346. [DOI] [PubMed] [Google Scholar]
- 9.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, Hooks DP, Cowan LS, Plikaytis BB, Posey JE. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:2032–2041. doi: 10.1128/AAC.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Q, Dai G, Long Q, Yu X, Dong L, Huang H, Xie J. 2013. Mycobacterium tuberculosis rrs A1401G mutation correlates with high-level resistance to kanamycin, amikacin, and capreomycin in clinical isolates from mainland China. Diagn Microbiol Infect Dis 77:138–142. doi: 10.1016/j.diagmicrobio.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Feuerriegel S, Cox HS, Zarkua N, Karimovich HA, Braker K, Rusch-Gerdes S, Niemann S. 2009. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob Agents Chemother 53:3353–3356. doi: 10.1128/AAC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC. 2012. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275. doi: 10.1371/journal.pone.0033275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Via LE, Cho SN, Hwang S, Bang H, Park SK, Kang HS, Jeon D, Min SY, Oh T, Kim Y, Kim YM, Rajan V, Wong SY, Shamputa IC, Carroll M, Goldfeder L, Lee SA, Holland SM, Eum S, Lee H, Barry CE III. 2010. Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean patients with drug-resistant tuberculosis. J Clin Microbiol 48:402–411. doi: 10.1128/JCM.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y, Liu S, Wang Q, Wang L, Tang S, Wang J, Lu W. 2013. Rapid diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol using genotype MTBDRsl assay: a meta-analysis. PLoS One 8:e55292. doi: 10.1371/journal.pone.0055292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albert H, Bwanga F, Mukkada S, Nyesiga B, Ademun JP, Lukyamuzi G, Haile M, Hoffner S, Joloba M, O'Brien R. 2010. Rapid screening of MDR-TB using molecular Line Probe Assay is feasible in Uganda. BMC Infect Dis 10:41. doi: 10.1186/1471-2334-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling DI, Zwerling AA, Pai M. 2008. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J 32:1165–1174. doi: 10.1183/09031936.00061808. [DOI] [PubMed] [Google Scholar]
- 17.Hoshide M, Qian L, Rodrigues C, Warren R, Victor T, Evasco HB II, Tupasi T, Crudu V, Douglas JT. 2014. Geographical differences associated with single-nucleotide polymorphisms (SNPs) in nine gene targets among resistant clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol 52:1322–1329. doi: 10.1128/JCM.00857-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, Shen Y, Fan X, Diao N, Wang F, Wang S, Weng X, Zhang W. 2013. Underestimation of the resistance of Mycobacterium tuberculosis to second-line drugs by the new GenoType MTBDRsl test. J Mol Diagn 15:44–50. doi: 10.1016/j.jmoldx.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Ajbani K, Shetty A, Mehta A, Rodrigues C. 2011. Rapid diagnosis of extensively drug-resistant tuberculosis by use of a reverse line blot hybridization assay. J Clin Microbiol 49:2546–2551. doi: 10.1128/JCM.02511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Luo T, Li J, Mei J, Gao Q. 2013. Triplex real-time PCR melting curve analysis for detecting Mycobacterium tuberculosis mutations associated with resistance to second-line drugs in a single reaction. J Antimicrob Chemother 68:1097–1103. doi: 10.1093/jac/dks509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vadwai V, Shetty A, Rodrigues C. 2012. Multiplex allele specific PCR for rapid detection of extensively drug resistant tuberculosis. Tuberculosis (Edinb) 92:236–242. doi: 10.1016/j.tube.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Yadav R, Sethi S, Mewara A, Dhatwalia SK, Gupta D, Sharma M. 2012. Rapid detection of rifampicin, isoniazid and streptomycin resistance in Mycobacterium tuberculosis clinical isolates by high-resolution melting curve analysis. J Appl Microbiol 113:856–862. doi: 10.1111/j.1365-2672.2012.05379.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhao B, Huang H, Zhu Y, Peng J, Dai G, Jiang G, Liu L, Zhao Y, Jin Q. 2013. Co-occurrence of amikacin-resistant and -susceptible Mycobacterium tuberculosis isolates in clinical samples from Beijing, China. J Antimicrob Chemother 68:1537–1542. doi: 10.1093/jac/dkt082. [DOI] [PubMed] [Google Scholar]
- 24.Zimenkov DV, Antonova OV, Kuz'min AV, Isaeva YD, Krylova LY, Popov SA, Zasedatelev AS, Mikhailovich VM, Gryadunov DA. 2013. Detection of second-line drug resistance in Mycobacterium tuberculosis using oligonucleotide microarrays. BMC Infect Dis 13:240. doi: 10.1186/1471-2334-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravorty S, Aladegbami B, Thoms K, Lee JS, Lee EG, Rajan V, Cho EJ, Kim H, Kwak H, Kurepina N, Cho SN, Kreiswirth B, Via LE, Barry CE III, Alland D. 2011. Rapid detection of fluoroquinolone-resistant and heteroresistant Mycobacterium tuberculosis by use of sloppy molecular beacons and dual melting-temperature codes in a real-time PCR assay. J Clin Microbiol 49:932–940. doi: 10.1128/JCM.02271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravorty S, Kothari H, Aladegbami B, Cho EJ, Lee JS, Roh SS, Kim H, Kwak H, Lee EG, Hwang SH, Banada PP, Safi H, Via LE, Cho SN, Barry CE III, Alland D. 2012. Rapid, high-throughput detection of rifampin resistance and heteroresistance in Mycobacterium tuberculosis by use of sloppy molecular beacon melting temperature coding. J Clin Microbiol 50:2194–2202. doi: 10.1128/JCM.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jnawali HN, Hwang SC, Park YK, Kim H, Lee YS, Chung GT, Choe KH, Ryoo S. 2013. Characterization of mutations in multi- and extensive drug resistance among strains of Mycobacterium tuberculosis clinical isolates in Republic of Korea. Diagn Microbiol Infect Dis 76:187–196. doi: 10.1016/j.diagmicrobio.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Armstrong DT, Ssengooba W, Park JA, Yu Y, Mumbowa F, Namaganda C, Mboowa G, Nakayita G, Armakovitch S, Chien G, Cho SN, Via LE, Barry CE III, Ellner JJ, Alland D, Dorman SE, Joloba ML. 2014. Sensititre MYCOTB MIC plate for testing Mycobacterium tuberculosis susceptibility to first- and second-line drugs. Antimicrob Agents Chemother 58:11–18. doi: 10.1128/AAC.01209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engstrom A, Perskvist N, Werngren J, Hoffner SE, Jureen P. 2011. Comparison of clinical isolates and in vitro selected mutants reveals that tlyA is not a sensitive genetic marker for capreomycin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 66:1247–1254. doi: 10.1093/jac/dkr109. [DOI] [PubMed] [Google Scholar]
- 30.Rigouts L, Gumusboga M, de Rijk WB, Nduwamahoro E, Uwizeye C, de Jong B, Van Deun A. 2013. Rifampin resistance missed in automated liquid culture system for Mycobacterium tuberculosis isolates with specific rpoB mutations. J Clin Microbiol 51:2641–2645. doi: 10.1128/JCM.02741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Deun A, Aung KJ, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC. 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 51:2633–2640. doi: 10.1128/JCM.00553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maus CE, Plikaytis BB, Shinnick TM. 2005. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob Agents Chemother 49:3192–3197. doi: 10.1128/AAC.49.8.3192-3197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaunbrecher MA, Sikes RD Jr, Metchock B, Shinnick TM, Posey JE. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 106:20004–20009. doi: 10.1073/pnas.0907925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves AZ, Campbell PJ, Sultana R, Malik S, Murray M, Plikaytis BB, Shinnick TM, Posey JE. 2013. Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob Agents Chemother 57:1857–1865. doi: 10.1128/AAC.02191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice JE, Reis AH Jr, Rice LM, Carver-Brown RK, Wangh LJ. 2012. Fluorescent signatures for variable DNA sequences. Nucleic Acids Res 40:e164. doi: 10.1093/nar/gks731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirgel FA, Tait M, Warren RM, Streicher EM, Bottger EC, van Helden PD, Gey van Pittius NC, Coetzee G, Hoosain EY, Chabula-Nxiweni M, Hayes C, Victor TC, Trollip A. 2012. Mutations in the rrs A1401G gene and phenotypic resistance to amikacin and capreomycin in Mycobacterium tuberculosis. Microb Drug Resist 18:193–197. doi: 10.1089/mdr.2011.0063. [DOI] [PubMed] [Google Scholar]
- 37.Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PK, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, Liu Z, Li C, Galwey N, Deng J, Zhou Y, Zhu Y, Gao Y, Wang S, Huang Y, Wang M, Zhong Q, Zhou L, Chen T, Zhou J, Yang R, Zhu G, Hang H, Zhang J, Li F, Wan K, Wang J, Zhang XE, Bi L. 2013. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet 45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]