Abstract
Mycoplasma pneumoniae is a leading cause of respiratory infections, including community-acquired pneumonia (CAP). Currently, pathogen-specific testing is not routinely performed in the primary care setting, and the United States lacks a systematic surveillance program for M. pneumoniae. Documentation of individual cases and clusters typically occurs only when severe illness and/or failure to improve with empirical antibiotic therapy is observed. Outbreaks, some lasting for extended periods and involving a large number of cases, occur regularly. However, many more likely go unrecognized due to the lack of diagnostic testing and structured reporting. We reviewed data from 17 investigations of cases, small clusters, and outbreaks of M. pneumoniae infections that were supported by the Centers for Disease Control and Prevention (CDC) between 2006 and 2013. We examined 199 M. pneumoniae-positive specimens collected during this time period in order to identify trends in antimicrobial resistance and circulating types. Overall, macrolide resistance was identified in approximately 10% of M. pneumoniae infections occurring during this time period. Typing of strains revealed cocirculation of multiple multilocus variable-number tandem-repeat analysis (MLVA) and P1 types throughout this period, including diversity in types detected within individual outbreaks. Three MLVA types (4572, 3562, and 3662) accounted for 97% of the infections during the study period. A systematic surveillance program is necessary to understand the burden of M. pneumoniae disease in the United States, facilitate case and outbreak identification, and inform appropriate therapeutic and infection control strategies.
INTRODUCTION
Mycoplasma pneumoniae is a common respiratory pathogen in adults and children worldwide and is a leading cause of community-acquired pneumonia (CAP). The majority of infections are self-limiting, and empirical treatment without pathogen-specific diagnostic testing is common in the primary care setting. In fact, the current treatment guidelines do not recommend diagnostic testing for suspected CAP of atypical bacterial etiology in adults in the outpatient setting unless a change in the patient treatment regimen is anticipated (1). The recommendation to perform diagnostic testing for M. pneumoniae infection in children is classified as a weak recommendation and likely not routinely performed, in part due to a lack of available diagnostic tests in the primary care setting and at state and local public health laboratories (2). The estimates of the actual number of cases occurring annually are inexact due to the lack of systematic surveillance and reporting. Although historically noted to occur in 3- to 7-year cycles, outbreaks of M. pneumoniae are common and may last for several months as a result of the long incubation period and prolonged carriage after resolution of symptoms (3–5).
Excessive or inappropriate antibiotic use provides selective pressure for the development of antimicrobial resistance. In M. pneumoniae infections, macrolide resistance is an emerging threat worldwide, and in some parts of the world, >90% of M. pneumoniae infections are caused by resistant strains (6, 7). In the United States, the incidence of infection with macrolide-resistant M. pneumoniae strains is not well defined, although the trait has been consistently reported over the past decade (8–10). Macrolide-resistant M. pneumoniae infection has been associated with increased febrile period, increased duration of persistent cough, and extended antibiotic therapy compared to macrolide-sensitive strains (11–16).
Methods for molecular typing of M. pneumoniae strains have been developed, including discrimination of two major types and variants based on the sequence of the P1 adhesion molecule gene and identification of multiple types using multilocus variable-number tandem-repeat analysis (MLVA) to examine four or five polymorphic loci. Despite development of these sophisticated typing methods, characterization of M. pneumoniae strains is even rarer than detection and is only performed by a few specialized laboratories in the United States. As a result, the majority of information on circulating M. pneumoniae strains, including the prevalence of macrolide susceptibility, originates from investigations of outbreaks and individual cases or clusters of severe disease. Here, we review the sporadic cases, small clusters, and outbreaks of M. pneumoniae infection that were investigated by the Centers for Disease Control and Prevention (CDC) between 2006 and 2013 and examine trends in strain types and macrolide resistance during this period.
MATERIALS AND METHODS
Respiratory specimens (n = 199), including nasopharyngeal (NP) and/or oropharyngeal (OP) swabs (n = 135), sputum (n = 1), bronchoalveolar lavage (BAL) fluid (n = 8), and respiratory aspirates (nasal, NP, or OP) (n = 55) submitted to the Pneumonia Response and Surveillance Laboratory (PRSL) at the CDC between 2006 and 2013 were included in this analysis. When multiple specimens from the same patient were available, only one was included in the analysis. Each investigation was classified based on the number of M. pneumoniae-positive specimens as either an individual case (n = 1), a cluster (n ≤ 3), or an outbreak (n > 3).
All respiratory specimens were tested using either a singleplex quantitative PCR (qPCR) assay for the specific detection of M. pneumoniae (17) and/or a Clinical Laboratory Improvement Amendments (CLIA)-approved multiplex assay for detection of M. pneumoniae, Chlamydophila pneumoniae, Legionella spp., and human nucleic acid (18). Sputa specimens were treated with dithiothreitol (DTT) for 30 min prior to extraction. Total nucleic acid was extracted from each specimen using the MagNA Pure Compact or MagNA Pure LC instrument (Roche Applied Science, Indianapolis, IN) with a total nucleic acid isolation kit I. Cultures were attempted for all M. pneumoniae-positive specimens as previously described (19). The recovered isolates were confirmed using a singleplex qPCR assay for M. pneumoniae (17).
MLVA was performed on all PCR-positive specimens having a sufficient volume of original material and all isolates using previously described methods (20). MLVA types were reported using four loci (Mpn13, Mpn14, Mpn15, and Mpn16) as previously suggested (20–22). High-resolution melt (HRM) analysis for identification of the macrolide susceptibility genetic profile was performed with a slight modification of the original procedure (9); the modification consisted of using a nested PCR approach to allow standardization of input material into the HRM reactions. The HRM profiles for clinical specimens were compared to those of the macrolide-resistant and macrolide-sensitive reference strains which were included in each run; the reference strains had been previously verified by sequencing and determination of MICs. P1 typing was performed using HRM analysis as previously described (23). The isolates were classified as type 1, type 2, or variant based upon comparison of melting profiles to that of the type 1 reference strain (M129) and the type 2 reference strain (FH) included in each run.
RESULTS
Characteristics of recent investigations.
Between 2006 and 2013, the CDC PRSL assisted in 17 investigations of M. pneumoniae infections, including individual cases (n = 4), small clusters (n = 3), and outbreaks (n = 10) in 14 different states (Table 1). The number of investigations in a single year ranged from zero to eight during this time period with the highest number of investigations (n = 8, including four outbreaks) occurring in 2013. Of the 10 outbreaks during this period, seven were associated with a school, school district, or university. Patient ages were available for 149 (75%) specimens; the average patient age was 13 years (range, 0 to 50 years). Extrapulmonary manifestations, including encephalitis and dermatological complications (Stevens-Johnson syndrome [SJS]) occurred in two outbreaks. Three outbreaks involved official requests for epidemiological assistance (Epi-Aid) by the CDC, which included retrospective and prospective case findings and investigation of household contacts. The number of M. pneumoniae-positive specimens identified in each outbreak investigation varied widely (range, 4 to 72). More specimens were collected, and more positive specimens were identified during Epi-Aid investigations than for other outbreaks.
TABLE 1.
Characteristics of the CDC-assisted investigations of M. pneumoniae infections, 2006–2013
| Characteristic | No. (%) of investigations |
|---|---|
| Total CDC-assisted investigations (2006–2013) | 17 |
| Sporadic cases | 4 (23.5) |
| Clustera | 3 (17.6) |
| Outbreak | 10 (58.8) |
| Epi-Aid response | 3 (18) |
| Associated with educational institution | 7 (41) |
| Evidence of household transmissionb | 5 (29) |
| Extrapulmonary involvementc | 2 (12) |
| Macrolide resistance detectedc | 8 (47) |
Two to three cases with an epidemiological connection.
Not evaluated in all investigations.
Documented in at least one patient.
Macrolide resistance.
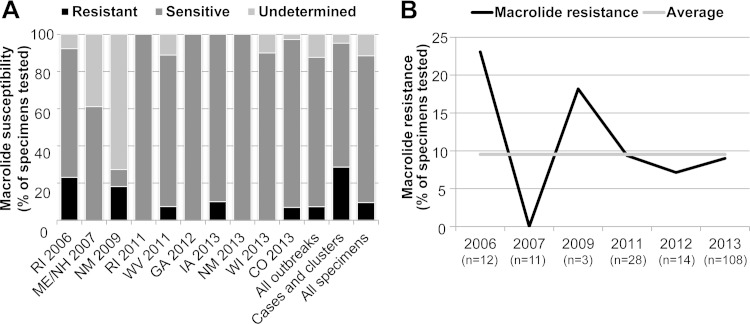
Macrolide-resistant M. pneumoniae strains were detected in at least one patient specimen during eight (47%) of 17 investigations, including 5 (50%) of 10 outbreaks (Table 1). The proportion of macrolide-resistant M. pneumoniae strains detected in qPCR-positive specimens varied between individual outbreaks and among nonoutbreak specimens (sporadic cases and clusters) over the time period studied (Fig. 1). Macrolide-resistant M. pneumoniae strains were identified in 0 to 23% of specimens in a given outbreak (Fig. 1A). Across all outbreaks, the average proportion of macrolide-resistant M. pneumoniae specimens detected was 7%, whereas 29% of specimens not associated with an outbreak were identified as resistant (Fig. 1A). Overall, 10% of M. pneumoniae-positive specimens tested at the CDC during this time period displayed genetic profiles consistent with resistance to macrolides (Fig. 1). Variations in macrolide-resistant specimens over time generally reflected the pattern observed by examining individual outbreaks in order of occurrence (Fig. 1).
FIG 1.
Macrolide resistance in M. pneumoniae specimens in the United States, 2006 to 2013. (A) Proportions of macrolide-resistant and sensitive M. pneumoniae strains identified in individual outbreaks. The proportions of sensitive and resistant strains in all outbreak specimens (n = 156), sporadic cases and clusters (n = 20), and all specimens (n = 176) are also shown. Macrolide susceptibility could not be determined for 23 (12%) of 199 specimens due to the lack of an available specimen or inadequate amplification of the target region. (B) Proportion of macrolide-resistant M. pneumoniae infections identified over the study period. Nineteen resistant strains (10%) were identified during this time period. The number of M. pneumoniae-positive specimens with macrolide susceptibility results is shown for each year; no positive specimens were available from 2008 or 2010.
Molecular typing.
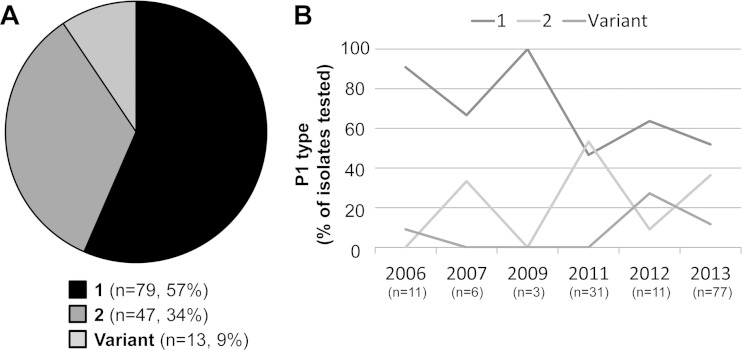
P1 typing, historically used to differentiate strains based on the genetic sequence encoding the P1 adhesion molecule, was performed only on isolates. During the 8-year study period, 57% of M. pneumoniae isolates were identified as type 1 and 34% as type 2 (Fig. 2A). Nine percent of isolates were determined to be variants of the type 2 strain, differing slightly in melting profile from that of either reference strain. The proportion of each type varied over the study period (Fig. 2B). Type 1 isolates accounted for the majority of M. pneumoniae isolates in 2006, but dropped to 50% of isolates in 2013. Type 2 and variant strains each increased between 2011 and 2013.
FIG 2.
P1 typing of M. pneumoniae isolates, 2006 to 2013. (A) Proportion of each P1 type identified among all isolates (n = 139). (B) Proportions of P1 types during each year of the study period. The number of M. pneumoniae isolates for each year is shown; no isolates were available from 2008 or 2010.
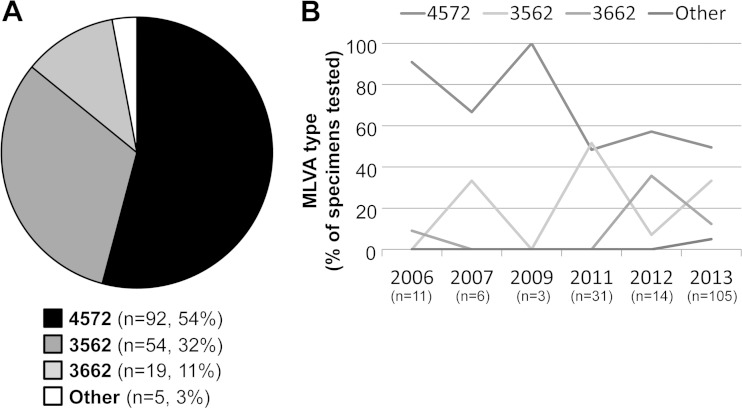
The primary MLVA types identified during the study period were 4572 (54%), 3562 (32%), and 3662 (11%) (Fig. 3A). MLVA types identified in recovered isolates matched the types identified from primary specimens in each case (data not shown). Other rare types, including 3572 (n = 1), 4472 (n = 1), and 4672 (n = 3) accounted for only 3% of all specimens tested and were observed in 2013 only. Multiple MLVA types were detected during 8 (80%) of 10 outbreak investigations. A single MLVA type was observed in two outbreaks. Six different MLVA types were observed in a single outbreak investigation; this outbreak involved the highest number of M. pneumoniae-positive specimens (n = 72). MLVA types 4572 and 3562 each were the predominant MLVA pattern identified in five outbreaks. The pattern of MLVA type distribution over the study period approximately mirrored the P1 typing distribution, consistent with a previously observed correlation between P1 and MLVA types (22).
FIG 3.
MLVA typing of M. pneumoniae specimens and/or isolates, 2006 to 2013. Results for four loci (Mpn13 to Mpn16) are reported. (A) Proportion of each MLVA type identified among specimens/isolates (n = 170). MLVA type could not be determined for 29 (15%) of 199 specimens due to the lack of an available specimen/isolate or inadequate amplification of all four loci. Other types detected include 3572 (n = 1), 4472 (n = 1), and 4672 (n = 3). (B) Proportions of MLVA types during each year of the study period. The number of M. pneumoniae-positive specimens with available MLVA results is shown for each year; no positive specimens were available from 2008 or 2010.
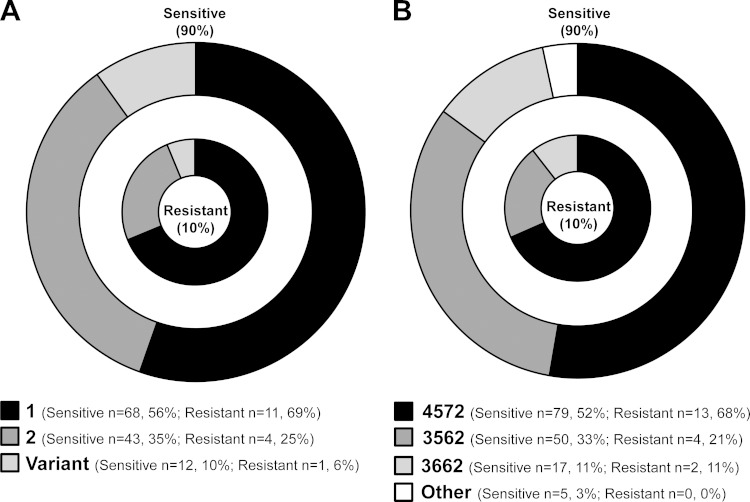
Notably, multiple cocirculating types, as determined by P1 and MLVA typing, were identified in 8 (80%) of 10 outbreaks. The two outbreaks in which only a single P1 or MLVA type was identified consisted of only two or three M. pneumoniae-positive specimens each. Furthermore, the 19 macrolide-resistant isolates were identified as different P1 types, including type 1 (n = 13), type 2 (n = 4), and type 2 variant (n = 2). The resistant isolates were similarly distributed among the predominant MLVA profiles, including 4572 (n = 13), 3562 (n = 4), and 3662 (n = 2). The distributions of P1 types (Fig. 4A) and MLVA types (Fig. 4B) between resistant and sensitive M. pneumoniae strains were similar. There were no apparent differences in the distributions of MLVA types in sporadic cases and clusters compared to those in outbreaks (data not shown).
FIG 4.
Distribution of strain types in macrolide-resistant (inner circle) and macrolide-sensitive (outer circle) M. pneumoniae specimens. (A) P1 types in sensitive (n = 122) and resistant (n = 16) M. pneumoniae isolates (n = 138). (B) MLVA types in sensitive (n = 151) and resistant (n = 19) M. pneumoniae-positive specimens (n = 170).
DISCUSSION
In the absence of national surveillance for M. pneumoniae, some information can be gleaned from a comprehensive examination of recent investigations of M. pneumoniae illness. We examined M. pneumoniae specimens from 17 investigations of individual cases, small disease clusters, and outbreaks of various sizes that occurred in the United States between 2006 and 2013 in which the CDC led or assisted in a laboratory and/or epidemiological investigation. The CDC became involved in investigating individual cases or small clusters when the etiology could not be identified in the clinical or state public health laboratory or when a patient failed to improve despite appropriate antibiotic therapy. In some cases, the CDC assisted in identifying M. pneumoniae as the causative agent, while in other outbreaks, the CDC provided confirmatory laboratory testing and further characterization. More than 40% of outbreaks were associated with an educational institution, which reflects the fact that M. pneumoniae is a leading cause of CAP in children and adolescents (24). Transmission of M. pneumoniae infections between family members is common (8, 25, 26); multiple cases within a household were documented in approximately one-third of the clusters or outbreaks in this investigation, although evidence of household transmission was not evaluated in all of the clusters and outbreaks.
A macrolide-resistant M. pneumoniae strain was identified in at least one patient specimen in approximately half of the investigations included in the current study. We identified an overall rate of macrolide resistance of 10% among specimens collected between 2006 and 2013. Substantial variations in the rates of macrolide resistance in individual outbreaks were noted, ranging from 0 to 23%. The proportion of infections in which macrolide resistance was identified was higher in individual cases and clusters than in outbreaks. The most likely reason for this observation is that requests for assistance from the CDC to investigate individual cases and clusters are more likely when patients fail to improve despite antibiotic treatment and/or when the clinician suspects antibiotic resistance. The introduction of commercially available tests to determine macrolide susceptibility in the primary care setting might facilitate identification of macrolide-resistant M. pneumoniae infections and help prevent further transmission by informing antibiotic therapy decisions.
Studies describing the rates of macrolide resistance in M. pneumoniae strains in the United States are limited. Yamada and colleagues previously reported a rate of 8.2% macrolide-resistant M. pneumoniae among specimens collected between 2010 and 2012 (10). This is similar to the prevalence of resistance observed in the current study. However, the rate of resistance in an individual outbreak can be substantially higher. For instance, 23% of the M. pneumoniae-positive specimens were identified as resistant to macrolides in a single outbreak in the current study. This may truly indicate a higher proportion of resistant specimens in this particular outbreak or simply be a function of testing a limited number of patient specimens. In recent years, several reports have documented low levels of macrolide resistance (≤3%) in M. pneumoniae infections across Europe, including Denmark, France, and Germany, during an active epidemic period (27–29). A recent report by Ferguson and colleagues documented the first macrolide-resistant M. pneumoniae infections in the United Kingdom in 6 of 32 (19%) patients studied (30). The rates of macrolide resistance observed in the United States and Europe contrast dramatically with those in reports from Asia where 80 to 90% of M. pneumoniae infections are resistant (6, 7). Improved surveillance and monitoring are necessary to manage the threat of emerging resistance to the first-line therapy for M. pneumoniae infection in the United States.
The current analysis of 8 years of M. pneumoniae infections in the United States reveals trends in the circulating strain types. Notably, we observed cocirculation of both type 1 and 2 strains during this time period. Multiple MLVA types were detected in 80% of discrete outbreaks. The greatest diversity in MLVA types in a single outbreak setting was observed in the investigation having the highest number of specimens. The appearance of clonality in the remaining two outbreaks was likely due to testing of too few specimens to obtain an accurate representation of the diversity of circulating strains, as has been suggested previously (20, 22). Cocirculation of P1 types in both endemic and epidemic settings has been documented (22, 31). Three MLVA types (4572, 3562, and 3662) accounted for 97% of the M. pneumoniae infections in this study. In 2013, several other MLVA types were infrequently detected; this small increase in strain diversity may simply reflect a more accurate picture of the repertoire of circulating strains due to the testing of a higher number of specimens in that particular year or may indicate the emergence of an increasingly diverse repertoire of M. pneumoniae strains in the community. The same three types identified most frequently during this period were also identified by Sun and colleagues as the predominant MLVA types circulating worldwide between 2009 and 2012 (21). The predominance of these types was revealed only after removal of the Mpn1 locus from the original MLVA typing scheme due to instability observed at this variable-number tandem repeat (VNTR) region (20–22). Although limited to fewer possible types, the modified four-loci scheme appears to provide a more accurate classification of M. pneumoniae strains.
The distribution of P1 and MLVA types did not differ between macrolide-resistant and -sensitive M. pneumoniae strains, suggesting that there is no association of an individual strain type with the resistant genotype. Previous reports have also concluded that no correlation exists between P1 or MLVA type and macrolide resistance (20, 21, 32, 33). Taken together, the cocirculation of multiple strain types and the lack of association of any type with macrolide resistance, disease severity, or the potential to cause outbreaks suggests that current typing schemes may be inadequate to reveal clonality or chains of transmission. A deeper exploration of the genomes of diverse M. pneumoniae types, including both sensitive and resistant isolates, is warranted.
The observation of multiple types of cocirculating M. pneumoniae strains, particularly within a defined outbreak, is difficult to reconcile with traditional epidemiological models of infectious disease outbreaks in which the cases are linked by the characteristics of the bacterial strain, indicating a common exposure or person-to-person transmission. Over the time period studied here, multiple types were present at any given time in the population, even in a closed setting. The predominance of type 1 or 2 strains in the population has historically been documented, and the cyclic pattern of type-specific dominance was attributed to the development of temporary immunity to one type, yielding an opportunity for the alternate type to reemerge (34). The reasons for the more recently observed cocirculation of type strains in the global population and even within a defined outbreak are unclear, and further investigation is warranted to explain this phenomenon. Although current strain typing methods provide some insight into the epidemiology of M. pneumoniae, these methods only allow for classification of strains into a few unique types. Implementation of whole-genome sequencing might provide vastly more information and improve our ability to compare and classify M. pneumoniae strains and to identify bacterial factors which may contribute to severe disease and poor patient outcomes.
There are several limitations to this study. First, the data described here are limited to investigations in which the CDC was invited to participate. There was substantial variation in the number of specimens tested in individual outbreaks. For example, the three Epi-Aid responses were conducted for larger outbreaks and included both retrospective and prospective case findings, resulting in the collection of a large number of specimens and, therefore, identification of more M. pneumoniae-positive specimens than for other outbreaks. This retrospective analysis likely includes only a fraction of the actual infections attributable to this pathogen. The lack of a comprehensive and systematic approach prevents estimation of incidence of individual cases or outbreaks and could conceal geographic or temporal trends in circulating types or macrolide resistance both endemically and during localized outbreaks. Second, this study is limited to 8 years, which may be long enough to observe changes in circulating types, but spanning of a longer time period would be necessary in order to identify broader trends occurring over time and to identify a classical cyclic pattern (34).
Establishment of a systematic surveillance program for M. pneumoniae will be critical for providing the necessary data to fully understand the biology and epidemiology of this organism in the United States. The potential for macrolide resistance to grow to levels observed in other parts of the world is also particularly concerning, especially considering the widespread administration of azithromycin in the United States. Monitoring M. pneumoniae and the prevalence of macrolide resistance is critical for providing appropriate therapy, recognizing outbreaks, and identifying the emergence of hypervirulent or drug-resistant strains.
ACKNOWLEDGMENTS
We thank the laboratorians and epidemiologists at the clinical microbiology and public health laboratories who contributed specimens to this collection and participated in these investigations.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM, Musher DM, Niederman MS, Torres A, Whitney CG. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 44:S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley JS, Byington CL, Shah SS, Alverson B, Carter ER, Harrison C, Kaplan SL, Mace SE, McCracken GH, Moore MR, St Peter SD, Stockwell JA, Swanson JT. 2011. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis 53:e25–e76. doi: 10.1093/cid/cir531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson TP, Balish MF, Waites KB. 2008. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev 32:956–973. doi: 10.1111/j.1574-6976.2008.00129.x. [DOI] [PubMed] [Google Scholar]
- 4.Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winchell JM. 2013. Mycoplasma pneumoniae—a national public health perspective. Curr Pediatr Rev 9:324–333. doi: 10.2174/15733963113099990009. [DOI] [Google Scholar]
- 6.Liu Y, Ye X, Zhang H, Xu X, Li W, Zhu D, Wang M. 2009. Antimicrobial susceptibility of Mycoplasma pneumoniae isolates and molecular analysis of macrolide-resistant strains from Shanghai, China. Antimicrob Agents Chemother 53:2160–2162. doi: 10.1128/AAC.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin D, Mi Z, Han X, Qin L, Li J, Wei T, Chen X, Ma S, Hou A, Li G, Shi D. 2009. Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob Agents Chemother 53:2158–2159. doi: 10.1128/AAC.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newell L, Burke P, Woods J, Posey R, Smith B, Ibrahim SM, Bixler D, Haddy L, Shwe T, Radcliffe R, Benitez A, Thurman KA, Diaz MH, Wolff BJ, Warner A, Winchell JM, Conklin LM, Barbour K, Fleming-Dutra KE, Steinhardt LC. 2012. Mycoplasma pneumoniae respiratory illness—two rural counties, West Virginia, 2011. Morb Mortal Wkly Rep 61:834–838. [PubMed] [Google Scholar]
- 9.Wolff BJ, Thacker WL, Schwartz SB, Winchell JM. 2008. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high resolution melt analysis. Antimicrob Agents Chemother 52:3542–3549. doi: 10.1128/AAC.00582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada M, Buller R, Bledsoe S, Storch GA. 2012. Rising rates of macrolide-resistant Mycoplasma pneumoniae in the central United States. Pediatr Infect Dis J 31:409–411. doi: 10.1097/INF.0b013e318247f3e0. [DOI] [PubMed] [Google Scholar]
- 11.Cao B, Zhao CJ, Yin YD, Zhao F, Song SF, Bai L, Zhang JZ, Liu YM, Zhang YY, Wang H, Wang C. 2010. High prevalence of macrolide resistance in Mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin Infect Dis 51:189–194. doi: 10.1086/653535. [DOI] [PubMed] [Google Scholar]
- 12.Cardinale F, Chironna M, Chinellato I, Principi N, Esposito S. 2013. Clinical relevance of Mycoplasma pneumoniae macrolide resistance in children. J Clin Microbiol 51:723–724. doi: 10.1128/JCM.02840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsubara K, Morozumi M, Okada T, Matsushima T, Komiyama O, Shoji M, Ebihara T, Ubukata K, Sato Y, Akita H, Sunakawa K, Iwata S. 2009. A comparative clinical study of macrolide-sensitive and macrolide-resistant Mycoplasma pneumoniae infections in pediatric patients. J Infect Chemother 15:380–383. doi: 10.1007/s10156-009-0715-7. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka M, Narita M, Okazaki N, Ohya H, Yamazaki T, Ouchi K, Suzuki I, Andoh T, Kenri T, Sasaki Y, Horino A, Shintani M, Arakawa Y, Sasaki T. 2004. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother 48:4624–4630. doi: 10.1128/AAC.48.12.4624-4630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morozumi M, Iwata S, Hasegawa K, Chiba N, Takayanagi R, Matsubara K, Nakayama E, Sunakawa K, Ubukata K, the Acute Respiratory Diseases Study Group . 2008. Increased macrolide resistance of Mycoplasma pneumoniae in pediatric patients with community-acquired pneumonia. Antimicrob. Agents Chemother 52:348–350. doi: 10.1128/AAC.00779-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki S, Yamazaki T, Narita M, Okazaki N, Suzuki I, Adoh T, Matsuoka M, Kenri T, Arakawa Y, Sasaki T. 2006. Clinical evaluation of macrolide-resistant Mycoplasma pneumoniae. Antimicrob Agents Chemother 50:709–712. doi: 10.1128/AAC.50.2.709-712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS. 2008. Evaluation of three real-time PCR assays for the detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 46:3116–3118. doi: 10.1128/JCM.00440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thurman KA, Warner AK, Cowart KC, Benitez AJ, Winchell JM. 2011. Detection of Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella spp. in clinical specimens using a single-tube multiplex real-time PCR assay. Diagn Microbiol Infect Dis 70:1–9. doi: 10.1016/j.diagmicrobio.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thurman KA, Walter ND, Schwartz SB, Mitchell SL, Dillon MT, Deutscher M, Fulton JP, Tongren JE, Hicks LA, Winchell JM. 2009. Comparison of laboratory diagnostic procedures for detection of Mycoplasma pneumoniae in community outbreaks. Clin Infect Dis 48:1244–1249. doi: 10.1086/597775. [DOI] [PubMed] [Google Scholar]
- 20.Benitez AJ, Diaz MH, Wolff BJ, Pimentel G, Njenga MK, Estevez A, Winchell JM. 2012. Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J Clin Microbiol 50:3620–3626. doi: 10.1128/JCM.01755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun H, Xue G, Yan C, Li S, Cao L, Yuan Y, Zhao H, Feng Y, Wang L, Fan Z. 2013. Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens and proposal for amendment of MLVA nomenclature. PLoS One 8:e64607. doi: 10.1371/journal.pone.0064607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waller JL, Diaz MH, Petrone BL, Benitez AJ, Wolff BJ, Edison L, Tobin-D'Angelo M, Moore A, Martyn A, Dishman H, Drenzek CL, Turner K, Hicks LA, Winchell JM. 2014. Detection and characterization of Mycoplasma pneumoniae during an outbreak of respiratory illness at a university. J Clin Microbiol 52:849–853. doi: 10.1128/JCM.02810-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz SB, Thurman KA, Mitchell SL, Wolff BJ, Winchell JM. 2009. Genotyping of Mycoplasma pneumoniae isolates using real-time PCR and high-resolution melt analysis. Clin Microbiol Infect 15:756–762. doi: 10.1111/j.1469-0691.2009.02814.x. [DOI] [PubMed] [Google Scholar]
- 24.Jain S, Ampofo K, Arnold SR, Williams DJ, Anderson E, Bramley AM, Hymas W, Kaufman RA, Self WH, Erdman D, Winchell JM, Carvalho G, Lindstrom S, Chappell JD, Patel A, Hillyard D, Grijalva CG, Schneider E, Hicks L, Wunderink RG, Edwards KM, McCullers JA, Pavia AT, Finelli L. 2012. Etiology of community-acquired pneumonia among hospitalized children in the United States: preliminary data from the CDC Etiology of Pneumonia in the Community (EPIC) Study, abstr 168. 49th Annu Meet Infectious Diseases Society of America, Boston, MA. [Google Scholar]
- 25.Foy HM, Grayston JT, Kenny G, Alexander E, McMahan R. 1966. Epidemiology of Mycoplasma pneumoniae infection in families. JAMA 197:859–866. doi: 10.1001/jama.1966.03110110083019. [DOI] [PubMed] [Google Scholar]
- 26.Walter ND, Grant GB, Bandy U, Alexander NE, Winchell JM, Jordan HT, Sejvar JJ, Hicks LA, Gifford DR, Alexander NT, Thurman KA, Schwartz SB, Dennehy PH, Khetsuriani N, Fields BS, Dillion MT, Erdman DD, Whitney CG, Moore MR. 2008. Community outbreak of Mycoplasma pneumoniae infection: school-based cluster of neurologic disease associated with household transmission of respiratory illness. J Infect Dis 198:1365–1374. doi: 10.1086/592281. [DOI] [PubMed] [Google Scholar]
- 27.Dumke R, von Baum H, Luck PC, Jacobs E. 2010. Occurrence of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Clin Microbiol Infect 16:613–616. doi: 10.1111/j.1469-0691.2009.02968.x. [DOI] [PubMed] [Google Scholar]
- 28.Pereyre S, Charron A, Renaudin H, Bebear CM, Bébéar CM. 2007. First report of macrolide-resistant strains and description of a novel nucleotide sequence variation in the P1 adhesin gene in Mycoplasma pneumoniae clinical strains isolated in France over 12 years. J Clin Microbiol 45:3534–3539. doi: 10.1128/JCM.01345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uldum SA, Bangsborg JM, Gahrn-Hansen B, Ljung R, Molvadgaard M, Fons Petersen R, Wiid Svarrer C. 2012. Epidemic of Mycoplasma pneumoniae infection in Denmark, 2010 and 2011. Euro Surveill 17(5):pii=20073 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20073. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson GD, Gadsby NJ, Henderson SS, Hardie A, Kalima P, Morris AC, Hill AT, Cunningham S, Templeton KE. 2013. Clinical outcomes and macrolide resistance in Mycoplasma pneumoniae infection in Scotland, UK. J Med Microbiol 62:1876–1882. doi: 10.1099/jmm.0.066191-0. [DOI] [PubMed] [Google Scholar]
- 31.Pereyre S, Charron A, Hidalgo-Grass C, Touati A, Moses AE, Nir-Paz R, Bebear C. 2012. The spread of Mycoplasma pneumoniae is polyclonal in both an endemic setting in France and in an epidemic setting in Israel. PLoS One 7:e38585. doi: 10.1371/journal.pone.0038585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dégrange S, Cazanave C, Charron A, Renaudin H, Bebear C, Bebear CM. 2009. Development of multiple-locus variable-number tandem-repeat analysis for the molecular typing of Mycoplasma pneumoniae. J Clin Microbiol 47:914–923. doi: 10.1128/JCM.01935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Ye X, Zhang H, Xu X, Wang M. 2012. Multiclonal origin of macrolide-resistant Mycoplasma pneumoniae isolates as determined by multilocus variable-number tandem-repeat analysis. J Clin Microbiol 50:2793–2795. doi: 10.1128/JCM.00678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lind K, Benzon MW, Jensen S, Clyde WA Jr. 1997. A seroepidemiological study of Mycoplasma pneumoniae infections in Denmark over the 50-year period 1946 to 1995. Eur J Epidemiol 13:581–586. doi: 10.1023/A:1007353121693. [DOI] [PubMed] [Google Scholar]