Abstract
Outbreaks and pseudo-outbreaks of infection related to bronchoscopy typically involve Gram-negative bacteria, Mycobacterium species or Legionella species. We report an unusual bronchoscopy-related pseudo-outbreak due to Actinomyces graevenitzii. Extensive epidemiological and microbiological investigation failed to identify a common source. Strain typing revealed that the cluster was comprised of heterogeneous strains of A. graevenitzii. A change in laboratory procedures for Actinomyces cultures was coincident with the emergence of the pseudo-outbreak, and we determined that A. graevenitzii isolates more readily adopted a white, dry, molar tooth appearance on anaerobic colistin nalidixic acid (CNA) agar which likely facilitated its detection and identification in bronchoscopic specimens. This unusual pseudo-outbreak was related to frequent requests of bronchoscopists for Actinomyces cultures combined with a change in microbiology laboratory practices.
INTRODUCTION
Actinomyces graevenitzii is infrequently identified in clinical specimens, and its pathogenicity is not well defined. A. graevenitzii is a component of the oropharyngeal microbiota and has been reported to cause pneumonia, lung abscesses, osteitis of the jaw, and bacteremia (1–8). In early December 2013, an increase in the recovery of A. graevenitzii from specimens from patients who underwent bronchoscopy was noted by microbiology and infection control staff. Our investigation revealed that the cluster of A. graevenitzii isolates represented a pseudo-outbreak associated with a change in laboratory practices.
MATERIALS AND METHODS
Setting.
A 1,500-bed tertiary care university-affiliated teaching hospital was the setting for this study.
Epidemiological investigation.
The microbiology laboratory's computerized database was queried to obtain the number of bronchoscopy cultures performed, the number of Actinomyces cultures ordered on bronchoscopy specimens, and the number of those cultures ordered that were positive for Actinomyces from January 2011 through December 2013. Medical records of patients whose bronchoscopy specimens yielded Actinomyces were reviewed, and the following variables were recorded: date of bronchoscopy, results of bronchoscopic specimen cultures for routine respiratory pathogens and for Actinomyces, bronchoscope used, bronchoscopist, endoscopy personnel involved in the procedure, procedure room number, and medications administered during bronchoscopy procedures. Only three bronchoscopes were routinely used, and almost all bronchoscopies were performed in procedure room 3. Therefore, we focused our investigation on room 3, the three bronchoscopes, and the “wash room” in which all bronchoscopes and endoscopes undergo high-level disinfection using one of three automated endoscope reprocessors (AERs) (DSDs Medivators, Minneapolis, MN). Findings of cytologic examination and bronchial biopsy specimens were also recorded.
Environmental sources of samples collected for culture in the endoscopy suite included unopened bottles of lidocaine, atomizer, cotton balls, an ice bucket, a nasal cannula, suction tubing, oxygen mask, the inside of a supply cabinet, sterile syringes, a supply table, syringe caps, a faucet, a sink, and tap water. Samples collected from the wash room included samples from working enzymatic cleaner (Enzol; Advanced Sterilization Products, Irvine, CA), washer basins, connectors, working solutions of ortho-phthalaldehyde (Cidex OPA, Advanced Sterilization Products, Irvine, CA), tap water, alcohol containers, special bronchoscope connectors, a tip protector, and metal drains of AERs.
Two of the three bronchoscopes used on case patients that were available for culturing, and one other randomly selected endoscope were cultured by injecting sterile water through the suctions channels and biopsy ports. The third bronchoscope was used on only a single case patient. Fluid cultures were filtered through a 0.45-μm filter (Thermo Scientific, Rochester, NY), and the filter was placed on a CDC anaerobic blood agar plate and incubated in an anaerobic chamber for 10 days at 36°C. Other environmental samples that were collected on swabs were cultured for Actinomyces on anaerobic blood agar plates and thioglycolate broth (Remel Inc., Norcross, GA) and incubated in an anaerobic chamber for 10 days at 36°C. Swab samples of throats and fingers of personnel common to most case patients and the 2 personnel performing the high-level disinfection process on the bronchoscopes were also cultured for Actinomyces. The microbiology laboratory retrieved any recent Actinomyces isolates that had been stored in an ultralow freezer and saved all Actinomyces isolates going forward.
Culture methods of bronchoscopic specimens.
Bronchoscopic specimens were quantitatively and semiquantitatively cultured for routine respiratory pathogens using standard methods (9). Prior to late May 2013, specimens for Actinomyces cultures were plated on CDC anaerobic blood agar (Remel, Lenexa, KS). After May 2013, specimens for Actinomyces cultures were inoculated onto anaerobic colistin nalidixic acid (CNA) reducible blood agar (Remel). Cultures on either plate were incubated anaerobically at 36°C for 10 days.
Identification of Actinomyces isolates.
All Actinomyces cultures were examined by experienced medical technologists, and the decision to work up colonies suggestive of Actinomyces was at the discretion of the technologist in accordance with laboratory procedures. Colonies were initially identified using RapID ANA II kits (Remel) or matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) (Vitek MS, bioMérieux, Durham, NC) using the Saramis v4.10 database (prior to mid-November 2013) or Vitek MS IVD database (after mid-November 2013). Colonies were directly smeared onto the target plate. Isolates failing to give an acceptable identification underwent 16S rRNA sequencing using the MicroSeq 500-bp 16S kit (Life Technologies, Foster City, CA) according to the manufacturer's instructions. Species identifications were made by comparison of sequences to MicroSeq Bacterial Database 2.2, and if identification with this database was not possible, comparisons were made to the BLAST 16S database and/or BLAST nucleotide database. The criteria described in CLSI document MM18-A for identification were used (10).
Strain typing.
Nucleic acid was extracted with the UltraClean kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions, and strain typing was performed using repetitive-sequence PCR (rep-PCR) (DiversiLab, bioMérieux, Durham, NC) according to the manufacturer's instructions. Relatedness was determined using Kullback-Leibler analysis in the Diversilab Software v3.4, and further comparisons were made using the criteria of Fluit et al. for Gram-positive bacteria (11). Briefly, isolates with < 96% identity were considered unrelated. Pairwise comparisons were made for isolates with 96% to 99% similarities to assess relatedness.
Comparison of growth on anaerobic CNA and CDC blood agars.
Ten A. graevenitzii isolates and seven Actinomyces isolates of species other than A. graevenitzii (A. neuii [1], A. odontolyticus [3], and A. schaalii [3]) were plated to both CDC and anaerobic CNA agar, and colony appearance was assessed at 48 h, 72, h, and 96 h. Colonies were classified as either “wet” appearing, “dry” appearing, or “molar tooth,” and the relative abundance of each colony morphology was estimated.
Case-control study.
A case patient was defined as any patient who had a bronchoscopy specimen culture-positive for Actinomyces during the period from May to December 2013. A random sample of 33 control patients who had bronchoscopy specimen cultures negative for Actinomyces during the period from May through December 2013 was generated using statistical analysis software (SPSS. IBM, Armonk, NY). Variables recorded included day of the week that bronchoscopy was performed, the time of the day, the room in which the procedure was performed, bronchoscopist and other endoscopy personnel involved in the procedure, the bronchoscope used, the AER used to disinfect the bronchoscope prior to its use, whether the patient was an inpatient or outpatient, results of routine, mycobacterial, and fungal cultures, and the occurrence of any previous bronchoscopies.
Statistical analysis.
Differences in proportions were analyzed using chi-square or Fisher's exact tests. Differences in colony counts between agar types were compared by two-way analysis of variance (ANOVA) (GraphPad, San Diego, CA). A P value of <0.05 was considered significant.
RESULTS
Epidemiological investigation.
The microbiological findings for the 18 cases are shown in Table 1. All 18 case patients had abnormalities on chest radiography. None of these patients had cytologic or transbronchial biopsy findings typical of pulmonary actinomycosis (e.g., sulfur granules or organisms with typical morphology on silver stains). One patient had histologic findings compatible with organizing pneumonia. Two immunocompromised patients were treated with high-dose penicillin for possible actinomycosis, but infectious disease consultants questioned this diagnosis. One of the two patients had histologic evidence of mucormycosis and was also treated with antifungal therapy.
TABLE 1.
Microbiological findings of cultures positive for Actinomyces during the pseudo-outbreaka
| Case | Specimen | Normal flora (CFU/ml) | Primary stain | Other organisms | Actinomyces culture result | DivseriLab strain | Original clinical ID method | % MALDI-TOF |
|---|---|---|---|---|---|---|---|---|
| 1 | BAL | 75–100,000 | 2+ WBC 2+ GPC Pairs 2+ GNR | None | 3+ A. graevenitzii | 4 | 16S | No ID (Saramis) |
| 2 | Lung tissue | 3+ mixed | 2+ WBC 3+ GPC pairs 1+ GNR | None | 2+ A. graevenitzii | 6 | 16S | No ID (Saramis) |
| BAL | 25–49,000 | 1+ WBC 2+ mixed | 1 CFU of S. aureus | Not detected | NA | NA | NA | |
| 3 | Bronchial washing | 10–24,000 | 1+ WBC NOS | None | 1+ Actinomyces sp., not A. israelii | NT | RapID ANA | Not tested |
| Lung biopsy sample | None | No WBC NOS | None | 3 CFU Actinomyces sp., not A. israelii | NT | RapID ANA + Saramis | A. odontolyticus, 77.5% | |
| 4 | BAL | 9,500 | No WBC NOS | None | 2+ Actinomyces sp., not A. israelii | NT | Saramis | A. odontolyticus, 77.5% |
| 5 | BAL | 10–24,000 | 2+ WBC NOS | 4+ A. graevenitzii | NT | 16S | No ID (Saramis) | |
| 6 | Bronchial, NS | 10–24,000 | 1+ WBC NOS | None | 3+ A. graevenitzii | 4 | 16S | No ID (Saramis) |
| 7 | Lung biopsy sample | 1+ mixed | 1+ WBC NOS | 1+ MAC | 1+ A. odontolyticus | NT | RapID ANA + Saramis | 77.5% (Saramis) |
| Bronchial washing | >100,000 | 2+ WBC 1+ mixed 2+ AFB | 3+ MAC | Not detected | NA | NA | NA | |
| 8 | BAL | 10–24,000 | 1+ WBC 2+ mixed | 100 CFU/ml of S. aureus | 2+ A. graevenitzii | NT | 16S | No ID (Saramis) |
| 9 | BAL | 50–74,000 | 2+ WBC NOS | 10–24,000 CFU/ml of P. aeruginosa | 2+ A. odontolyticus | NT | RapID ANA + Saramis | 80.6% (Saramis) |
| 10 | BAL | 4,000 | No WBC NOS | None | 1 CFU A. graevenitzii | 2 | 16S | No ID (Vitek MS IVD) |
| 11 | Bronchial, NS | 10–24,000 | 1+ WBC 1+ mixed | None | 2+ A. graevenitzii | 7 | 16S | No ID (Vitek MS IVD) |
| 12 | Bronchial washing | 50–74,000 | 1+ WBC NOS | None | 2+ A. odontolyticus | NT | Vitek MS IVD | 99.9% |
| 13 | BAL | 100 | 1+ WBC NOS | None | 1 CFU A. graevenitzii | 3 | 16S | No ID (Vitek MS IVD) |
| 14 | BAL | 10–24,000 | 2+ WBC 2+ RBC 2+ Epi 2+ mixed | None | 3+ A. graevenitzii | 1 | 16S | No ID (Vitek MS IVD) |
| 15 | BAL | 200 | 1+ WBC NOS | None | 2 CFU A. odontolyticus 2 CFU A. graevenitzii | NT | Vitek MS IVD 16S | 99.9% no ID (Vitek MS IVD) |
| 16 | BAL | >100,000 | 1+ WBC 1+ mixed | 1+ MAC | 3+ A. graevenitzii | NT | 16S | Not tested |
| Bronchial brushing | 400 CFU/ml | 1+ WBC NOS | None | None | NA | NA | Not tested | |
| 17 | BAL | 10–24,000 | 1+ WBC 1+ mixed | 400 CFU of GAS | 1+ A. graevenitzii | 5 | 16S | Not tested |
| 18 | BAL | 5,000 | 1+ WBC NOS | None | 1+ A. graevenitzii | 4 | 16S | No ID (Vitek MS IVD) |
Abbreviations: ID, identification; MALDI-TOF, matrix-assisted laser desorption ionization–time of flight; BAL, bronchoalveolar lavage fluid; WBC, white blood cells; NA, not applicable; NOS, no organisms seen; NS, not specified; MAC, Mycobacterium avium complex; MS, mass spectrometry; RBC, red blood cells; GAS, group A streptococcus; GPC, Gram-positive cocci; GNR, Gram-negative rod; AFB, acid-fast bacteria; EPI, epithelial cells.
Eight cultures obtained from personnel (two staff members involved in bronchoscopy of 50% of case patients and two staff members who had performed high-level disinfection on the bronchoscopes) and all 62 environmental cultures were negative for Actinomyces.
Case-control study.
Case patients were not more likely than control patients to have undergone previous bronchoscopy or pulmonary function tests. Only seven case patients had inhaled medications on their list of current medications at the time of admission. There was no bronchoscope that was used significantly more frequently for case patients than for control patients. Similarly, there were no statistically significant differences between cases and controls with respect to the bronchoscopist, scrub nurse or circulator involved, the proportion of procedures performed in room 3, the day of the week or time of day the procedure was performed, or the patients' inpatient status. None of the 4 bays in the 2 AERs were implicated as a common source related to case patients. Finally, there were no statistical differences based on other types of cultures and results that were collected for each patient.
Strain typing.
A total of nine A. graevenitzii isolates were available for rep-PCR analysis (Fig. 1). Seven different patterns were found with <96% similarity among bacterial strains. Three isolates were >98% similar, with two band differences among all three bacteria, leading these to be considered closely related. However, these three isolates were widely temporally separated, having been recovered in May, September, and December. Based on these results, we concluded that the A. graevenitzii recovered from these patients did not arise from a common source.
FIG 1.

Results for DiversiLab RepPCR strain typing for selected Actinomyces graevenitzii isolates. DiversiLab (DL) rep-PCR strain typing was performed on available Actinomyces graevenitzii isolates. The dendrogram shows relatedness determined by Kullback-Leibler analysis grouped according to DL pattern. The vertical dashed line indicates a 96% similarity of the DL profiles. The case number corresponds to that listed in Table 1, and the month during which the culture was obtained is indicated.
Patterns of test ordering and laboratory protocols.
For the years 2011, 2012, and 2013, the number of bronchoscopies performed, the number for which routine bacterial cultures and cultures for Actinomyces were ordered, and the number which yielded Actinomyces spp. are shown in Table 2. A significantly greater proportion of Actinomyces cultures were positive in 2013 than in 2011 or 2012 (P = 0.0005 and P = 0.0001, respectively).
TABLE 2.
Frequency of bronchoscopies, with routine cultures, cultures for Actinomyces, and number positive for Actinomycesa
| Year | No. of bronchoscopies performed | No. (%) of bronchoscopies with routine culture | No. (%) of bronchoscopies with Actinomyces culture | No. (%) of Actinomyces cultures positive |
|---|---|---|---|---|
| 2011 | 668 | 214 (32.0) | 170 (25.4) | 1 (0.6) |
| 2012 | 1,054 | 379 (36.0) | 307 (29.1) | 1 (0.3) |
| 2013 | 864 | 349 (40.4) | 227 (26.2) | 18 (7.9)* |
*, 2013 versus 2012, P = 0.0001; 2013 versus 2011, P = 0.0005.
Among the specimens submitted in 2013, no positive Actinomyces cultures from bronchial specimens were noted until late May (Fig. 2). A review of laboratory procedures revealed that the laboratory modified its Actinomyces culture procedure in late May 2013 as part of a laboratory consolidation process. Previously, Actinomyces cultures were plated to CDC anaerobic blood agar, but the modified protocol specified the use of anaerobic CNA agar.
FIG 2.
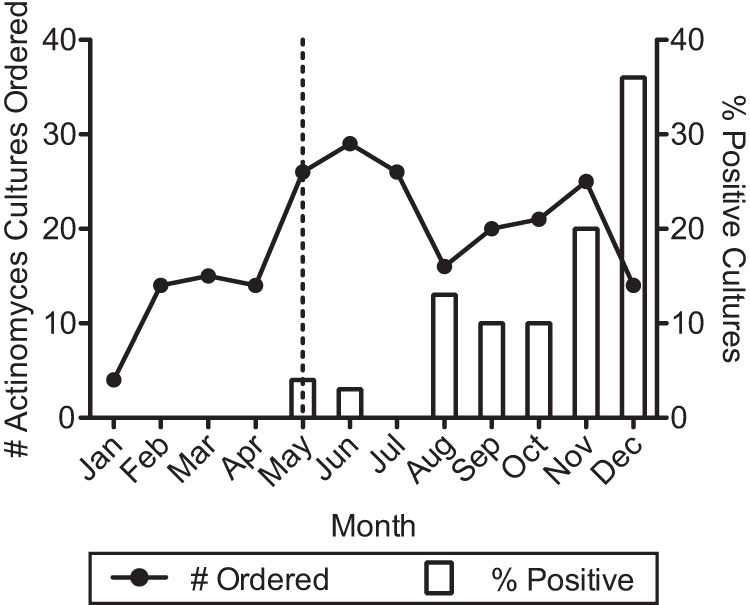
Actinomyces culture orders and results for 2013. Laboratory records were reviewed for Actinomyces cultures ordered and those with positive results for calendar year 2013. The vertical dashed line indicates the month during which the laboratory plating protocol for Actinomyces cultures was changed. Prior to that month, anaerobic CDC blood agar was used, and afterwards reducible CNA agar was used.
Comparison of growth on anaerobic CNA and CDC blood agars.
The temporal relationship between the start of the pseudo-outbreak and the change in laboratory procedures for the plating of Actinomyces cultures led us to hypothesize that differences in the growth of Actinomyces in general and A. graevenitzii in particular between anaerobic CNA and CDC anaerobic blood agar contributed to the pseudo-outbreak. We developed three hypotheses to explain these findings: (i) greater suppression of oropharyngeal microbiota by CNA agar leading to the greater apparent abundance of Actinomyces, (ii) anaerobic CNA agar better supporting the growth of Actinomyces such that greater numbers of colonies would grow, and (iii) qualitative differences in the growth of Actinomyces leading to their greater apparent abundance.
We excluded possibilities 1 and 2 through quantitative plating and other microbiological studies (data not shown). We compared the colony morphologies of Actinomyces isolates on both CDC and anaerobic CNA agar at 48 h, 72h, and 96 h (Fig. 3). Among the seven non-A. graevenitzii isolates, only a single A. odontolyticus isolate produced a small number of molar tooth colonies, and those only appeared on the anaerobic CNA agar. In contrast, 8 of 10 A. graevenitzii isolates produced molar tooth colonies as early as 48 h after inoculation. The molar tooth colonies were admixed with both wet- and dry-appearing colonies, and the relative proportion of dry and molar tooth colonies was significantly greater for the anaerobic CNA agar at all time points examined (P < 0.001).
FIG 3.

Morphology of A. graevenitzii on anaerobic CDC and reducible CNA agars. The same A. graevenitzii isolate growing on anaerobic CNA (A) and anaerobic CDC blood agar (B) at 72 h is shown. The proportion of dry- or molar-tooth-appearing colonies for 10 different A. graevenitzii isolates on anaerobic CNA (▲) or anaerobic CDC blood agar (●) was assessed at the indicated time points. The proportion of dry or molar-tooth colonies was significantly greater for anaerobic CNA agar (P < 0.001) at all time points.
DISCUSSION
Our epidemiological investigation failed to identify a common source for the Actinomyces recovered from bronchoscopic specimens. Furthermore, the fact that species identification of isolates using 16S rRNA sequencing identified two different species (A. graevenitzii and A. odontolyticus) and that rep-PCR strain typing established that A. graevenitizii isolates represented seven different strains revealed that the cluster of cases represented a pseudo-outbreak, not a common-source outbreak caused by bronchoscopic equipment or procedures contaminated by a single strain. Numerous outbreaks and pseudo-outbreaks related to bronchoscopy have been reported (12), but none involved Actinomyces spp. Identification of A. graevenitzii as the predominant species recovered was also unique, since this species is less frequently recovered from clinical specimens than A. odontolyticus, A. turicensis, and A. radingae. (13, 14) The use of 16S rRNA sequencing facilitated recognition of A. graevenitzii, since this species is not in the RapID ANA II, or MS databases, consistent with previous reports (13, 15). A. odontolyticus is contained in both the Saramis RUO and Vitek MS IVD databases, facilitating its identification.
The lack of reported outbreaks related to Actinomyces and other anaerobes may be related to the fact that current guidelines recommend against routine anaerobic culture of bronchial specimens, since most will be contaminated with respiratory flora (9). In fact, all specimens growing Actinomyces during the pseudo-outbreak were contaminated with respiratory flora. One of the contributing causes of this pseudo-outbreak was the inappropriate requests by bronchoscopists for cultures for Actinomyces on bronchoalveolar lavage specimens. Bronchoscopic specimens should not be cultured for anaerobes, including Actinomyces, unless specimens are collected via a protected sheath, which was not the case during the pseudo-outbreak (9). The proportion of bronchoscopic specimens for which Actinomyces cultures were requested in 2013 was similar to those in prior years. Thus, additional factors likely contributed to the significant increase in the proportion of cultures growing Actinomyces spp.
After extensive investigation, we concluded that the significant increase in recovery of Actinomyces spp. was related to a change in culture methods. While a change in laboratory methods is an unusual cause of bronchoscopy-related pseudo-outbreaks, it has been previously described (16). We found that A. graevenitzii plated onto anaerobic CNA agar more readily formed white, dry, molar tooth colonies characteristic of Actinomyces. While colonies with this appearance were seen on CDC anaerobic agar, they were not the predominant morphotype. Additionally, Actinomyces spp. other than A. graevenitzii were much less likely to adopt this morphology. We concluded that the different growth characteristics among Actinomyces spp. on the different plates contributed to the increased recovery of Actinomyces spp. following the protocol change.
Gram-negative organisms were seen on Gram stain from several specimens, and their suppression by CNA agar may have enhanced the recovery of Actinomyces spp. However, Gram-negative organisms are not the predominant components of the upper airway microbiota, and the selective effects of CNA agar would not be expected to contribute substantially to this pseudo-outbreak.
The base composition of CNA agar differs from that of CDC agar, and it was possible that the nutrient requirements of Actinomyces spp. are better met by the CNA agar, leading to greater numbers of Actinomyces colonies. However, robust growth of several different Actinomyces spp. was seen on both agar types. No differences in colony counts were seen between the two media, and Actinomyces colonies appeared slightly larger on the CDC agar.
A. graevenitzii is most commonly recovered from oral or respiratory specimens and is often present in low numbers (6, 13, 14). This is not surprising since A. graevenitzii, like other species of Actinomyces, is a colonizer of the oropharynx (7, 8). However, A. graevenitzii is recovered less frequently than A. odontolyticus, A. naeslundii, and A. israelii (7). We are unaware of data regarding the prevalence of A. graevenitzii as a colonizer of the oropharynx in adult populations such as the one served by our hospital.
In summary, we conclude that the unusual pseudo-outbreak of Actinomyces associated with bronchoscopy was due to frequent requests by pulmonologists for Actinomyces cultures on bronchoscopic lavage and transbronchial biopsy specimens and a change in culture media which favored detection and identification of Actinomyces by the microbiology laboratory. The availability of 16S rRNA sequencing and rep-PCR methods established that the cluster was due to a variety of A. graevenitzii strains and A. odontolyticus that likely represented contamination of bronchoscopy specimens by the normal oral flora. Investigation of this pseudo-outbreak could likely have been avoided through judicious ordering of Actinomyces cultures and proper collection using a protected catheter sheath.
ACKNOWLEDGMENTS
We thank Vincent Piscitelli of the Clinical Microbiology Laboratory for his assistance.
REFERENCES
- 1.Alfaro TM, Bernardo J, Garcia H, Alves F, Carvalho L, Caseiro AF, Robalo CC. 2011. Organizing pneumonia due to actinomycosis: an undescribed association. Respiration 81:433–436. doi: 10.1159/000321247. [DOI] [PubMed] [Google Scholar]
- 2.Cohen R, Bowie W, Enns R, Flint J, Fitzgerald M. 2009. Pulmonary actinomycosis complicating infliximab therapy for Crohn disease. BMJ Case Rep 2009:bcr11.2008.1262. doi: 10.1136/bcr.11.2008.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita Y, Iikura M, Horio Y, Ohkusu K, Kobayashi N. 2012. Pulmonary Actinomyces graevenitzii infection presenting as organizing pneumonia diagnosed by PCR analysis. J Med Microbiol 61:1156–1158. doi: 10.1099/jmm.0.040394-0. [DOI] [PubMed] [Google Scholar]
- 4.Hwang SS, Park SD, Jang IH, Uh Y, Yoon KJ, Kim HY. 2011. Actinomyces graevenitzii bacteremia in a patient with alcoholic liver cirrhosis. Anaerobe 17:87–89. doi: 10.1016/j.anaerobe.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Nagaoka K, Izumikawa K, Yamamoto Y, Yanagihara K, Ohkusu K, Kohno S. 2012. Multiple lung abscesses caused by Actinomyces graevenitzii mimicking acute pulmonary coccidioidomycosis. J Clin Microbiol 50:3125–3128. doi: 10.1128/JCM.00761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramos CP, Falsen E, Alvarez N, Akervall E, Sjoden B, Collins MD. 1997. Actinomyces graevenitzii sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol 47:885–888. doi: 10.1099/00207713-47-3-885. [DOI] [PubMed] [Google Scholar]
- 7.Sarkonen N, Kononen E, Eerola E, Kononen M, Jousimies-Somer H, Laine P. 2005. Characterization of Actinomyces species isolated from failed dental implant fixtures. Anaerobe 11:231–237. doi: 10.1016/j.anaerobe.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Sarkonen N, Kononen E, Summanen P, Kanervo A, Takala A, Jousimies-Somer H. 2000. Oral colonization with Actinomyces species in infants by two years of age. J Dent Res 79:864–867. doi: 10.1177/00220345000790031301. [DOI] [PubMed] [Google Scholar]
- 9.York MK, Gilligan P, Church DL. 2010. Aerobic bacteriology, lower respiratory tract cultures, p 3.11.2.1–3.11.2.17. In Garcia LS, Isenberg HD (ed), Clinical microbiology procedures handbook, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Fluit AC, Terlingen AM, Andriessen L, Ikawaty R, van Mansfeld R, Top J, Cohen Stuart JW, Leverstein-van Hall MA, Boel CH. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J Clin Microbiol 48:3979–3989. doi: 10.1128/JCM.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber DJ, Rutala WA. 2012. Lessons learned from outbreaks and pseudo-outbreaks associated with bronchoscopy. Infect Control Hosp Epidemiol 33:230–234. doi: 10.1086/664495. [DOI] [PubMed] [Google Scholar]
- 13.Clarridge J E III, Zhang Q. 2002. Genotypic diversity of clinical Actinomyces species: phenotype, source, and disease correlation among genospecies. J Clin Microbiol 40:3442–3448. doi: 10.1128/JCM.40.9.3442-3448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall V, Talbot PR, Stubbs SL, Duerden BI. 2001. Identification of clinical isolates of actinomyces species by amplified 16S ribosomal DNA restriction analysis. J Clin Microbiol 39:3555–3562. doi: 10.1128/JCM.39.10.3555-3562.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, Ginocchio C, Jennemann R, Manji R, Procop GW, Richter S, Rychert J, Sercia L, Westblade L, Lewinski M. 9 August 2013. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK MS system. Clin Microbiol Infect doi: 10.1111/1469-0691.12317. [DOI] [PubMed] [Google Scholar]
- 16.Weinstein RA, Stamm WE. 1977. Pseudoepidemics in hospital. Lancet ii:862–864. [DOI] [PubMed] [Google Scholar]


