Abstract
Real-time PCR (rt-PCR) is an important diagnostic tool for the identification of Bordetella pertussis, Bordetella holmesii, and Bordetella parapertussis. Most U.S. public health laboratories (USPHLs) target IS481, present in 218 to 238 copies in the B. pertussis genome and 32 to 65 copies in B. holmesii. The CDC developed a multitarget PCR assay to differentiate B. pertussis, B. holmesii, and B. parapertussis and provided protocols and training to 19 USPHLs. The 2012 performance exercise (PE) assessed the capability of USPHLs to detect these three Bordetella species in clinical samples. Laboratories were recruited by the Wisconsin State Proficiency Testing program through the Association of Public Health Laboratories, in partnership with the CDC. Spring and fall PE panels contained 12 samples each of viable Bordetella and non-Bordetella species in saline. Fifty and 53 USPHLs participated in the spring and fall PEs, respectively, using a variety of nucleic acid extraction methods, PCR platforms, and assays. Ninety-six percent and 94% of laboratories targeted IS481 in spring and fall, respectively, in either singleplex or multiplex assays. In spring and fall, respectively, 72% and 79% of USPHLs differentiated B. pertussis and B. holmesii and 68% and 72% identified B. parapertussis. IS481 cycle threshold (CT) values for B. pertussis samples had coefficients of variation (CV) ranging from 10% to 28%. Of the USPHLs that differentiated B. pertussis and B. holmesii, sensitivity was 96% and specificity was 95% for the combined panels. The 2012 PE demonstrated increased harmonization of rt-PCR Bordetella diagnostic protocols in USPHLs compared to that of the previous survey.
INTRODUCTION
Clinical laboratories increasingly depend on real-time PCR (rt-PCR) to diagnose pertussis, a respiratory disease caused by Bordetella pertussis. Similar disease symptoms are also caused by Bordetella parapertussis and sometimes by Bordetella holmesii or Bordetella bronchiseptica. Many rt-PCR assays target an insertion element, IS481, present in high copy number (218 to 238 copies) in B. pertussis (1, 2), low copy number (32 to 65 copies) in B. holmesii (3), and some strains of B. bronchiseptica (4). If IS481 is the sole target, the laboratory cannot report a specific result for B. pertussis and will miss B. parapertussis infections. The CDC multitarget rt-PCR assay to detect and identify B. pertussis, B. holmesii, and B. parapertussis is a multiplex assay targeting IS481, IS1001 in B. parapertussis (pIS1001), and the IS1001-like element in B. holmesii (hIS1001) (5). As confirmation of B. pertussis, the assay also includes a singleplex rt-PCR for ptxS1, which encodes the first subunit of the pertussis toxin. PCR inhibition is identified with a singleplex assay for human rnaseP. After CDC laboratory validation with isolates and clinical specimens, the multitarget assay was offered to U.S. public health laboratories (USPHLs) along with a series of onsite trainings, webinars, and additional communications.
With the increase in rt-PCR diagnostics, public health, hospital, and commercial laboratories have adopted many protocols that differ in DNA extraction methods, rt-PCR platforms, and protocols. It is important, therefore, to monitor and assess the ability of laboratories to demonstrate consistency in diagnostic accuracy and precision. Several surveys and performance exercises have been performed in Europe and one of each in the United States (6–9). For instance, eight hospital laboratories in France in 2006 to 2007 participated in a performance evaluation provided by the French National Centre of Reference of Pertussis and other Bordetelloses (6). All eight laboratories used rt-PCR targeting IS481, with resulting sensitivity and specificity of 92.2% and 94.3%, respectively. Some laboratories also included targets to confirm B. pertussis or other species, including B. parapertussis. In a 2010 survey of pertussis diagnostics in 27 European countries, 74% of countries reported using serology, followed by 67% using PCR, and 63% using culture (7). This contrasts with a previous survey in the United States in which 123 public health, commercial, and hospital laboratories reported that 7% perform serologic diagnostics, 54% perform PCR, and 71% perform culture (8). Along with the survey, 41 laboratories participated in a performance exercise that included 13 public health laboratories. Laboratories employing various extraction and rt-PCR methods reported B. pertussis with a sensitivity of 92% (8). In a 2011 European PCR performance exercise involving 24 national reference laboratories from 19 countries, 100% of laboratories detected B. pertussis in high concentrations within the samples of extracted DNA provided to them. However, 14 out of 24 laboratories (58%) misidentified B. holmesii as B. pertussis because they were targeting IS481 only, with no target to distinguish B. holmesii from B. pertussis (9).
With a high dependence on PCR for pertussis diagnostics and evidence that extraction, PCR platform, protocol, and PCR targets vary among laboratories (7, 8), it is important to provide external assessment through performance exercises to analyze the consistency in PCR results across laboratories. We report here the results of the 2012 rt-PCR performance exercise for USPHLs for the detection of B. pertussis, B. parapertussis, and B. holmesii.
MATERIALS AND METHODS
Laboratory recruitment, testing, and reporting.
State and local public health laboratories were recruited by the Wisconsin State Laboratory of Hygiene (WSLH) Proficiency Testing program through the Association of Public Health Laboratories (APHL), in partnership with the CDC. Two panels of bacteria were sent in the spring and fall of 2012 that contained 12 samples each of Bordetella and non-Bordetella species in saline at 1 × 103 to 1 × 106 CFU/ml in the spring and 1 × 104 to 1 × 107 CFU/ml in the fall (Table 1). DNA from human A549 cells was included in all the samples at a concentration that would result in a crossing cycle threshold (CT) value of 30 to 34 when amplifying rnaseP. Samples were aliquoted and stored frozen at −80°C until shipped on wet ice to participating laboratories. Laboratories were instructed to use their routine nucleic acid extraction and PCR procedures and to complete a worksheet to record when samples were received, thawed, and analyzed. The extraction method, rt-PCR platform, PCR targets, and number of replicates were also reported. Results included CT values, melting point (MP) value (if applicable), target interpretation, and a final interpretation for the sample. Participants entered answers of positive, negative, equivocal, indeterminate, or not tested for B. pertussis, B. parapertussis, B. holmesii, and B. pertussis/holmesii undifferentiated (BpBh) for each sample. The BpBh response choice was intended for laboratories that performed a single target assay, i.e., IS481, that prevented them from distinguishing between B. pertussis and B. holmesii. When laboratories misunderstood the directions to complete the worksheet and filled in answers for the B. pertussis or B. holmesii species response in addition to the BpBh choice, only one answer was chosen based on the PCR targets that the lab reported, to avoid counting the laboratory's answers twice.
TABLE 1.
Fall and spring panels, each containing 12 samples of Bordetella pertussis, B. holmesii, B. parapertussis, and non-Bordetella species
| Panel and sample no. | Organism | Sample bacterial concn (CFU/ml) |
|---|---|---|
| Spring | ||
| BP12-1-1 | Bordetella pertussis | 1.0 × 106 |
| BP12-1-2 | Pseudomonas aeruginosa | 1.0 × 105 |
| BP12-1-3 | B. parapertussis | 1.0 × 103 |
| BP12-1-4 | B. holmesii | 1.0 × 103 |
| BP12-1-5 | B. pertussis | 1.0 × 103 |
| BP12-1-6 | B. holmesii | 1.0 × 106 |
| BP12-1-7 | Haemophilus influenzae | 1.0 × 105 |
| BP12-1-8 | B. parapertussis | 1.0 × 104 |
| BP12-1-9 | B. pertussis | 1.0 × 105 |
| BP12-1-10 | B. pertussis | 1.0 × 103 |
| BP12-1-11 | B. holmesii | 1.0 × 103 |
| BP12-1-12 | B. parapertussis | 1.0 × 104 |
| Fall | ||
| BP12-2-1 | B. pertussis | 1.0 × 105 |
| BP12-2-2 | B. parapertussis | 1.0 × 104 |
| BP12-2-3 | B. holmesii | 1.0 × 106 |
| BP12-2-4 | B. parapertussis | 1.0 × 106 |
| BP12-2-5 | B. pertussis | 1.0 × 104 |
| BP12-2-6 | B. holmesii | 1.0 × 106 |
| BP12-2-7 | B. parapertussis | 1.0 × 106 |
| BP12-2-8 | B. pertussis | 1.0 × 107 |
| BP12-2-9 | P. aeruginosa | 1.0 × 105 |
| BP12-2-10 | B. holmesii | 1.0 × 105 |
| BP12-2-11 | B. pertussis | 1.0 × 105 |
| BP12-2-12 | H. influenzae | 1.0 × 105 |
Statistics.
For each panel and sample, lab-reported results were compared with previously determined results to establish the accuracy and precision of the tests. Statistical summaries of performance were tabulated for completeness of testing and reporting and for performance metrics. Sensitivity and specificity were calculated using known positive and negative samples. Positive samples for each species included all respective species-positive samples in each panel. Since negative samples must be included to compute meaningful specificity estimates, and since negative samples for a given species have no concentration, samples that were positive for some other species at the same respective concentration were included as the negative samples. Thus, a uniform set of positive and negative samples was identified for each species at each concentration, allowing for consistent computations of sensitivity and specificity at each concentration and across all concentrations. For those computations, responses of indeterminate and equivocal were counted as positive.
For counts of tests reported and for counts of positive and negative detections, all relevant tests were used. For example, a given lab might have tested all specimens in a given panel for B. pertussis but only some of the specimens for B. parapertussis. Similarly, a lab might have distinguished between B. holmesii and B. pertussis in some specimens but not in others. Finally, a lab might have reported or tested the spring and fall panels differently. In such cases, the number of tests for the reported statistics may be different for different pathogens and may be greater than the number of labs. Other differences between counting tests and counting labs appear in the figure and tables and are designated where not apparent from context.
In addition to the qualitative positive/negative analyses which produced estimates of sensitivity and specificity, a quantitative analysis of the CTs was performed. Pearson's correlation coefficients were calculated relating CT values of frequently reported rt-PCR targets to sample bacterial concentrations (CFU/ml).
RESULTS
Fifty USPHLs representing 41 states participated in the spring PE, and 53 USPHLs from 42 states took part in the fall PE. USPHLs included state and local public health laboratories, resulting in participation by multiple laboratories in some states.
Three methods for nucleic acid extraction were reported. The most common method was automated magnetic-based extraction, reported by 44% and 53% of laboratories in the spring and fall, respectively. The MagNA Pure platform (Roche Applied Science, Indianapolis, IN) was the most frequently reported automated system, employed by 38% and 45% of USPHLs in the spring and fall, respectively, followed by NucliSENS EasyMAG (bioMérieux, Durham, NC). The other methods employed were extraction with a manual spin column kit (Qiagen, Valencia, CA), reported by 30% and 28% of laboratories, and heat lysis, reported by 16% and 9%, respectively. Five laboratories from each panel did not provide their extraction methods.
Laboratories reported five rt-PCR platforms. The majority of laboratories employed the AB7500 fast machine (Applied Biosystems, Foster City, CA), reported by 58% and 53% in the spring and fall, respectively. The other platforms reported were the SmartCycler (Cepheid, Sunnyvale, CA), employed by 26% and 24% USPHLs; LightCycler (Roche Applied Science), reported by 10% and 17%; iCycler iQ (Bio-Rad, Hercules, CA), employed by 4% in each panel; and the FilmArray (BioFire Diagnostics, Salt Lake City, UT), employed by 2% (one laboratory) in each panel.
For both panels, 94% of laboratories targeted IS481 either as a single target or as part of a multitarget assay. In the spring and fall, respectively, 76% and 83% of USPHLs tested multiple targets, including 46% and 47% that performed the CDC assay. Laboratories were considered to have performed the CDC assay if they included the multitarget assay, IS481, pIS1001, and hIS1001, with or without ptxS1 confirmation or internal control. In the spring and fall, respectively, 46% and 53% included ptxS1 as a target to confirm B. pertussis. When considering all targets and protocols, 72% and 79% of USPHLs detected B. pertussis (note that these percentages are not sensitivities because they include in the denominator opportunities for testing when no test was reported or, presumably, performed), 52% and 57% detected B. holmesii, and 68% and 72% detected B. parapertussis, reporting results at least once per panel. Those reporting in only the BpBh category decreased slightly from 18% in the spring to 16% in the fall. Nine Bordetella targets were reported: IS481, hIS1001, pIS1001, IS1663, ptxS1, ptxP, BP485, recA, and prn. One laboratory did not provide target identities.
A total of 84% and 83% of USPHLs in the spring and fall, respectively, included an internal control. Although various targets were included as internal controls, human rnaseP was the most prevalent, utilized by 73.8% and 75% of the laboratories. Other targets included the gene encoding beta-actin, pCB1, the 16S rRNA gene, QC-DNA SSC from a commercial kit (Cepheid), recA, and the beta-globin gene.
In both spring and fall, one laboratory reported either MP or categorical data (i.e., positive or negative) only. All other USPHLs provided CT values. Two USPHLs in the spring and four in the fall included MP data along with CT values.
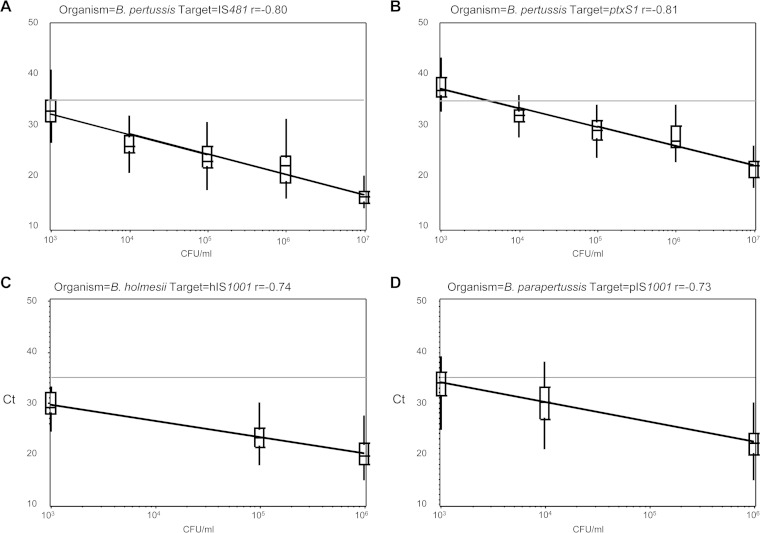
Figure 1 demonstrates that CT values correlated with concentrations across all labs and both panels (spring and fall combined) for four targets: IS481 (n = 386) and ptxS1 (n = 177) for eight B. pertussis samples, hIS1001 (n = 147) for six B. holmesii samples, and pIS1001 (n = 198) for six B. parapertussis samples. The coefficient of variation (CV) values varied from 8.3% to 28.0% across concentrations for these four targets (Table 2). All but one value was <25%. For target IS481, the results for the B. pertussis 1 × 107-CFU/ml sample included one false-negative value listed with an assigned nominal-negative CT value of 45, skewing the CV to 28%.
FIG 1.
Correlation of cycle threshold (CT) values with sample concentrations for four real-time PCR targets, combining results from the spring and fall performance panels. Pearson's correlation coefficient (r) is shown above each plot. (A) Eight B. pertussis samples, IS481 target (n = 386). (B) Eight B. pertussis samples, ptxS1 target (n = 177). (C) Six B. holmesii samples, hIS1001 target (n = 147). (D) Six B. parapertussis samples, pIS1001 target (n = 198). Each horizontal line indicates the positive cutoff value for IS481 in the CDC multitarget assay (CT < 35).
TABLE 2.
Coefficient of variation for PCR targets IS481 and ptxS1 (B. pertussis samples), hIS1001 (B. holmesii samples), and pIS1001 (B. parapertussis samples), fall and spring samples combined
| PCR target | CV (%) for indicated target at a concn (CFU/ml) of: |
||||
|---|---|---|---|---|---|
| 1 × 103a | 1 × 104 | 1 × 105 | 1 × 106 | 1 × 107 | |
| IS481 | 12.0 | 10.2 | 14.2 | 17.2 | 28.0 |
| ptxS1 | 8.4 | 8.3 | 11.8 | 12.8 | 11.9 |
| hIS1001 | 10.8 | —a | 14.3 | 20.7 | — |
| pIS1001 | 10.0 | 13.6 | — | 19.2 | — |
—, samples not provided at this concentration.
From the 50 labs for spring and 53 labs for fall combined, 330 tests of undifferentiated B. pertussis or B. holmesii were reported, including tests of positive and negative samples; 907 tests of B. pertussis, specifically, and 844 tests of B. holmesii were reported. Results are also reported by concentration; for 1 × 103 CFU/ml of any pathogen, for example, 170 tests were reported for B. pertussis. For sample BP12-1-5, 36 USPHLs reported tests for B. pertussis specifically, and 32 tests were positive. Thirty-seven USPHLs reported B. pertussis tests for sample BP12-1-10, and 31 tests were positive. Counts for all combinations of pathogens and concentrations, along with sensitivities and specificities, are reported in Table 3.
TABLE 3.
Sensitivity and specificity for the subset of tests performed by USPHLs in B. pertussis, B. parapertussis, B. holmesii, and B. pertussis/B. holmesii (undifferentiated)
| Organism | Sample bacterial concn (CFU/ml) | na | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) |
|---|---|---|---|---|
| B. pertussis | All | 907 | 96.1 (93.2–97.9) | 95.1 (93.2–96.8) |
| 1 × 103 | 170 | 88.4 (78.4–94.9) | 89.1 (81.3–94.4) | |
| 1 × 104 | 154 | 97.6 (87.1–99.9) | 99.1 (95.2–100) | |
| 1 × 105 | 312 | 97.4 (92.7–99.5) | 96.9 (93.4–98.9) | |
| 1 × 106 | 230 | 100 (90.3–100) | 94.3 (90.1–97.1) | |
| 1 × 107 | 41 | 100 (91.4–100) | —b | |
| B. parapertussis | All | 844 | 98.6 (95.9–99.7) | 99.7 (98.9–100) |
| 1 × 103 | 170 | 97.1 (84.7–99.9) | 99.3 (96.0–100) | |
| 1 × 104 | 141 | 99.0 (94.8–100) | 100 (90.3–100) | |
| 1 × 105 | 284 | — | 99.6 (98.1–100) | |
| 1 × 106 | 213 | 98.6 (92.7–100) | 100 (97.4–100) | |
| 1 × 107 | 36 | — | 100 (90.3–100) | |
| B. holmesii | All | 666 | 98.2 (94.7–99.6) | 97.8 (96.1–98.9) |
| 1 × 103 | 130 | 96.2 (86.8–99.5) | 98.7 (93.1–100) | |
| 1 × 104 | 111 | — | 96.4 (91.0–99.0) | |
| 1 × 105 | 227 | 100 (88.1–100) | 97.5 (94.2–99.2) | |
| 1 × 106 | 168 | 98.8 (93.5–100) | 98.8 (93.6–100) | |
| 1 × 107 | 30 | — | 100 (88.4–100) | |
| B. pertussis/B. holmesii | All | 330 | 97.6 (94.4–99.2) | 96.0 (90.9–98.7) |
| 1 × 103 | 78 | 93.7 (84.5–98.2) | 100 (78.2–100) | |
| 1 × 104 | 52 | 100 (73.5–100) | 95.0 (83.1–99.4) | |
| 1 × 105 | 105 | 100 (93.5–100) | 100 (92.9–100) | |
| 1 × 106 | 83 | 100 (94.3–100) | 85.0 (62.1–96.8) | |
| 1 × 107 | 12 | 91.7 (61.5–99.8) | — |
n, number of tests reported for each organism, including tests of positive and negative samples.
—, incalculable values due to empty cells, e.g., no false negatives with 95% CI.
The USPHLs reported >95% sensitivities and specificities for all four diagnostic categories, B. pertussis, B. parapertussis, B. holmesii, and BpBh (Table 3), calculated for the subset of USPHLs that provided results in each category. Eight laboratories (three in the spring, two in the fall, and three in both panels) filled in answers for the B. pertussis or B. holmesii categories in addition to BpBh, and only one category was counted. Another eight laboratories reported a false-positive result for B. pertussis in at least one B. holmesii sample (four in the spring, two in the fall, and two in both panels; data not shown). Since negative results for samples of the same concentration were included in the specificity calculation, false-positive results caused by misidentification of B. holmesii as B. pertussis were a major contributor to reduced B. pertussis specificity. For example, B. pertussis specificity was highest in the 1 × 104 samples, and the panels did not contain a B. holmesii sample at 1 × 104. B. pertussis specificity was lowest in the 1 × 103 and 1 × 106 samples, concentrations at which two and six B. holmesii samples, respectively, were provided (Tables 1 and 3). Few USPHLs reported false-positive B. pertussis identification for samples other than B. holmesii: one laboratory misidentified Pseudomonas aeruginosa as B. pertussis in the spring panel (2% of laboratories), and three other laboratories each reported one false-positive B. pertussis for B. parapertussis (two laboratories) or P. aeruginosa (one laboratory) in the fall (5.7% of laboratories).
Examining the effect of the DNA extraction method on the results, sensitivity ranged from 94.3% to 100%, and specificity ranged from 88.8% to 100% in B. pertussis, B. parapertussis, B. holmesii, and BpBh results (Table 4). USPHLs that extracted DNA with the EasyMAG reported results with sensitivity and specificity higher than results obtained after DNA extraction with the MagNA Pure system, except for equivalent specificity for B. parapertussis samples and lower specificity for B. holmesii samples. However, very few USPHLs used the EasyMAG, making comparison difficult. No other trend emerged for a particular DNA extraction method.
TABLE 4.
Sensitivity, specificity, and confidence interval for the subset of tests performed by USPHLs in all diagnostic categories, by DNA extraction method
| Organism | DNA extraction method | na | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) |
|---|---|---|---|---|
| B. pertussis | All | 907 | 96.1 (93.2–97.9) | 95.2 (93.2–96.8) |
| Automated, EasyMAG | 062 | 100 (83.2–100) | 100 (91.6–100) | |
| Automated, MagNA Pure | 371 | 94.3 (88.6–97.7) | 95.6 (92.2–97.8) | |
| Crude boil, heat lysis | 84 | 100 (87.7–100) | 100 (93.6–100) | |
| Spin column, Qiagen | 270 | 94.6 (87.9–98.2) | 94.9 (90.6–97.6) | |
| Not specified | 120 | 100 (91.2–100) | 88.8 (79.7–94.7) | |
| B. parapertussis | All | 844 | 98.6 (95.9–99.7) | 99.7 (98.9–100) |
| Automated, EasyMAG | 77 | 100 (83.9–100) | 100 (93.6–100) | |
| Automated, MagNA Pure | 348 | 98.8 (93.8–100) | 100.0 (98.6–100) | |
| Crude boil, heat lysis | 84 | 95.2 (76.2–99.9) | 98.4 (91.5–100) | |
| Spin column, Qiagen | 286 | 98.6 (92.5–100) | 99.5 (97.4–100) | |
| Not specified | 49 | 100 (73.5–100) | 100 (90.5–100) | |
| B. holmesii | All | 666 | 98.2 (94.7–99.6) | 97.8 (96.1–98.9) |
| Automated, EasyMAG | 60 | 100 (78.2–100) | 93.3 (81.7–98.6) | |
| Automated, MagNA Pure | 311 | 97.4 (91.0–99.7) | 99.6 (97.6–100) | |
| Crude boil, heat lysis | 72 | 94.4 (72.7–99.9) | 96.3 (87.3–99.5) | |
| Spin column, Qiagen | 175 | 100 (91.4–100) | 97.0 (92.5–99.2) | |
| Not specified | 48 | 100 (73.5–100) | 97.2 (85.5–99.9) | |
| B. pertussis/B. holmesii (undifferentiated) | All | 330 | 97.6 (94.4–99.2) | 96.0 (90.9–98.7) |
| Automated, EasyMAG | 19 | 100 (76.8–100) | 100 (47.8–100) | |
| Automated, MagNA Pure | 144 | 94.4 (87.4–98.2) | 90.9 (80.0–97.0) | |
| Crude boil, heat lysis | 72 | 100 (91.6–100) | 100 (88.4–100) | |
| Spin column, Qiagen | 92 | 100 (93.7–100) | 100 (90.0–100) | |
| Not specified | 3 | 100 (29.2–100) |
n, number of tests reported for each organism, including tests of positive and negative samples.
DISCUSSION
The 2012 pertussis rt-PCR performance exercise for USPHLs demonstrated improvement over the previous exercise in several areas, including increased participation (50 [spring] and 53 [fall] versus 41 total laboratories, including 13 USPHLs, in the 2010 performance exercise) and a higher percentage of laboratories that include multiple targets in their assays (76% versus 61%) (8). Sensitivity of B. pertussis detection improved from 92% to 96%. This comparison was made with the understanding that laboratories were recruited differently in the previous exercise and included commercial and hospital laboratories and public health laboratories (8). Qualitative improvement was observed between the 2012 spring and fall panels for the B. pertussis results. The majority of B. holmesii samples misidentified as B. pertussis were reported by six laboratories in the spring, and only two laboratories reported false-positive B. pertussis for B. holmesii samples in both panels. Among the laboratories in the spring panel, two reported equivocal or indeterminate results for B. pertussis, which were converted to positive results in this analysis. This conversion was chosen in case of nucleic acid loss due to shipping. One laboratory added the hIS1001 target to its protocol for the fall panel and subsequently did not report equivocal or indeterminate results. Two USPHLs that reported B. pertussis false-positive results only in the spring made no change to their rt-PCR targets, so improvement must have occurred for other reasons. The four B. pertussis false-positive results for B. parapertussis or P. aeruginosa samples (one in spring and three in fall) may indicate cross-contamination. If so, each of the four laboratories had a single instance of contamination.
Although there is no assurance or expectation that the CTs across laboratories would be consistent, there was in fact a high inverse correlation between CT values and specimen concentration, as one might ideally expect (Fig. 1 and Table 2). This is a reassuring finding in regard to the common use of a CT cutoff, e.g., <35 for target IS481 in the CDC assay, but warrants further study in regard to harmonization.
The expansion of the U.S. performance exercise to include two other Bordetella species demonstrated high sensitivity and specificity of B. parapertussis and B. holmesii detection by laboratories with species-level detection capacity. A future target for improvement is a higher percentage of USPHLs that correctly detect all three Bordetella species, such as that seen in the 2013 external quality assurance exercise for Bordetella identification reported by the European CDC (10).
As anticipated, B. pertussis detection sensitivity was lower in samples with the lowest B. pertussis concentration (1 × 103 CFU/ml), suggesting that the samples were close to the limit of detection of the PCR assays (Table 3). Sensitivity was higher in low-concentration samples of B. parapertussis and B. holmesii than in low-concentration B. pertussis samples. This may be due to the need to confirm B. pertussis diagnoses by a second PCR when using the IS481 target, most commonly targeting a single-copy gene, such as ptxS1, which is a slightly less sensitive assay (5). All six USPHLs that submitted a false-negative result for a B. pertussis 1 × 103-CFU/ml sample included ptxS1 as one of their targets, and five out of six of the laboratories employed the MagNA Pure system for DNA extraction (data not shown). The most common targets to identify B. parapertussis and B. holmesii (pIS1001 and hIS1001, respectively) occur in multiple copies per genome and do not require additional confirmation. The CDC assay, designed for outbreak investigations, includes an upper-limit CT of <35 for IS481 to avoid false-positive results (5). In contrast, the CDC has set the upper limit for pIS1001 and hIS1001 at a CT of <40 (5). It is important to have accurate species-level diagnostics of pertussis and pertussis-like infections for public health response, vaccine efficacy, and epidemiological studies (11). Inclusion of additional targets such as ptxS1 in pertussis PCR diagnostics is important to avoid misidentification of B. holmesii as B. pertussis, to prevent other false-positive results, and to detect the rare coinfection with B. pertussis and a second Bordetella species (12–14).
The lower B. pertussis sensitivities achieved for some samples by laboratories that extract DNA with MagNA Pure or manual Qiagen spin columns suggest that these two methods may be less efficient at DNA extraction than the EasyMAG platform or simple heat lysis. A disadvantage of using heat lysis to release DNA for PCR is that it does not remove potential PCR inhibitors. Although human DNA was added to the bacterial saline suspensions to simulate clinical specimens, actual specimens may contain inhibiting components. An additional concern with manual extraction or heat lysis is the increased possibility for DNA cross-contamination (15). An efficacy comparison of DNA extraction kits to detect Bordetella species in nasopharyngeal swabs or aspirates without the added complication of multiple laboratories and PCR protocols will be required to definitively resolve this question.
Limitations of the 2012 performance exercise included confusion for using the BpBh category, which led to a few results that were reported in two categories. Bias may have been introduced into the analysis by choosing one result per sample to avoid double counting. Another possible source of bias is the choice to include indeterminate and equivocal results in the positive category. The current performance exercise did not include a detailed survey of laboratory protocols or validation of assays and did not request that USPHLs provide positive cutoff CT values for their assays. The analysis was based solely on the result interpretations made by each USPHL. The previous performance exercise found that most participating laboratories took appropriate precautions to avoid contamination, such as having separate areas for working with the PCR master mix, template, and positive controls (8), indicating that additional surveys of laboratory protocols were not necessary at this time.
In conclusion, the 2012 pertussis rt-PCR performance exercise demonstrated that USPHLs detect Bordetella species with high sensitivity and specificity, despite the continued variety of DNA extraction methods, PCR platforms, targets, and PCR protocols adopted by the laboratories. The majority of participating laboratories differentiate between B. pertussis, B. parapertussis, and B. holmesii; and 47% have adopted the CDC rt-PCR assay. Intensive training and communication with USPHLs increased harmonization of the rt-PCR assay protocol and improved sensitivity for pertussis diagnostics. Suggestions for additional improvement include encouraging all USPHLs to expand their rt-PCR assays to detect B. pertussis, B. parapertussis, and B. holmesii. All assays should include an internal control, and, where feasible, DNA extraction should be automated to avoid potential cross-contamination of specimens.
ACKNOWLEDGMENTS
We thank the Association of Public Health Laboratories for providing funding for the 2012 testing panels. We also thank the participating laboratories.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeno-Tarraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 2.Harvill ET, Goodfield LL, Ivanov Y, Meyer JA, Newth C, Cassiday P, Tondella ML, Liao P, Zimmerman J, Meert K, Wessel D, Berger J, Dean JM, Holubkov R, Burr J, Liu T, Brinkac L, Kim M, Losada L. 2013. Genome sequences of 28 Bordetella pertussis U.S. outbreak strains dating from 2010 to 2012. Genome Announc 1:pii=e01075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvill ET, Goodfield LL, Ivanov Y, Smallridge WE, Meyer JA, Cassiday PK, Tondella ML, Brinkac L, Sanka R, Kim M, Losada L. 2014. Genome sequences of nine Bordetella holmesii strains isolated in the United States. Genome Announc 2:pii=e00438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. 2001. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol 39:1963–1966. doi: 10.1128/JCM.39.5.1963-1966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatti KM, Sparks KN, Boney KO, Tondella ML. 2011. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol 49:4059–4066. doi: 10.1128/JCM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caro V, Guiso N, Alberti C, Liguori S, Burucoa C, Couetdic G, Doucet-Populaire F, Ferroni A, Papin-Gibaud S, Grattard F, Reglier-Poupet H, Raymond J, Soler C, Bouchet S, Charreau S, Couzon B, Leymarie I, Tavares N, Choux M, Bingen E, Bonacorsi S. 2009. Proficiency program for real-time PCR diagnosis of Bordetella pertussis infections in French hospital laboratories and at the French National Reference Center for Whooping Cough and other Bordetelloses. J Clin Microbiol 47:3197–3203. doi: 10.1128/JCM.00817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Q, Barkoff AM, Mertsola J, Glismann S, Bacci S. 2012. High heterogeneity in methods used for the laboratory confirmation of pertussis diagnosis among European countries, 2010: integration of epidemiological and laboratory surveillance must include standardisation of methodologies and quality assurance. Euro Surveill 17:pii=20239 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20239. [DOI] [PubMed] [Google Scholar]
- 8.Tatti KM, Martin SW, Boney KO, Brown K, Clark TA, Tondella ML. 2013. Qualitative assessment of pertussis diagnostics in United States laboratories. Pediatr Infect Dis J 32:942–945. doi: 10.1097/INF.0b013e3182947ef8. [DOI] [PubMed] [Google Scholar]
- 9.Dalby T, Fry NK, Krogfelt KA, Jensen JS, He Q. 2013. Evaluation of PCR methods for the diagnosis of pertussis by the European surveillance network for vaccine-preventable diseases (EUVAC.NET). Eur J Clin Microbiol Infect Dis 32:1285–1289. doi: 10.1007/s10096-013-1874-0. [DOI] [PubMed] [Google Scholar]
- 10.European Centre for Disease Prevention and Control. 2012. External quality assurance scheme on PCR for Bordetella pertussis, 2012: on behalf of the EUpert-labnet network. ECDC, Stockholm, Sweden. [Google Scholar]
- 11.Loeffelholz M. 2012. Towards improved accuracy of Bordetella pertussis nucleic acid amplification tests. J Clin Microbiol 50:2186–2190. doi: 10.1128/JCM.00612-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention (CDC). 2007. Outbreaks of respiratory illness mistakenly attributed to pertussis–New Hampshire, Massachusetts, and Tennessee, 2004-2006. MMWR Morb Mortal Wkly Rep 56:837–842. [PubMed] [Google Scholar]
- 13.Mandal S, Tatti KM, Woods-Stout D, Cassiday PK, Faulkner AE, Griffith MM, Jackson ML, Pawloski LC, Wagner B, Barnes M, Cohn AC, Gershman KA, Messonnier NE, Clark TA, Tondella ML, Martin SW. 2012. Pertussis pseudo-outbreak linked to specimens contaminated by Bordetella pertussis DNA from clinic surfaces. Pediatrics 129:e424–e430. doi: 10.1542/peds.2011-1710. [DOI] [PubMed] [Google Scholar]
- 14.Rodgers L, Martin SW, Cohn A, Budd J, Marcon M, Terranella A, Mandal S, Salamon D, Leber A, Tondella ML, Tatti K, Spicer K, Emanuel A, Koch E, McGlone L, Pawloski L, Lemaile-Williams M, Tucker N, Iyer R, Clark TA, Diorio M. 2013. Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis–Ohio, 2010-2011. Clin Infect Dis 56:322–331. doi: 10.1093/cid/cis888. [DOI] [PubMed] [Google Scholar]
- 15.Lievano FA, Reynolds MA, Waring AL, Ackelsberg J, Bisgard KM, Sanden GN, Guris D, Golaz A, Bopp DJ, Limberger RJ, Smith PF. 2002. Issues associated with and recommendations for using PCR to detect outbreaks of pertussis. J Clin Microbiol 40:2801–2805. doi: 10.1128/JCM.40.8.2801-2805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]