Abstract
Yersinia enterocolitica and Yersinia pseudotuberculosis, the two Yersinia species that are enteropathogenic for humans, are distributed worldwide and frequently cause diarrhea in inhabitants of temperate and cold countries. Y. enterocolitica is a major cause of foodborne disease resulting from consumption of contaminated pork meat and is further associated with substantial economic cost. However, investigation of enteropathogenic Yersinia species is infrequently performed routinely in clinical laboratories because of their specific growth characteristics, which make difficult their isolation from stool samples. Moreover, current isolation procedures are time-consuming and expensive, thus leading to underestimates of the incidence of enteric yersiniosis, inappropriate prescriptions of antibiotic treatments, and unnecessary appendectomies. The main objective of the study was to develop fast, sensitive, specific, and easy-to-use immunoassays, useful for both human and veterinary diagnosis. Monoclonal antibodies (MAbs) directed against Y. enterocolitica bioserotypes 2/O:9 and 4/O:3 and Y. pseudotuberculosis serotypes I and III were produced. Pairs of MAbs were selected by testing their specificity and affinity for enteropathogenic Yersinia and other commonly found enterobacteria. Pairs of MAbs were selected to develop highly sensitive enzyme immunoassays (EIAs) and lateral flow immunoassays (LFIs or dipsticks) convenient for the purpose of rapid diagnosis. The limit of detection of the EIAs ranged from 3.2 × 103 CFU/ml to 8.8 × 104 CFU/ml for pathogenic serotypes I and III of Y. pseudotuberculosis and pathogenic bioserotypes 2/O:9 and 4/O:3 of Y. enterocolitica and for the LFIs ranged from 105 CFU/ml to 106 CFU/ml. A similar limit of detection was observed for artificially contaminated human feces.
INTRODUCTION
The genus Yersinia belongs to the family of Enterobacteriaceae and is composed of three human-pathogenic species: Yersinia pestis, the causative agent of the plague, and Yersinia enterocolitica and Y. pseudotuberculosis, responsible for human enteric yersiniosis. Enteric yersiniosis is a foodborne disease caused by consumption of contaminated food or water (1) and can be transmitted between humans through the fecal-oral route. The disease is usually characterized by a self-limiting acute infection beginning in the intestine and is often limited to the ileocecal junction for Y. enterocolitica. In contrast, Y. pseudotuberculosis often disseminates deeply to the mesenteric lymph nodes. Clinical presentation is characterized by enterocolitis (diarrhea, abdominal pain, fever, and sometimes vomiting) (2), which predominates in young children and is often self-limiting. However, diarrhea is a predominant symptom of Y. enterocolitica infection whereas abdominal pain is more usual in Y. pseudotuberculosis infection. Moreover, Y. pseudotuberculosis can also cause different clinical symptoms such as scarlatinoid rash, conjunctivitis, acute organ failure, and toxic shock syndrome often reported in Far East (3). For both enteropathogenic Yersinia species, more-serious infections and sepsis can also occur, particularly in new-born, elderly, and immunocompromised patients. Sometimes, the infection appears as a pseudoappendicular syndrome in which mesenteric lymph nodes are involved, thus possibly leading to unnecessary appendectomies (4). Some secondary complications such as reactive arthritis and erythema nodosum are sometimes observed (5, 6). Rarely, Y. enterocolitica is responsible for a serious sepsis incident after transfusion of contaminated red blood cell preparations (7).
Y. enterocolitica and Y. pseudotuberculosis are widespread worldwide, with a higher incidence in cold and temperate regions. Most Y. enterocolitica strains associated with human yersiniosis belong to bioserotypes 2/O:9, 4/O:3, 2/O:5,27, 3/O:3, and 1B/O:8 (8). In France and worldwide, serotypes 2/O:9 and 4/O:3 and Y. pseudotuberculosis serotypes I and III are the prevailing isolated strains (9). The incidence of human enteric yersiniosis has been estimated to be 16, 1.65, and 0.35 per 100,000 inhabitants in France (10), Europe (11), and the United States (12), respectively, but is probably largely underestimated for many reasons. Y. enterocolitica is the third greatest causative agent of diarrhea of bacterial origin in France and Europe after Campylobacter and Salmonella (11). Even when the incidence of Y. pseudotuberculosis is lower, it represents a major public health problem in some countries such as Japan or Russia, where it causes a particular and severe infection known as Far East scarlet-like fever or Izumi fever (13, 14), and in Finland, where multiple outbreaks were observed (15). In France, a sudden onset of Y. pseudotuberculosis infections was reported between 2004 and 2005 (16).
Nowadays, diagnosis of enteric yersiniosis is performed by a direct isolation of enteropathogenic Yersinia from stool cultures together with an enrichment in a specific broth before isolation on a semiselective medium known as cefsulodin-irgasan-novobiocin medium (CIN). Since Yersinia strains differ by a lower growth rate and a different optimal growth temperature (28°C instead of 37°C) from other enterobacteria, stool cultures performed at 37°C for 24 h (optimal conditions for most enterobacteria) are not efficient for recovering Yersinia colonies in the commensal flora. Moreover, isolation, even performed on selective media, needs time-consuming enrichment steps and is poorly successful for Y. pseudotuberculosis (17). Finally, detection of enteropathogenic bacteria is generally not required by physicians due to the lack of knowledge about these pathogens. However, personnel in clinical laboratories are becoming more and more conscious of the enteropathogenic Yersinia issues and are disposed to perform systematic analysis on feces samples.
After a bacterial colony is isolated, identification of the Yersinia species is achieved by a biochemical characterization with commercial systems such as API 20E or 50CH (bioMérieux). For Y. enterocolitica, determination of the biotype is necessary to assess the pathogenicity of the strain and is based on a biotype scheme involving supplementary biochemical characterizations (18). For Y. pseudotuberculosis, as all strains are considered pathogenic, determination of the biotype is not done. Furthermore, the characterization of serotypes for both enteropathogenic Yersinia species can be achieved by seroagglutination of strains. However, this technique is available only in specialized laboratories and serotypes are not necessary related to the pathogenicity of Y. enterocolitica (19). Some molecular techniques such as DNA colony hybridization, PCR, real-time PCR, multilocus sequence typing (MLST), and pulsed-field gel electrophoresis (PFGE) have been developed, but only PCR techniques are used for detection (20). However, most of these techniques need isolation of the bacteria or an enrichment step to avoid inhibition due to the complex composition of stool samples (21) and may also require specific devices possibly not present in clinical laboratories (8, 22). Until now, there has been no available standard procedure for identification of all enteropathogenic Yersinia enterobacteria.
New diagnostic tools are needed to complete the currently gold standard tests, i.e., the microbiological methods that are quite time-consuming. Ideally, these tests should be rapid, sensitive, specific, efficient when performed on fecal samples, inexpensive, and user-friendly. Among the analytical methods, immunoassays, particularly lateral flow immunoassays, are simple to perform. Some lateral flow immunoassays have already been successfully developed for Yersinia pestis (23, 24), Vibrio cholerae (25), and other enterobacteria such as Shigella spp. (26, 27). The aim of this study was thus to develop sensitive and specific immunoassays using either the enzyme immunoassay (EIA) format or the lateral flow immunoassay (LFI or dipstick) format, targeting enteropathogenic Yersinia, for rapid and simple detection in human samples. Monoclonal antibodies (MAbs) were thus raised against two Y. enterocolitica bioserotypes (2/O:9 and 4/O:3) and two Y. pseudotuberculosis serotypes (I and III).
MATERIALS AND METHODS
Ethics statement.
All experiments were performed in compliance with the French and European regulations on care and protection of laboratory animals (European Community [EC] Directive 86/609, French Law 2001-486, 6 June 2001) and with agreement no. 91-416 delivered to S. Simon by the French Veterinary Services and CEA agreement D-91-272-106 from the Veterinary Inspection Department of Essonne (France).
No ethics approval or written consent was necessary because the human sample was recovered from the first author of the article. Verbal informed consent was obtained from the first author.
Reagents.
Biotin N-hydroxysuccinimide ester, streptavidin, gold chloride solution, and N-succinimidyl-S-acetyl-thioacetate (SATA) were from Sigma-Aldrich. Goat anti-mouse IgG and IgM polyclonal antibodies were from Jackson ImmunoResearch. Proteinase K was from Bio-Rad. Stabilized goat anti-mouse IgG (H+L)-conjugated horseradish peroxidase (HRP) was from Thermo Fisher. The polyvinylidene difluoride (PVDF) membrane for Western blotting was from Amersham Biosciences. Luminata Forte Western HRP substrate and protein A Sepharose were from Millipore (ProsepA). EIA was performed with MaxiSorp 96-well microtiter plates (Nunc), and all reagents were diluted in EIA buffer (0.1 M phosphate buffer [pH 7.4] containing 0.15 M NaCl, 0.1% bovine serum albumin [BSA], and 0.01% sodium azide). Plates coated with proteins were saturated in EIA buffer (18 h at 4°C) and washed with washing buffer (0.01 M potassium phosphate [pH 7.4] containing 0.05% Tween 20).
Strains and growth conditions.
The bacterial strains used in this study are listed in Table 1 and Table 2. All enterobacteria (Yersinia, Escherichia coli, Shigella, and Salmonella) were grown in Luria Bertani broth (LB), and Brucella species were grown in Trypticase soy agar (TSA) with 5% sheep blood (bioMérieux). Growth temperatures were 28°C or 37°C for Yersinia strains and 37°C for other bacteria. Before the assays were performed, all strains were grown overnight in 5 ml LB broth using agitation at 28°C or 37°C. Bacteria were subcultured with 1:100 of the first culture for 4 h under the same conditions.
TABLE 1.
Specificity and comprehensiveness of the lateral flow immunoassay for Yersinia enterocolitica detectiona
| Species | Bioserotype or characteristic(s) | Tested concn (CFU/ml) | Country | Origin | Strain no. | 2/O:9 LFI result | 4/O:3 LFI result |
|---|---|---|---|---|---|---|---|
| Yersinia enterocolitica | 2/O:9 | 5 × 106 | Belgium | Clinical | IP00383 | + | − |
| 2/O:9 | 5 × 106 | France | Clinical | IP28114 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP29717 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP29193 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP29476 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP29523 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP29944 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP33498 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP33617 | + | − | |
| 2/O:9 | 5 × 106 | France | Animal | IP33949 | + | − | |
| 2/O:9 | 5 × 106 | France | Clinical | IP34070 | + | − | |
| 2/O:9 | 5 × 106 | Netherland | Animal | Ye21 | + | − | |
| 2/O:9 | 5 × 106 | Belgium | Clinical | IP4294 | + | − | |
| 4/O:3 | 106 | Sweden | Clinical | IP00134 | − | + | |
| 4/O:3 | 106 | Greece | Clinical | IP08896 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP10393 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP28096 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP28164 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP28983 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP29001 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP29310 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP29534 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP29610 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP33526 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP33550 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP33563 | − | + | |
| 4/O:3 | 106 | France | Clinical | IP34075 | − | + | |
| 2/O:5 | 5 × 106 | France | Clinical | IP34120 | − | − | |
| 1A/NAG | 5 × 106 | France | Clinical | IP25166 | − | − | |
| 1A/O:7,8-8-8,19 | 5 × 106 | France | Animal | IP26014 | − | − | |
| 1A/O:10,34 | 5 × 106 | India | Environment | IP26309 | − | − | |
| 1A/O:5 | 5 × 106 | Italy | Food | IP26618 | − | − | |
| 1A/O:6,30-6,31 | 5 × 106 | France | Clinical | IP29463 | − | − | |
| 1A/O:41,42-41,43 | 5 × 106 | France | Clinical | IP29465 | − | − | |
| 1A/O:12,25-12,26-25,35-35 | 5 × 106 | France | Clinical | IP29469 | − | − | |
| 1A/NAG | 5 × 106 | France | Clinical | IP29845 | − | − | |
| 1A/O:12,25-12,26 | 5 × 106 | France | Clinical | IP33764 | − | − | |
| 1A/O:3 | 5 × 106 | France | Clinical | IP27875 | − | + | |
| 1A/NAG | 5 × 107 | France | Clinical | IP33592 | − | − | |
| 1B/O:8 | 5 × 107 | USA | Clinical | CIP80.27 | − | − | |
| 2/O:5,27 | 5 × 107 | United Kingdom | Animal | CIP106676 | − | − | |
| Yersinia pseudotuberculosis | I | 5 × 107 | Cuba | Animal | IP31629 | − | − |
| III | 5 × 107 | France | Clinical | IP33434 | − | − | |
| V | 5 × 107 | Sweden | Animal | CIP55.88 | − | − | |
| Escherichia coli | BL21(DE3) | 5 × 107 | Invitrogen | NA | − | − | |
| DH5 alpha | 5 × 107 | Invitrogen | NA | − | − | ||
| Serotype O26 | 5 × 107 | CIP52.172 | − | − | |||
| Serotype O55 | 5 × 107 | CIP52.170 | − | − | |||
| Shigella sonnei | Lysotype 4 | 5 × 107 | CIP67.63 | − | − | ||
| Shigella flexneri | Serotype 2 | 5 × 107 | CIP106236 | − | − | ||
| Salmonella enterica | Serotype Typhimurium | 5 × 107 | CIP104474 | − | − | ||
| Serotype Enteritidis | 5 × 107 | CIP105150 | − | − | |||
| Serotype Paratyphi A | 5 × 107 | CIP55.155 | − | − | |||
| Serotype Senftenberg | 5 × 107 | CIP105343 | − | − | |||
| Erwinia pyrifoliae | 5 × 107 | CIP106111 | − | − | |||
| Brucella abortus | Biotype 4 | 5 × 107 | NCTC10503 | − | − | ||
| Brucella melitensis | Biotype 2 | 5 × 107 | NCTC10508 | − | − | ||
| Biotype 3 | 5 × 107 | NCTC10509 | − | − |
The bacterial suspensions were grown at 28°C for Yersinia or 37°C for other bacteria. +, positive test line estimated by eye; −, negative test line estimated by eye; NA, not applicable; NAG, nonagglutinable; IP, strains from the collection of the Yersinia Research Unit/National Reference Laboratory; CIP, strains from the Collection of the Institut Pasteur; NCTC, strains from the National Counterterrorism Center.
TABLE 2.
Specificity and comprehensiveness of the lateral flow immunoassay for Yersinia pseudotuberculosis detectiona
| Species | Serotype or characteristic(s) | Tested concn (CFU/ml) | Country | Origin | Strain no. | Yps LFI result |
|---|---|---|---|---|---|---|
| Yersinia pseudotuberculosis | I | 106 | Morocco | Animal | IP30636 | + |
| I | 106 | Tunisia | Animal | IP30642 | + | |
| I | 106 | Cuba | Animal | IP31629 | + | |
| I | 106 | Poland | Animal | IP32080 | + | |
| I | 106 | Hungary | Animal | IP32414 | + | |
| I | 106 | former Czechoslovakia | Animal | IP32575 | + | |
| I | 106 | Russia | Animal | IP33178 | + | |
| I | 107 | Hungary | Animal | IP30842 | + | |
| I | 107 | Cuba | Animal | IP31630 | + | |
| I | 107 | Chile | Clinical | IP32654 | + | |
| I | 107 | former Yugoslavia | Animal | IP32665 | + | |
| I | 107 | England | Animal | IP32670 | + | |
| I | 107 | Switzerland | Clinical | IP32730 | + | |
| I | 107 | Switzerland | Clinical | IP32907 | + | |
| I | 107 | France | Clinical | IP32953 | + | |
| I | 107 | Switzerland | Clinical | IP32989 | + | |
| I | 107 | Russia | Animal | IP33242 | + | |
| I | 107 | Russia | Clinical | IP33247 | + | |
| I | 107 | France | Animal | IP33424 | + | |
| I | 107 | France | Clinical | IP33438 | + | |
| I | 107 | former Yugoslavia | Animal | IP32651 | − | |
| I | 107 | France | Clinical | IP32777 | − | |
| I | 107 | Italy | Animal | IP32784 | − | |
| I | 107 | Italy | Animal | IP32800 | − | |
| I | 107 | Switzerland | Clinical | IP32906 | − | |
| I | 107 | France | Clinical | IP32950 | − | |
| I | 107 | France | Clinical | IP32953 | − | |
| I | 107 | USA | unknown | IP33035 | − | |
| I | 107 | France | Clinical | IP33053 | − | |
| I | 107 | France | Clinical | IP33109 | − | |
| I | 107 | France | Clinical | IP33285 | − | |
| I | 107 | France | Animal | IP33427 | − | |
| II | 107 | France | Clinical | IP32554 | + | |
| II | 107 | Spain | Animal | IP32584 | + | |
| II | 107 | New Zealand | Clinical | IP32589 | + | |
| II | 107 | France | Clinical | IP32598 | + | |
| II | 107 | France | Animal | IP32870 | + | |
| II | 107 | France | Clinical | IP32951 | + | |
| II | 107 | France | Clinical | IP33006 | + | |
| II | 107 | France | Clinical | IP33047 | + | |
| II | 107 | France | Animal | IP33098 | + | |
| II | 107 | France | Clinical | IP33306 | + | |
| III | 5 × 106 | Spain | Clinical | IP32666 | + | |
| III | 5 × 106 | Italy | Animal | IP32787 | + | |
| III | 5 × 106 | Argentina | Animal | IP32976 | + | |
| III | 5 × 106 | Australia | Animal | IP32990 | + | |
| III | 5 × 106 | France | Animal | IP33049 | + | |
| III | 5 × 106 | Argentina | Animal | IP33104 | + | |
| III | 5 × 106 | Russia | Clinical | IP33185 | + | |
| III | 5 × 106 | Russia | Clinical | IP33250 | + | |
| III | 5 × 106 | Argentina | Animal | IP33297 | + | |
| III | 5 × 106 | France | Clinical | IP33377 | + | |
| III | 5 × 106 | France | Clinical | IP33434 | + | |
| IV | 5 × 106 | Japan | Animal | Ryster | + | |
| IV | 5 × 106 | Japan | Animal | IP30103 | + | |
| IV | 5 × 106 | former USSR | Animal | IP30290 | + | |
| IV | 5 × 106 | former USSR | Animal | IP30291 | + | |
| IV | 5 × 106 | England | Clinical | IP30298 | + | |
| IV | 5 × 106 | Denmark | Animal | IP31411 | + | |
| IV | 5 × 106 | England | Clinical | IP31830 | + | |
| IV | 5 × 106 | England | Animal | IP31833 | + | |
| IV | 5 × 106 | France | Animal | IP32687 | + | |
| IV | 5 × 106 | Russia | Animal | IP33234 | + | |
| V | 107 | Sweden | Animal | CIP55.88 | + | |
| V | 107 | Switzerland | Animal | IP32463 | + | |
| V | 107 | France | Animal | IP32699 | + | |
| V | 107 | France | Animal | IP32727 | + | |
| V | 107 | Japan | Unknown | IP32814 | + | |
| V | 107 | Japan | Unknown | IP32817 | + | |
| V | 107 | France | Clinical | IP32821 | + | |
| V | 107 | France | Clinical | IP32843 | + | |
| V | 107 | Germany | Animal | IP33061 | + | |
| V | 107 | Russia | Clinical | IP33278 | + | |
| V | 107 | France | Clinical | IP33397 | + | |
| Yersinia similis | O:1c | 107 | Japan | Environment | Kuratani-2 | − |
| O:6 | 107 | Germany | Animal | CIP109846 | − | |
| O:6 | 107 | Japan | Animal | R116 | − | |
| O:7 | 107 | Japan | Animal | R2091-2 | − | |
| O:11 | 107 | Japan | Animal | R2031 | − | |
| Yersinia wautersii | O:4a | 107 | Japan | Clinical | #51 | − |
| O:4a | 107 | Germany | Animal | Y428 | − | |
| O:11 | 107 | Korea | Environment | WP-930601 | − | |
| O:11 | 107 | Korea | Environment | WP-931205 | − | |
| O:15 | 107 | Korea | Clinical | 12-219N1 | − | |
| Yersinia enterocolitica | 2/O:9 | 5 × 107 | Belgium | Clinical | IP00383 | − |
| 4/O:3 | 5 × 107 | Sweden | Clinical | IP00134 | − | |
| 1A/NAG | 5 × 107 | France | Clinical | IP33592 | − | |
| 1B/O:8 | 5 × 107 | USA | Clinical | CIP80.27 | − | |
| 2/O:5,27 | 5 × 107 | United Kingdom | Animal | CIP106676 | − | |
| Escherichia coli | BL21(DE3) | 5 × 107 | Invitrogen | NA | − | |
| DH5 alpha | 5 × 107 | Invitrogen | NA | − | ||
| Serotype O26 | 5 × 107 | CIP52.172 | − | |||
| Serotype O55 | 5 × 107 | CIP52.170 | − | |||
| Shigella sonnei | Lysotype 4 | 5 × 107 | CIP67.63 | − | ||
| Shigella flexneri | Serotype 2 | 5 × 107 | CIP106236 | − | ||
| Salmonella enterica | Serotype Typhimurium | 5 × 107 | CIP104474 | − | ||
| Serotype Enteritidis | 5 × 107 | CIP105150 | − | |||
| Serotype Paratyphi A | 5 × 107 | CIP55.155 | − | |||
| Serotype Senftenberg | 5 × 107 | CIP105343 | − | |||
| Erwinia pyrifoliae | 5 × 107 | CIP106111 | − | |||
| Brucella abortus | Biotype 4 | 5 × 107 | NCTC10503 | − | ||
| Brucella melitensis | Biotype 2 | 5 × 107 | NCTC10508 | − | ||
| Biotype 3 | 5 × 107 | NCTC10509 | − |
The bacterial suspensions were grown at 28°C for Yersinia or 37°C for other bacteria. +, positive test line estimated by eye; −, negative test line estimated by eye; NA, not applicable; NAG, nonagglutinable; IP, strains from the collection of the Yersinia Research Unit/National Reference Laboratory; CIP, strains from the Collection of the Institut Pasteur. Y. wautersii strain Y428 was kindly provided by the Thüringer Landesamt für Verbraucherschutz (TLV), Bad Langensalza, Germany. Other Y. similis and Y. wautersii strains were from the collection of the Shimane Prefectural Institute of Public Health and Environmental Science, Matsue, Japan.
Production of monoclonal antibodies.
Ten-week-old female Biozzi mice were immunized monthly for 3 months by injection of 109 heat-inactivated Yersinia enterobacteria cultured at 37°C into the footpad to simulate the temperature conditions before an infection. Mice were bled before the first immunization (S0, used as the negative control) and 2 weeks after each injection (S1, S2, and S3). The immune polyclonal response was evaluated by enzyme-linked immunosorbent assay (ELISA) using enteropathogenic Yersinia strains as coated antigens (see “Enzyme immunoassays” below). The two mice presenting the highest ELISA titer were selected for preparation of MAbs and given two intravenous booster injections of 109 CFU of heat-killed bacteria 2 months after the last immunization. Two days after the last boost, spleen cells from mice were fused with myeloma NS1 cells as previously described (28). The hybridoma culture supernatants were screened for the presence of anti-Yersinia antibodies by ELISA (see “Enzyme immunoassays” below). Selected hybridomas were subsequently cloned by limiting dilution, and MAbs were obtained after inducing ascitic fluid in BALB/c mice. MAbs were further purified by affinity chromatography using protein A and dialyzed in 0.05 M phosphate buffer (pH 7.4). Purity was assessed by SDS-PAGE and Coomassie blue staining.
In this study, MAbs obtained after immunization with Y. enterocolitica bioserotype 2/O:9 or bioserotype 4/O:3 were called Ye, and MAbs obtained after immunization with Y. pseudotuberculosis serotype I or serotype III were called Yps. MAbs called Yp were obtained from a previous study (unpublished work) after immunization with Y. pestis.
Enzyme immunoassays. (i) Evaluation of polyclonal response and screening of MAbs in hybridoma supernatants.
Anti-Yersinia antibodies were detected in sera of immunized mice or hybridoma culture supernatants using ELISA. Briefly, 50 μl of 2 × 108 CFU/ml of enterobacteria in sterile water was added to each well of microtiter plates and allowed to dry overnight (ON) at room temperature (RT). The plates were then saturated with 300 μl/well of EIA buffer ON at 4°C. After a washing cycle was performed with the washing buffer, the plates were incubated with 100 μl/well of each hybridoma culture supernatant or serial dilutions of mouse sera for 2 h at RT. The plates were then washed before the addition of 100 μl of acetylcholinesterase (AChE; EC 3.1.1.7)-labeled anti-mouse IgG and IgM conjugate (2 Ellman units [EU]/ml [29]) to each well (for AChE labeling details, see below). After 2 h of incubation at RT followed by three washing cycles, 200 μl of Ellman's reagent (30) was added, and the absorbance was measured at 414 nm after 30 min of reaction time.
(ii) Labeling with biotin.
A mixture of 0.67 nmol of MAb and 400 μl of borate buffer (0.1 M; pH 8.5) was incubated at a 1:20 molar ratio with biotin-N-hydroxysuccinimide ester (13.3 nmol) dissolved in 6 μl of anhydrous dimethylformamide (DMF). The reaction was stopped after 30 min at RT by adding 100 μl of 1 M Tris-HCl (pH 8) for 30 min. Finally, 500 μl of EIA buffer was added and the preparation was stored frozen at −20°C until use.
(iii) Labeling with AChE.
The labeling was performed as described in reference 31. In this procedure, MAbs of the IgG1 isotype (Ye18 and Yps104) were fragmented and reduced to Fab′ fragments in order to make thiol groups accessible, and MAbs of the IgG2b isotype (Ye300 and Yps2) were thiolated with SATA (N-succinimidyl-S-acetyl-thioacetate). Maleimide groups were introduced into the tetrameric form (G4) of AChE by reaction with SMCC [N-succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate]. MAbs were then coupled covalently to acetylcholinesterase (AChE) by the reaction of the thiol groups with the maleimide groups.
(iv) Evaluation of the best MAb pairs.
To select the best MAb pairs to develop two-site immunometric tests, a combinatorial analysis was carried out using each MAb as either a capture or a tracer MAb. Immobilization of the capture MAb in microtiter plates was performed overnight at RT after distributing 120 μl/well of the MAb at a concentration of 10 μg/ml in potassium phosphate buffer (0.05 M; pH 7.4). The plates were then emptied, saturated with 300 μl/well of EIA buffer, and kept at 4°C until use (within 1 year). Before use, saturated plates were washed once with washing buffer. Overnight cultures of Yersinia and other enterobacteria were adjusted to an optical density of 1 at 600 nm (corresponding to approximately 5 × 108 CFU/ml) in EIA buffer. Then, 50 μl/well of serial dilutions of bacteria and 50 μl/well of biotin-labeled MAb as a tracer (500 ng/ml) were distributed in duplicate in the microtiter plates coated with the various capture antibodies to be tested. EIA buffer was used as a negative control. After an ON incubation at 4°C and three washing cycles, 100 μl/well of AChE-labeled streptavidin (2 Ellman units/ml) was added for 2 h at RT. Plates were then washed 3 times before the addition of 200 μl/well of Ellman's reagent and measurement of the absorbance at 414 nm after 30 min at RT. For the following EIA, 100 μl of the bacterial sample and 100 μl of AChE-labeled MAb (2 or 5 EU/ml final concentration) were distributed in duplicate into the wells in order to obtain better sensitivity. Microtiter plates were optionally centrifuged for 5 min at 1,000 × g and incubated for 30 min or 3 h at RT. After three washing cycles, 200 μl/well of Ellman's reagent was added and the plates were kept at RT for 30 min before measurement of the absorbance at 414 nm was performed. Each measurement was independently performed three times. The five-parameter logistic fit (5-PL) function (GraphPad Prism 5) was used to fit the standard curve. The limit of detection (LoD) was calculated as the concentration providing a signal corresponding to the average background signal (as measured for 8 wells) plus 3 standard deviations.
Statistical analysis.
Statistical significance was assessed using an unpaired t test for comparisons between values of signals. Results were considered statistically significant when P values of <0.05 were obtained.
Western blot experiments.
Bacterial cultures of Yersinia were grown at 28°C to obtain similar immunogenicities of the in vivo lipopolysaccharide (LPS) (32). Cells were suspended in Laemmli buffer containing 2% SDS and separated into two samples. One sample was treated with proteinase K (80 μg/ml) during 30 min at 37°C. All samples were then denatured for 5 min at 95°C, and 107 to 108 CFU per well was subjected to SDS-PAGE for 1 h 30 min at 150 V in a 16% gel. Transfer onto a PVDF membrane was performed ON at RT at 25 V. For the saturation step, the membrane was blocked with Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST) with 5% skimmed dry milk during 30 min at RT. After two washes in TBST, specific MAbs (4 μg/ml in TBST containing 1% skimmed dry milk) were incubated for 30 min at RT with the membrane. After three washes in TBST, the membrane was incubated for 20 min at RT with HRP-labeled polyclonal goat anti-mouse immunoglobulins diluted to 1:5,000 in TBST containing 3% skimmed dry milk. After three washes in TBST and a brief wash in TBS, bands were detected by chemiluminescence.
Lateral flow immunoassays.
The colloidal-gold-labeled MAb was prepared by adding 25 μg of MAb to 1 ml of colloidal gold before mixing with 100 μl of 0.02 M borax buffer (pH 9.3) was performed. The reaction mixture was incubated for 1 h in the dark at RT. Then, 100 μl of 0.02 M borax buffer (pH 9.3) containing 1% BSA was added and the mixture was centrifuged at 15,000 × g for 50 min at 20°C. The supernatant was discarded, and the pellet was suspended in 250 μl of 0.002 M borax buffer (pH 9.3) containing 1% BSA, sonicated for a few seconds, and stored at 4°C in the dark until use. The strips (0.5 cm in width and 4.5 cm in length) were composed of 3 parts (33), (i) a sample pad (Standard 14; Whatman) (0.5 cm in length), (ii) a nitrocellulose membrane (Prima 40 or Hi-Flow Plus 75; Whatman or Millipore, respectively) (2.5 cm in length), and (iii) an absorption pad (Cellulose grade 470; Whatman) (1.5 cm in length), all attached to a backing card. The detection zone contained immobilized goat anti-mouse antibodies as a control line and an anti-Yersinia MAb as a test line (1 or 2 mg/ml in 0.05 M sodium phosphate buffer; pH 7.4) dispensed at 1 μl/cm using an automatic dispenser (Airjet XYZ 3050; BioDot). After drying for 1 h at 40°C in an air oven, the membrane was incubated with a blocking solution (PBS [pH 7.4] containing 0.5% BSA) for 30 min at RT. The membrane was washed twice with deionized water, incubated for 20 min at RT in a preserving solution (PBS containing 0.1% Tween 20 and 7.5% glucose), and then dried for 20 min at 40°C in an air oven. After the absorption pad and the sample pad were fixed to the top and the bottom of the membrane, respectively, the card was cut into strips 5 mm in width using an automatic programmable cutter (CM4000 Guillotine cutting system; BioDot). During the experiment, 100 μl of bacterial suspensions in analysis buffer (EIA containing 0.5% Tween 20) was mixed with 10 μl of colloidal-gold-labeled antibodies (20 μg/ml) in the wells of a 96-well microtiter plate. After 10 min of incubation of the mixture at RT, the strips were inserted into the wells. The capillary migration from the bottom of the sample pad to the absorption pad in the upper position lasted for about 30 min. The signal intensities of the test and control lines were visually estimated.
Detection in artificially spiked stool cultures.
A pea-size sample, corresponding to approximately 1 g of feces from a healthy individual, was diluted in 10 ml of EIA buffer for the enzyme immunoassay or in 10 ml of EIA buffer containing 0.5% Tween 20 (feces buffer) for the LFI. Overnight cultures of enteropathogenic Yersinia and other enterobacteria grown in LB broth at 28°C or 37°C were adjusted (based on optical density at 600 nm) to 5 × 107 CFU/ml in feces buffer and serially diluted in the same buffer either for EIA or for LFI just before analysis was performed using the conditions described above. The exact concentration of bacteria was then determined by enumeration of serial dilutions in EIA buffer.
RESULTS
Production and selection of anti-Yersinia MAbs.
After fusion of spleen cells with myeloma cells, hybridomas were screened by ELISA for the presence of specific antibodies directed against bioserotypes 2/O:9 and 4/O:3 of Y. enterocolitica and serotypes I and III of Y. pseudotuberculosis. As the aim of this study was to obtain antibodies specific for each of the prevailing species and bioserotypes of Yersinia strains, selection of antibodies was performed by differential-screening ELISA, using plates coated with different bacterial strains. A first selection of antibodies was achieved using a species/bioserotype screening (for example, Y. enterocolitica 2/0:9 was screened against Y. enterocolitica 4/O:3 and Y. pseudotuberculosis I and III) and a second one using a genus screening (against other Gram-negative enterobacteria: E. coli, Shigella sonnei, and Salmonella enterica serovar Typhimurium). These screenings led to the selection of 4 series of 15, 14, 14, and 17 specific MAbs against Y. enterocolitica 2/O:9, Y. enterocolitica 4/O:3, Y. pseudotuberculosis I, and Y. pseudotuberculosis III, respectively.
Combinatorial analyses were performed using biotinylated antibodies as tracer antibodies in a two-site immunometric format by testing (i) their specificities for the genus Yersinia compared to other enterobacteria (E. coli, S. sonnei, and S. enterica), (ii) their specificities in comparisons between the two enteropathogenic Yersinia species (Y. enterocolitica and Y. pseudotuberculosis), and (iii) their specificities in comparisons between the different bioserotypes of Y. enterocolitica strains (2/O:9 or 4/O:3 against 1A, 1B, and 2/O:5,27) and different serotypes of Y. pseudotuberculosis strains (I, III, and V). As shown in Table S1 in the supplemental material, among the 225 combinations tested, 56 detected Y. enterocolitica 2/O:9 but 8 appeared to lack specificity, with detection of E. coli also. For Y. enterocolitica 4/O:3 (as shown in Table S2 in the supplemental material), only a few (11 of 169) combinations were positive, all specific for the expected strain. As shown in Table S3 in the supplemental material, among the 196 Y. pseudotuberculosis I tests, 52 were shown to detect the strain but 13 exhibited cross-reactivity (CR) with unrelated strains such as other enterobacteria tested and Y. enterocolitica bioserotype 4/O:3. Finally, for Y. pseudotuberculosis III (shown in Table S4), most (265) of the 289 combinations strongly detected the expected strain and 16 of those were devoid of specificity, with nonspecific detection of the other enterobacteria tested. A second round of testing was performed for the positive and specific combinations by evaluating their sensitivity using serial dilutions of the related Yersinia strains (data not shown). For each MAb series (anti-Y. enterocolitica 2/O:9 anti-Y. enterocolitica 4/O:3, anti-Y. pseudotuberculosis I, and anti-Y. pseudotuberculosis III), the MAb tracers allowing the best sensitivity (Ye18*, Ye300*, Yps2*, and Yps104*) were chosen for further development.
Optimization of the EIA.
The 4 MAbs selected for tracer preparation were labeled with AChE to reduce the number of enzyme immunoassay (EIA) steps and to possibly improve the sensitivity of the tests. Combinatorial analyses using serial dilutions of the related Yersinia strains were performed to determine which capture MAbs provided the best sensitivity in association with these AChE-labeled MAbs. Four MAb pairs were finally retained: Ye4 and Ye18* for Y. enterocolitica 2/O:9, Ye300 and Ye300* for Y. enterocolitica 4/O:3, Yps2 and Yps2* for Y. pseudotuberculosis I, and Yps108 and Yps104* for Y. pseudotuberculosis III. The influence of different assay parameters (temperature, centrifugation step at the beginning of the incubation step, length of incubation) was evaluated, finally leading to an optimized assay that was performed for 3 h 30 min and included a centrifugation step compared to the initial 20-h format.
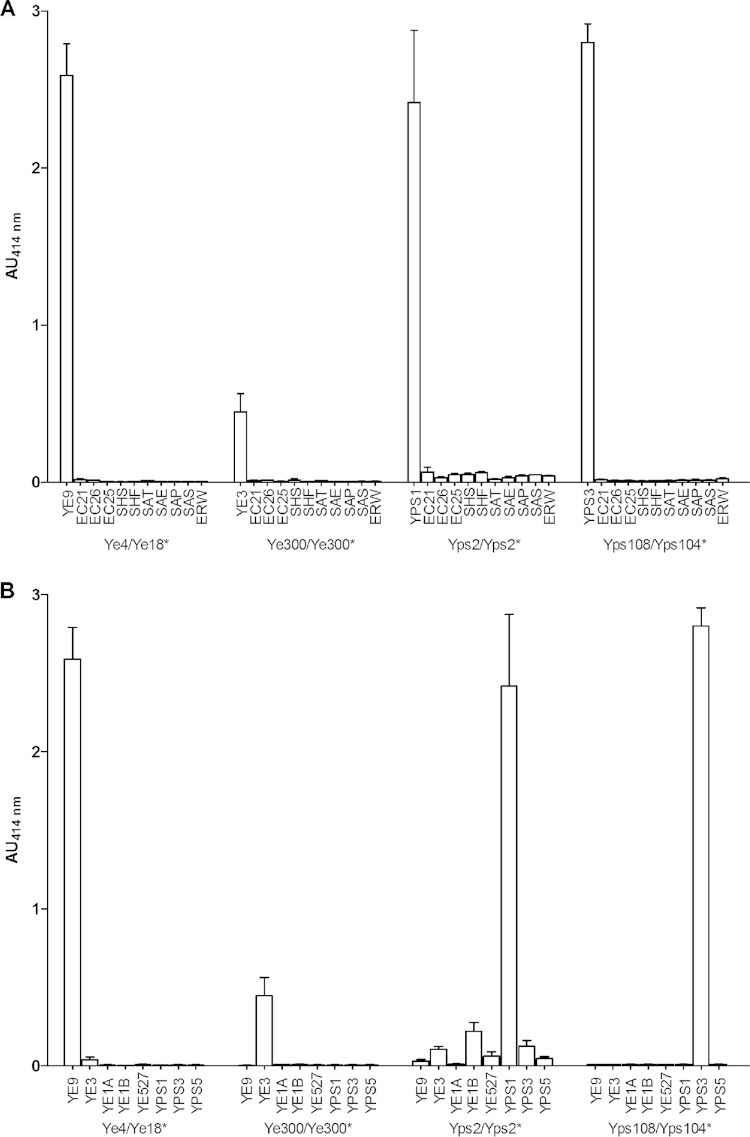
The specificity of the immunoassays was checked using a high concentration of enterobacteria (5 × 107 CFU/ml) cultured under their optimal growth conditions by comparing the results determined with their related Yersinia species to the results determined with other genera (Fig. 1A) and other species and bioserotype (Fig. 1B). As observed (Fig. 1), only the Y. pseudotuberculosis I EIA exhibited faint cross-reactivity (CR; calculated as the ratio of the absorbance unit values obtained for nonspecific and specific bacteria at the same concentration × 100) with unrelated E. coli strains (CR = <3%) and some Y. enterocolitica and Y. pseudotuberculosis strains (CR = <5%). The 3 other EIAs appeared to be totally specific, even though Y. enterocolitica 4/O:3 EIA provides a limited signal for this important concentration compared to the other EIAs, possibly indicating a difference of sensitivity. The optical signal was obtained for all MAb pairs selected.
FIG 1.
Specificity of Yersinia-optimized enzyme immunoassays. Yersinia enterobacteria were grown at 28°C for EIAs using Ye4/Ye18*, Ye300/Ye300*, and Yps108/Yps104* and at 37°C for an EIA using Yps2/Yps2* (the * indicates the tracer antibody). Other enterobacteria were grown at 37°C. Each strain was used at the concentration of 5 × 107 CFU/ml and was incubated during 3 h. Absorbance was measured at 414 nm after 30 min of incubation with Ellman's reagent. YE9, Y. enterocolitica 2/O:9; YE3, Y. enterocolitica 4/O:3; YE1A, Y. enterocolitica 1A; YE1B, Y. enterocolitica 1B; YE527, Y. enterocolitica O:5,27; YPS1, Y. pseudotuberculosis I; YPS3, Y. pseudotuberculosis III; YPS5, Y. pseudotuberculosis V; EC21, E. coli BL21; EC26, E. coli O26; EC25, E. coli O55, SHS, Shigella sonnei; SHF, Shigella flexneri; SAT, Salmonella enterica Typhimurium; SAE, Salmonella enterica Enteritidis; SAP, Salmonella enterica Paratyphi A; SAS, Salmonella enterica Senftenberg; ERW, Erwinia pyrifoliae. (A) Genus specificity of Yersinia compared to other enterobacteria. AU, absorbance units. (B) Species and biotype/serotype specificities compared to Y. enterocolitica strains (2/O:9, 4/O:3, 1A 1B, 2/O:5,27) and Y. pseudotuberculosis strains (I, III, V).
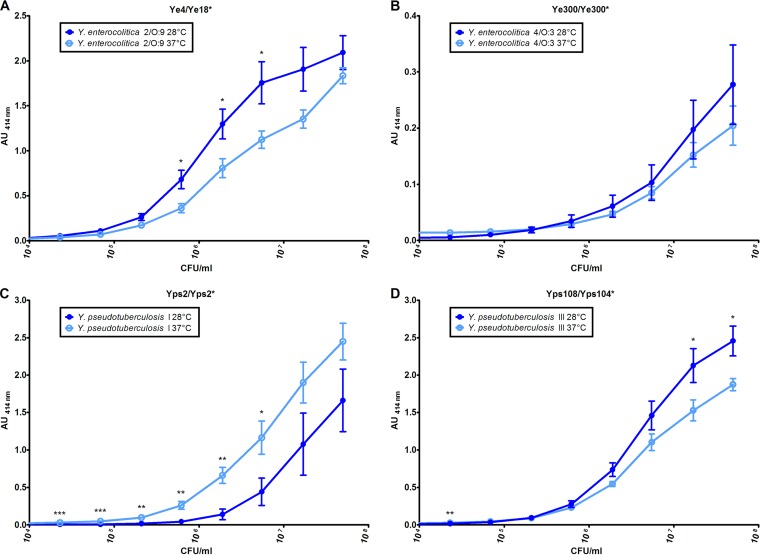
To evaluate both the sensitivity of these EIAs and the possible influence of the bacterial growth temperature, serial dilutions of the related Yersinia strain cultured at two temperatures (28°C and 37°C) were assayed. All the EIAs allowed sensitive detection in the 104 to 105 CFU/ml range. On the other hand, as expected, better signals were obtained with a growth temperature of 28°C for Y. enterocolitica 2/O:9 and Y. pseudotuberculosis III, while, surprisingly, 37°C appeared more favorable for Y. pseudotuberculosis I (Fig. 2). The signals were not significantly different for Y. enterocolitica 4/O:3 at the two temperatures. However, calculated LoD values were close for each of the targets, whatever the growth temperature, as illustrated in Table 3. It is worth noting that for the 37°C temperature setting, the optimized procedure provided an increase in sensitivity of at least 14-fold compared to the EIA performed under the initial conditions.
FIG 2.
Impact of the growth temperature on detection of Yersinia by optimized enzyme immunoassays. Yersinia species were grown at 28°C (dark-blue curve) and 37°C (light-blue curve), and 10-fold serial dilutions were performed before a 3-h incubation. (A) EIA Ye4/Ye18* for detection of Y. enterocolitica 2/O:9. (B) EIA Ye300/Ye300* for detection of Y. enterocolitica 4/O:3. (C) EIA using Yps2/Yps2* for detection of Y. pseudotuberculosis I. (D) EIA using Yps108/Yps104* for detection of Y. pseudotuberculosis III. Asterisks indicate values that are significantly different. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (for comparisons between signals).
TABLE 3.
Limits of detection of the enzyme immunoassays for Yersinia detection in EIA buffer or in artificially contaminated stool samplesa
| Strain | Limit of detection (CFU/ml) |
|||
|---|---|---|---|---|
| EIA buffer (n = 3) |
Stool samples (n = 3) |
|||
| 28°C | 37°C | 28°C | 37°C | |
| Y. enterocolitica 2/O:9 | 3.4 × 103 | 4.9 × 103 | 3.4 × 104 | 1.3 × 104 |
| Y. enterocolitica 4/O:3 | 7.8 × 104 | 1.4 × 105 | 5.2 × 104 | 7.5 × 104 |
| Y. pseudotuberculosis I | 2.0 × 105 | 1.2 × 103 | 5.4 × 106 | 2.6 × 105 |
| Y. pseudotuberculosis III | 1.9 × 104 | 1.1 × 104 | 4.7 × 104 | 3.3 × 104 |
Yersinia enterobacteria were grown at 28°C and 37°C.
Enzyme immunoassay with artificially contaminated stool cultures.
To validate these new EIAs, artificially contaminated stool samples were tested. The genus, the species, and the bioserotype specificities were checked as described above (see “Optimization of the EIA” above) using various Yersinia strains (Y. enterocolitica 2/O:9, 4/O:3, 1A 1B, and 2/O:5,27 and Y. pseudotuberculosis I, III, and V) and other enterobacteria (E. coli BL21, O26, and O55, S. sonnei, S. flexneri, S. enterica serovars Typhimurium, Enteritidis, Paratyphi A, and Senftenberg, and Erwinia pyrifoliae). As previously obtained with bacteria in simple EIA buffer, a specific signal was recovered for Y. enterocolitica 2/O:9, Y. enterocolitica 4/O:3, Y. pseudotuberculosis I, and Y. pseudotuberculosis III with MAb pairs Ye4 and Ye18*, Ye300 and Ye300*, Yps2 and Yps2*, and Yps108 and Yps104*, respectively.
Analyzing the signal intensity and the LoDs, the same influence of growth temperature was observed for the stool samples as was observed previously with samples in EIA buffer. As shown in Table 3, no clear change in sensitivity was induced by the stool medium, thus indicating the absence of an important matrix effect for the EIAs, except for the Y. enterocolitica 2/O:9 and Y. pseudotuberculosis I EIAs, for which the LoDs increased 20-fold and 35-fold, respectively, when bacteria were grown at 37°C.
Analysis of the nature of the antigens recognized by the antibodies by the use of Western blotting.
In order to determine if the recognized antigens were proteins, samples were treated or not with proteinase K prior to Western blot analysis (Fig. 3). The immunoblot analysis showed that only MAb Yps2 on the Y. pseudotuberculosis I extract recognized a band of approximately 70 kDa that was sensitive to proteinase K treatment, indicating that the MAb Yps2 bound protein epitope (lanes 7 and 8). For the other MAbs, Ye4 and Ye18 on Y. enterocolitica 2/O:9 extract and Ye300 on Y. enterocolitica 4/O:3 extract, proteinase K treatment did not induce any clear modification of the recognition results. For MAbs Yps104 and Yps108 on Y. pseudotuberculosis III extract, only the high molecular band around 100 kDa appeared partially sensitive to proteinase K treatment, with a signal diminution and a shift of the molecular mass to approximately 80 kDa (lanes 9 to 12). Moreover, a typical pattern profile of long-chain LPS without a ladder was observed on the immunoblot analysis using MAbs Ye4, Ye18, and Ye300 (lanes 1 to 6) (34). For MAbs Yps104 and Yps108, another typical pattern was observed, with signal for the core LPS and less signal on the long-chain LPS (lanes 9 to 12) (35). Therefore, all these samples presented a characteristic smear pattern profile of LPSs, indicating that these MAbs presumably interacted with LPSs of their related Yersinia strains.
FIG 3.
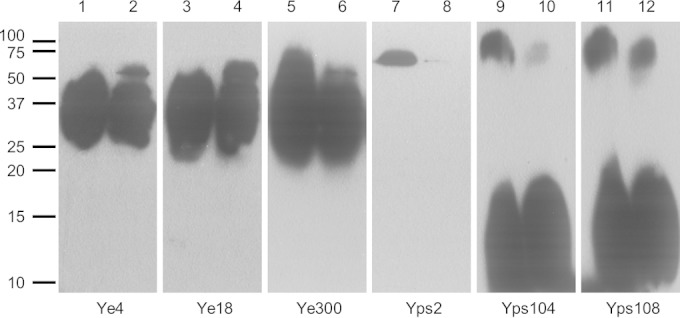
Recognition of protein and LPS epitopes with anti-Yersinia MAbs. Western blotting was performed with Ye4 (lanes 1 and 2) and Ye18 (lanes 3 and 4) against Y. enterocolitica 2/O:9 (107 CFU/well), with Ye300 against Y. enterocolitica 4/O:3 (107 CFU/well) (lanes 5 and 6), with Yps2 against Y. pseudotuberculosis I (107 CFU/well) (lanes 7 and 8), and with Yps 104 (lanes 9 and 10) and Yps108 (lanes 11 and 12) against Y. pseudotuberculosis III (108 CFU/well). Bacterial samples in lanes 2, 4, 6, 8, 10, and 12 were treated with proteinase K. Numbers on the left indicate the molecular mass markers (in kDa).
Development of LFIs.
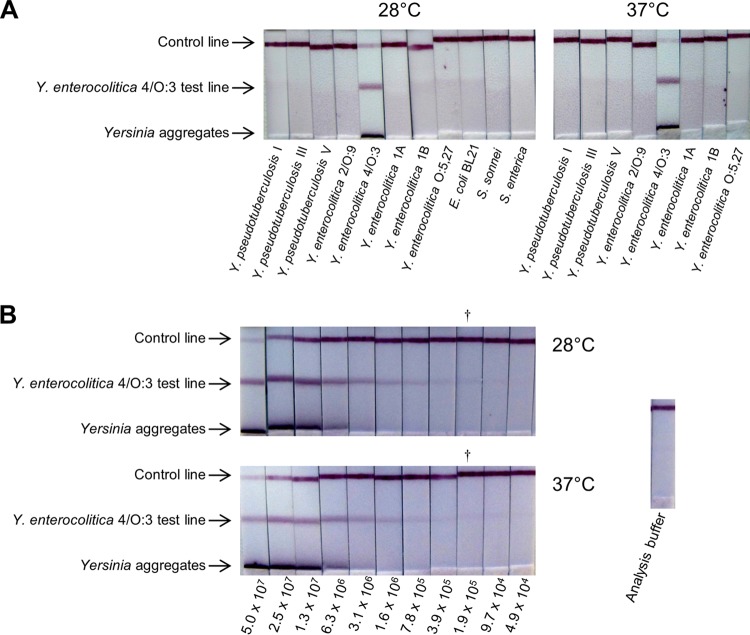
To set up an even faster and easier handling test, i.e., a lateral flow immunoassay (LFI), all MAb pairs displaying a specific signal for enteropathogenic Yersinia detection in the previous immunoenzymatic assay were reevaluated during the LFI development. As the format of the test can influence the specificity, we first checked the genus/species/bioserotype specificities, using 5 × 107 CFU/ml (5 × 106 CFU/dipstick) of various Yersinia and other enterobacterial species (E. coli BL21, O26, and O55, S. sonnei, S. flexneri, S. enterica serovars Typhimurium, Enteritidis, Paratyphi A, and Senftenberg, and Erwinia pyrifoliae). Then, the sensitivity of the selected specific LFI was evaluated using serial dilutions of the related Yersinia strains. The best MAb combinations for detecting Y. enterocolitica 2/O:9 and Y. enterocolitica 4/O:3 were the Ye4 and Ye18* pair (2/O:9 LFI) and the Ye300 and Ye300* pair (4/O:3 LFI), respectively. MAb pair Yp7 and Yp7* recognizing serotypes I, II, IV, and V of Y. pseudotuberculosis and MAb pair Yps103 and Yps104* recognizing serotype III were combined to achieve a single LFI allowing Y. pseudotuberculosis detection for serotypes I, II, III, IV, and V (Yps LFI). Since the results were similar for all the different LFIs, the results obtained for the 4/O:3 LFI are presented as an illustration (Fig. 4). The different LFIs proved to be as specific as the optimized EIA, and no cross-reactivity was observed with enterobacteria other than the related enteropathogenic Yersinia strains (see Fig. 4A for Ye4/O:3 LFI). Although Y. enterocolitica O:9 LPS exhibits a strong similarity to Brucella LPS (35), no cross-reactivity was observed for the Y. enterocolitica 2/O:9 LFI with Brucella strains (B. abortus and B. melitensis). The Ye4/O:3 LFI and Yps LFI were also tested with Brucella strains and provided the same negative result.
FIG 4.
Ye4O/3 lateral flow immunoassay. Bacteria were grown at 28°C and 37°C. The bacterial suspensions were incubated for 10 min with colloidal gold-labeled Ye300* MAb, and the Ye300 dipsticks were then dipped for 30 min into 100 μl of the bacterial suspensions for upward migration of the liquid. (A) Specificities of the Ye4/O:3 LFI. Each strain was used at the concentration of 5 × 107 CFU/ml. (B) Sensitivities of the Ye4/O:3 LFI for serial dilution of Y. enterocolitica 4/O:3. Numbers below the dipsticks indicate the number of CFU/ml. †, the last dipstick with a visible signal estimated by eye.
The sensitivities obtained for the different LFIs were 5 × 105 CFU/ml and 105 CFU/ml for detection of Y. enterocolitica 2/O:9 and Y. enterocolitica 4/O:3, respectively (see Fig. 4B for Ye4/O:3 LFI). The LoDs of the Yps LFI for Y. pseudotuberculosis detection were close to 105 CFU/ml for serotypes I, 5 × 105 CFU/ml for serotypes III and IV, and 106 CFU/ml for serotypes II and V. As expected, the LFIs appeared less sensitive than optimized EIA but no difference of sensitivity between the Yersinia strains grown at 28°C and those grown at 37°C was observed. It is worth noting that for all LFIs, a specific band was detected for high bacterial concentrations, in the lower part of the membrane, resulting from the presence of bacterial aggregates that were still recognized by the tracer MAbs but were too large to migrate along the membrane. At lower bacterial concentrations, the intensity of this band decreased (Fig. 4B).
All LFI were then tested with a large number of strains, including related Yersinia strains along with strains of unrelated Yersinia and other bacteria. All the related strains were tested at a concentration of 10× the LoD, and unrelated bacteria were tested at concentrations between 5 × 106 and 5 × 107 CFU/ml. For the 2/O:9 LFI, all 13 of the tested strains from bioserotype 2/O:9 were detected at 10× the LoD (5 × 106 CFU/ml), while there was no detection of 14 tested strains from biotype 4/O:3 (Table 1). For the 4/O:3 LFI, all 14 of the tested strains from bioserotype 4/O:3 proved to give positive results at 106 CFU/ml (10× the LoD), in contrast to the 13 tested strains from bioserotype 2/O:9 (Table 1). Except for Y. enterocolitica 1A/O:3, none of the other unrelated bacteria were recognized with the 4/O:3 LFI (Table 1). The biotype and serotype of this 1A/O:3 strain were verified before and after the experiment to confirm its characteristics. For Y. pseudotuberculosis, the Yps LFI detected 7 of 32 tested strains of serotype I at 106 CFU/ml (10× the LoD), and this number increased to 20 of 32 for a 107 CFU/ml concentration (Table 2). All the strains of serotypes II (10 strains), III (11 strains), IV (10 strains), and V (11 strains) were detected. Moreover, none of the other unrelated bacteria, including Y. enterocolitica, Yersinia similis, and Yersinia wautersii, were detected by the Yps LFI.
A multiplex LFI involving all the MAbs previously selected was designed with the aim to provide simultaneous detection of enteropathogenic Yersinia strains (Fig. 5). The sensitivity obtained with the multiplex LFI for the different bacterial strains proved to be similar to those described with the single LFIs.
FIG 5.
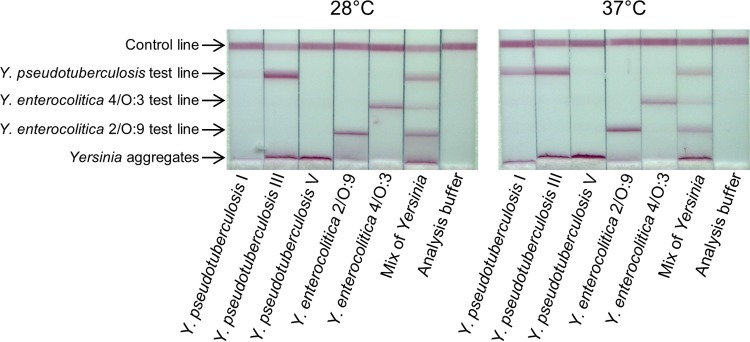
Multiplex lateral flow immunoassay. Bacteria were grown at 28°C and used at 10 times the limit of detection. The bacterial suspensions were incubated for 10 min with a mix of colloidal gold-labeled Ye18*, Ye300*, Yps 104*, and Yp7* MAbs, and the LFI dipsticks were then dipped for 30 min into 100 μl of the bacterial suspensions for upward migration of the liquid. The Ye4 and Ye18 MAbs were used for the test line of Y. enterocolitica 2/O:9 and Y. enterocolitica 4/O:3, respectively. A mix of Yps103 and Yp7 was used for the test line of Y. pseudotuberculosis.
Lateral flow immunoassays with artificially contaminated stool cultures.
As was performed for the EIA, LFIs were tested with the different bacteria spiked in feces and diluted in buffer before the analysis. The specificities and sensitivities of the different LFIs in feces were very similar to the results obtained in buffer. The LoD of the 2/O:9 LFI and 4/O:3 LFI was 5 × 105 CFU/ml for detection of Y. enterocolitica 2/O:9 and Y. enterocolitica 4/O:3, respectively. The LoDs of the Yps LFI for Y. pseudotuberculosis detection were close to 105 CFU/ml for serotype I, 5 × 105 CFU/ml for serotype III, and 106 CFU/ml for serotype V. Serotypes II and IV were not tested with artificially contaminated stool cultures. Before the LFI, bacterial samples in feces buffer were diluted 1:2 in EIA buffer containing 0.5% Tween 20.
DISCUSSION
The incidence of enteric yersiniosis caused by Y. enterocolitica and Y. pseudotuberculosis enteropathogenic bacteria is probably largely underestimated all around the world. One of the main reasons for this underestimation is the difficulty of recovery of Yersinia strains from bacterial flora present in stool samples. Because of their optimal growth temperature of 28°C, the conditions of culture suitable for most enterobacteria performed using nonselective medium at 37°C for 24 h do not allow optimal detection of Yersinia strains. This detection can be improved using enrichment procedures, but this requires additional time, money, and work. Even when some Yersinia-selective media were developed, allowing improvement of the isolation of Y. enterocolitica strains, the growth of Y. pseudotuberculosis was inhibited or not tested (36–39). Moreover, these selective media are not yet marketed and consequently cannot be routinely used by clinical laboratories. Nowadays, CIN can be used for isolation of Yersinia species but only at their optimal growth temperature of 28°C (40), thus requiring an additional device(s) for a clinical laboratory. Although CIN is marketed, this medium is not consistently used because of its additional cost. Moreover, CIN inhibits the growth of some Y. pseudotuberculosis strains while allowing the growth of nonpathogenic Yersinia. Given the lack of rapid and simple methods for detection of enteropathogenic Yersinia in human samples, the aim of this study was to develop two sensitive and specific immunoassay formats for the detection of the most frequent bioserotypes of Y. enterocolitica and Y. pseudotuberculosis.
We first produced a large set of MAbs against Y. enterocolitica and Y. pseudotuberculosis and screened them for their specificity and sensitivity. This selection was achieved for specific detection of each of the four prevailing isolated strains: Y. enterocolitica bioserotypes 2/O:9 and 4/O:3 and Y. pseudotuberculosis serotypes I and III. These MAbs led to the development of a one-step, 3-h-30-min EIA with excellent sensitivity and specificity and of a low-cost and user-friendly LFI with good sensitivity and specificity.
Compared to methods of isolation and characterization protocol involving CIN, these two immunoassays offer a great improvement with respect to the duration of Yersinia detection. Indeed, the LFI and the EIA allow characterizations of enteropathogenic Yersinia in 40 min and 3 h 30 min, respectively, while the CIN requires 48 h for isolation of bacterial colonies without further characterization. Moreover, LFI and EIA do not require complex sample preparation and are thus well suited for clinical laboratory applications. It has been shown that CIN is permissive for the growth of some bacterial species found in feces samples such as Acinetobacter, Aeromonas, Citrobacter, Enterobacter, Morganella, Pseudomonas, and Serratia (40, 41). Usually, the morphologies of the colonies of Y. enterocolitica and other bacteria can be distinguished, but colonies of Citrobacter freundii, Enterobacter agglomerans, and Serratia liquefaciens look similar to those of Y. enterocolitica, leading to possible misidentifications. Regarding the specificity of our immunoassays, none of the enteropathogenic bacteria such as S. sonnei and S. enterica serovars Typhimurium, Enteritidis, Paratyphi A, and Senftenberg usually searched for in stools of patients were detected. Moreover, the tests are robust enough to retain their sensitivity in complex matrices such as feces.
We developed a highly sensitive EIA with LoDs ranging from 1.2 × 103 to 2.0 × 105 CFU/ml for Y. enterocolitica bioserotypes 2/O:9 and 4/O:3 and Y. pseudotuberculosis serotypes I and III. For the Y. enterocolitica 2/O:9, Y. enterocolitica 4/O:3, and Y. pseudotuberculosis III EIAs, better signals were obtained when bacteria were grown at 28°C rather than 37°C. This result can be explained by the fact that the MAbs used in these EIA recognize LPS, production of which is higher at 28°C in vitro (42). For the Y. pseudotuberculosis serotype I EIA, the signal and the LoD were better for bacteria grown at 37°C. Since the MAb used in this EIA was targeting a protein, this higher temperature possibly enhanced its expression at 37°C in vitro. When stool samples were used to mimic clinical samples, the LoDs were slightly modified or left unmodified, except for the LoDs of the Y. enterocolitica 2/O:9 and Y. pseudotuberculosis I EIAs, which were 20 to 35 times higher when the bacteria were grown at 37°C.
Validation of LFI with a large number of bacteria showed 100% detection for Y. enterocolitica (bioserotypes 2/O:9 and 4/O:3) and for Y. pseudotuberculosis (serotypes II, III, IV, and V). For serotype I, 62.5% of the strains were detected at 107 CFU/ml. It is worth noting that CIN has been shown to exert an inhibitory effect on the growth of some Y. pseudotuberculosis strains (17); thus, our immunoassays could lead to better detection of this species from biological samples.
It has been previously reported that colonies of Yersinia exhibit similar aspects after isolation on CIN (17), making it impossible to distinguish between pathogenic and nonpathogenic strains. We thus tested a large number of nonpathogenic strains of Y. enterocolitica biotype 1A, which is ubiquitous in the environment and is the predominant biotype of Y. enterocolitica detected among Yersinia isolates from human clinical stool samples in some European countries (43, 44). We observed that only a single strain among 11 was detected with the 4/O:3 LFI. It appeared that this positive strain was of serotype O:3, which possesses the same LPS as Y. enterocolitica 4/O:3. This cross-reactivity can be explained by the epitope recognized by MAb Ye300, which involved the LPS. It is noteworthy that recovery of bioserotype 1A/O:3 from clinical samples is quite unusual (45), and only 4 strains from this exceptional bioserotype were present among 2,722 in the collection of the French National Reference Laboratory.
Although serotype O:9 shares epitopes with the O-antigen of Brucella (35), none of the three Brucella strains tested was detected by the 2/O:9 LFI, and the other LFIs gave similarly negative results. Our Yps LFI was also tested with Y. similis and Y. wautersii, two species that were recently described and are closely related to Y. pseudotuberculosis and (in the case of Y. wautersii) presumably pathogenic for human (46, 47), and gave negative results.
The LoDs of the LFIs ranged from 105 to 106 CFU/ml for detection of the four prevailing isolated strains: Y. enterocolitica bioserotypes 2/O:9 and 4/O:3 and Y. pseudotuberculosis serotypes I and III. Moreover, our Yps LFI was able to detect Y. pseudotuberculosis serotypes II, IV, and V. Thanks to the high LFI robustness, the corresponding LoDs were identical with those determined for the artificially contaminated stool samples.
To our knowledge, only one study has previously reported that the loads of Y. enterocolitica in stools of infected patients range from 105 to 108 CFU/g of feces (22). However, no experiment was done to support this affirmation. Using a procedure common for clinical laboratories, i.e., one pea-size sample of feces (corresponding to approximately 1 g, resuspended in 10 ml of buffer), the present EIA should clearly be useful, presenting a LoD value below that range. If this load of Y. enterocolitica in stools is correct, the LoDs of LFIs are above the lowest limit of 105 CFU/g of feces, but the tests should work properly in a large part of the concentration range expected in biological samples. Because we used stool samples from a healthy individual, dilution was necessary to obtain liquid samples. It could be interesting to determine if patient stool samples without or with a lower dilution can improve these LoDs. Compared to more-selective methods such as the use of CIN or real-time PCR with a LoD of ≥103 CFU/g of feces, our EIAs and LFIs provide very fast and economical detection and can be used by minimally trained personnel without additional devices being required. Moreover, our LFIs would be a useful tool to determine the exact load of enteropathogenic Yersinia in stools of diarrheal patients. Real-time PCR needs an enrichment step that is as long in duration as the isolation step on CIN and also requires the acquisition of expensive devices. The time saving of the approach described here can easily circumvent unadapted antibiotic treatments or unnecessary appendectomies.
Furthermore, it appears that the LFI can be easily handled under field conditions and consequently can be used for veterinary applications. First, serotype O:9 of Y. enterocolitica is often naturally isolated from cows, goats, sheep, or pigs but their antibodies against serotype O:9 cross-react with Brucella O-antigen and cause false-positive reactions in brucellosis serological diagnosis tests (48, 49). This represents a major economic problem for stock farmers because suspicion of brucellosis leads to the elimination of the animals in accordance with public health regulations. Our LFI can be used to discriminate between a Y. enterocolitica infection and a Brucella infection. Second, bioserotype 4/O:3 of Y. enterocolitica is frequently isolated from pigs on farms or in slaughterhouses, and consumption of pork meat is associated with Y. enterocolitica infections (50, 51). Our LFI can be used by the meat industry at different levels: on livestock, to control the absence of Y. enterocolitica and prevent contamination from positive herds, and in slaughterhouses, to exclude contaminated pig carcasses or for epidemiological studies. Additionally, infection by Y. pseudotuberculosis appears to be a recurrent veterinary issue with a significant economic burden in livestock and zoo animals (52). Our Yps LFI can be easily handled under field conditions and would be useful as a control test for the presence of the pathogen.
In conclusion, this report presents the first description of a very sensitive EIA and of a rapid LFI test suitable for detection of enteropathogenic Yersinia in stool samples which can be helpful to physicians for the diagnosis of patients. The EIA or the LFI would be useful as a first-line rapid test in clinical laboratories. They can be a great complementary tool to help clinical laboratories to focus the research of enteropathogenic Yersinia on positive stools before performing the gold standard test for confirmation, i.e., isolation of Yersinia with traditional culture methods. Therefore, we need to evaluate our immunoassays with pathogenic stool samples under the conditions encountered in clinical laboratories. Furthermore, both immunoassays, especially the easy-to-use LFI, have the potential to be used for veterinary applications for detection of Y. enterocolitica and Y. pseudotuberculosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marie-Claire Nevers, Audrey Rouaix, and Marc Plaisance for their technical support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02137-14.
REFERENCES
- 1.Galindo CL, Rosenzweig JA, Kirtley ML, Chopra AK. 2011. Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in human yersiniosis. J Pathog 2011:182051. doi: 10.4061/2011/182051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosner BM, Werber D, Hohle M, Stark K. 2013. Clinical aspects and self-reported symptoms of sequelae of Yersinia enterocolitica infections in a population-based study, Germany 2009–2010. BMC Infect Dis 13:236. doi: 10.1186/1471-2334-13-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukushima H, Matsuda Y, Seki R, Tsubokura M, Takeda N, Shubin FN, Paik IK, Zheng XB. 2001. Geographical heterogeneity between Far Eastern and Western countries in prevalence of the virulence plasmid, the superantigen Yersinia pseudotuberculosis-derived mitogen, and the high-pathogenicity island among Yersinia pseudotuberculosis strains. J Clin Microbiol 39:3541–3547. doi: 10.1128/JCM.39.10.3541-3547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perdikogianni C, Galanakis E, Michalakis M, Giannoussi E, Maraki S, Tselentis Y, Charissis G. 2006. Yersinia enterocolitica infection mimicking surgical conditions. Pediatr Surg Int 22:589–592. doi: 10.1007/s00383-006-1703-y. [DOI] [PubMed] [Google Scholar]
- 5.Kaasch AJ, Dinter J, Goeser T, Plum G, Seifert H. 2012. Yersinia pseudotuberculosis bloodstream infection and septic arthritis: case report and review of the literature. Infection 40:185–190. doi: 10.1007/s15010-011-0160-2. [DOI] [PubMed] [Google Scholar]
- 6.Yotsu R, Mii S, Hayashi R, Harada H, Furukawa K, Eto H. 2010. Erythema nodosum associated with Yersinia enterocolitica infection. J Dermatol 37:819–822. doi: 10.1111/j.1346-8138.2010.00892.x. [DOI] [PubMed] [Google Scholar]
- 7.Guinet F, Carniel E, Leclercq A. 2011. Transfusion-transmitted Yersinia enterocolitica sepsis. Clin Infect Dis 53:583–591. doi: 10.1093/cid/cir452. [DOI] [PubMed] [Google Scholar]
- 8.Fredriksson-Ahomaa M, Korkeala H. 2003. Low occurrence of pathogenic Yersinia enterocolitica in clinical, food, and environmental samples: a methodological problem. Clin Microbiol Rev 16:220–229. doi: 10.1128/CMR.16.2.220-229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottone EJ. 1997. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev 10:257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savin C, Leclercq A, Laurent E, Carniel E, Vaillant V. 2010. Enquête nationale sur le diagnostic des infections à Yersinia entéropathogènes en France métropolitaine en 2003. Bull Epid Hebd 29:307–311. http://www.invs.sante.fr/beh/2010/29/. [Google Scholar]
- 11.European Food Safety Authority European Centre for Disease Prevention and Control. 2011. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. European Food Safety Authority European Centre for Disease Prevention and Control, Parma, Italy. [Google Scholar]
- 12.Long C, Jones TF, Vugia DJ, Scheftel J, Strockbine N, Ryan P, Shiferaw B, Tauxe RV, Gould LH. 2010. Yersinia pseudotuberculosis and Y. enterocolitica infections, FoodNet, 1996–2007. Emerg Infect Dis 16:566–567. doi: 10.3201/eid1603.091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato K, Ouchi K, Taki M. 1983. Yersinia pseudotuberculosis infection in children, resembling Izumi fever and Kawasaki syndrome. Pediatr Infect Dis 2:123–126. doi: 10.1097/00006454-198303000-00011. [DOI] [PubMed] [Google Scholar]
- 14.Eppinger M, Rosovitz MJ, Fricke WF, Rasko DA, Kokorina G, Fayolle C, Lindler LE, Carniel E, Ravel J. 2007. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet 3:e142. doi: 10.1371/journal.pgen.0030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jalava K, Hallanvuo S, Nakari UM, Ruutu P, Kela E, Heinasmaki T, Siitonen A, Nuorti JP. 2004. Multiple outbreaks of Yersinia pseudotuberculosis infections in Finland. J Clin Microbiol 42:2789–2791. doi: 10.1128/JCM.42.6.2789-2791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent P, Leclercq A, Martin L, Yersinia Surveillance Network, Duez JM, Simonet M, Carniel E. 2008. Sudden onset of pseudotuberculosis in humans, France, 2004-05. Emerg Infect Dis 14:1119–1122. doi: 10.3201/eid1407.071339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savin C, Leclercq A, Carniel E. 2012. Evaluation of a single procedure allowing the isolation of enteropathogenic Yersinia along with other bacterial enteropathogens from human stools. PLoS One 7:e41176. doi: 10.1371/journal.pone.0041176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wauters G, Kandolo K, Janssens M. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol 9:14–21. [PubMed] [Google Scholar]
- 19.Wauters G, Aleksic S, Charlier J, Schulze G. 1991. Somatic and flagellar antigens of Yersinia enterocolitica and related species. Contrib Microbiol Immunol 12:239–243. [PubMed] [Google Scholar]
- 20.Garzetti D, Susen R, Fruth A, Tietze E, Heesemann J, Rakin A. 2014. A molecular scheme for Yersinia enterocolitica patho-serotyping derived from genome-wide analysis. Int J Med Microbiol 304:275–283. doi: 10.1016/j.ijmm.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Lantz PG, Knutsson R, Blixt Y, Al Soud WA, Borch E, Rådström P. 1998. Detection of pathogenic Yersinia enterocolitica in enrichment media and pork by a multiplex PCR: a study of sample preparation and PCR-inhibitory components. Int J Food Microbiol 45:93–105. doi: 10.1016/S0168-1605(98)00152-4. [DOI] [PubMed] [Google Scholar]
- 22.Zheng H, Sun Y, Lin S, Mao Z, Jiang B. 2008. Yersinia enterocolitica infection in diarrheal patients. Eur J Clin Microbiol Infect Dis 27:741–752. doi: 10.1007/s10096-008-0562-y. [DOI] [PubMed] [Google Scholar]
- 23.Chanteau S, Rahalison L, Ratsitorahina M, Mahafaly Rasolomaharo M, Boisier P, O'Brien T, Aldrich J, Keleher A, Morgan C, Burans J. 2000. Early diagnosis of bubonic plague using F1 antigen capture ELISA assay and rapid immunogold dipstick. Int J Med Microbiol 290:279–283. doi: 10.1016/S1438-4221(00)80126-5. [DOI] [PubMed] [Google Scholar]
- 24.Simon S, Demeure C, Lamourette P, Filali S, Plaisance M, Creminon C, Volland H, Carniel E. 2013. Fast and simple detection of Yersinia pestis applicable to field investigation of plague foci. PLoS One 8:e54947. doi: 10.1371/journal.pone.0054947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nato F, Boutonnier A, Rajerison M, Grosjean P, Dartevelle S, Guénolé A, Bhuiyan NA, Sack DA, Nair GB, Fournier JM, Chanteau S. 2003. One-step immunochromatographic dipstick tests for rapid detection of Vibrio cholerae O1 and O139 in stool samples. Clin Diagn Lab Immunol 10:476–478. doi: 10.1128/CDLI.10.3.476-478.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nato F, Phalipon A, Nguyen TL, Diep TT, Sansonetti P, Germani Y. 2007. Dipstick for rapid diagnosis of Shigella flexneri 2a in stool. PLoS One 2:e361. doi: 10.1371/journal.pone.0000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taneja N, Nato F, Dartevelle S, Sire JM, Garin B, Thi Phuong LN, Diep TT, Shako JC, Bimet F, Filliol I, Muyembe JJ, Ungeheuer MN, Ottone C, Sansonetti P, Germani Y. 2011. Dipstick test for rapid diagnosis of Shigella dysenteriae 1 in bacterial cultures and its potential use on stool samples. PLoS One 6:e24830. doi: 10.1371/journal.pone.0024830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grassi J, Frobert Y, Lamourette P, Lagoutte B. 1988. Screening of monoclonal antibodies using antigens labeled with acetylcholinesterase: application to the peripheral proteins of photosystem 1. Anal Biochem 168:436–450. doi: 10.1016/0003-2697(88)90341-7. [DOI] [PubMed] [Google Scholar]
- 29.Pradelles P, Grassi J, Maclouf J. 1985. Enzyme immunoassays of eicosanoids using acetylcholine esterase as label: an alternative to radioimmunoassay. Anal Chem 57:1170–1173. doi: 10.1021/ac00284a003. [DOI] [PubMed] [Google Scholar]
- 30.Ellman GL, Courtney KD, Andres V Jr, Feather-Stone RM. 1961. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol 7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 31.Grassi J, Frobert Y, Pradelles P, Chercuitte F, Gruaz D, Dayer JM, Poubelle PE. 1989. Production of monoclonal antibodies against interleukin-1 alpha and -1 beta. Development of two enzyme immunometric assays (EIA) using acetylcholinesterase and their application to biological media. J Immunol Methods 123:193–210. [DOI] [PubMed] [Google Scholar]
- 32.Kawaoka Y, Otsuki K, Tsubokura M. 1983. Serological evidence that Yersinia enterocolitica lipopolysaccharide produced during growth in vivo resembles that produced during growth in vitro at 25 degrees C. J Gen Microbiol 129:2749–2751. [DOI] [PubMed] [Google Scholar]
- 33.Khreich N, Lamourette P, Boutal H, Devilliers K, Creminon C, Volland H. 2008. Detection of Staphylococcus enterotoxin B using fluorescent immunoliposomes as label for immunochromatographic testing. Anal Biochem 377:182–188. doi: 10.1016/j.ab.2008.02.032. [DOI] [PubMed] [Google Scholar]
- 34.Chart H, Rowe B. 1991. Purification of lipopolysaccharide from strains of Yersinia enterocolitica belonging to serogroups 03 and 09. FEMS Microbiol Lett 61:341–345. [DOI] [PubMed] [Google Scholar]
- 35.Chart H, Cheasty T. 2006. The serodiagnosis of human infections with Yersinia enterocolitica and Yersinia pseudotuberculosis. FEMS Immunol Med Microbiol 47:391–397. doi: 10.1111/j.1574-695X.2006.00100.x. [DOI] [PubMed] [Google Scholar]
- 36.Blom M, Meyer A, Gerner-Smidt P, Gaarslev K, Espersen F. 1999. Evaluation of Statens Serum Institut enteric medium for detection of enteric pathogens. J Clin Microbiol 37:2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Zutter L, Le Mort L, Janssens M, Wauters G. 1994. Short-comings of irgasan ticarcillin chlorate broth for the enrichment of Yersinia enterocolitica biotype 2, serotype 9 from meat. Int J Food Microbiol 23:231–237. doi: 10.1016/0168-1605(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 38.Van Noyen R, Vandepitte J, Wauters G. 1980. Nonvalue of cold enrichment of stools for isolation of Yersinia enterocolitica serotypes 3 and 9 from patients. J Clin Microbiol 11:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wauters G, Goossens V, Janssens M, Vandepitte J. 1988. New enrichment method for isolation of pathogenic Yersinia enterocolitica serogroup O:3 from pork. Appl Environ Microbiol 54:851–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiemann DA. 1979. Synthesis of a selective agar medium for Yersinia enterocolitica. Can J Microbiol 25:1298–1304. doi: 10.1139/m79-205. [DOI] [PubMed] [Google Scholar]
- 41.Head CB, Whitty DA, Ratnam S. 1982. Comparative study of selective media for recovery of Yersinia enterocolitica. J Clin Microbiol 16:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skurnik M, Bengoechea JA. 2003. Biosynthesis and biological role of lipopolysaccharide O-antigens of pathogenic Yersiniae. Carbohydr Res 338:2521–2529. doi: 10.1016/S0008-6215(03)00305-7. [DOI] [PubMed] [Google Scholar]
- 43.Sihvonen LM, Haukka K, Kuusi M, Virtanen MJ, Siitonen A, YE study group . 2009. Yersinia enterocolitica and Y. enterocolitica-like species in clinical stool specimens of humans: identification and prevalence of bio/serotypes in Finland. Eur J Clin Microbiol Infect Dis 28:757–765. doi: 10.1007/s10096-008-0696-y. [DOI] [PubMed] [Google Scholar]
- 44.Fredriksson-Ahomaa M, Cernela N, Hächler H, Stephan R. 2012. Yersinia enterocolitica strains associated with human infections in Switzerland 2001–2010. Eur J Clin Microbiol Infect Dis 31:1543–1550. doi: 10.1007/s10096-011-1476-7. [DOI] [PubMed] [Google Scholar]
- 45.Sihvonen LM, Jalkanen K, Huovinen E, Toivonen S, Corander J, Kuusi M, Skurnik M, Siitonen A, Haukka K. 2012. Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties. BMC Microbiol 12:208. doi: 10.1186/1471-2180-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savin C, Martin L, Bouchier C, Filali S, Chenau J, Zhou Z, Becher F, Fukushima H, Thomson NR, Scholz HC, Carniel E. 2014. The Yersinia pseudotuberculosis complex: characterization and delineation of a new species, Yersinia wautersii. Int J Med Microbiol 304:452–463. doi: 10.1016/j.ijmm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Sprague LD, Scholz HC, Amann S, Busse HJ, Neubauer H. 2008. Yersinia similis sp. nov. Int J Syst Evol Microbiol 58:952–958. doi: 10.1099/ijs.0.65417-0. [DOI] [PubMed] [Google Scholar]
- 48.Chenais E, Bagge E, Lambertz ST, Artursson K. 11 December 2012, posting date Yersinia enterocolitica serotype O:9 cultured from Swedish sheep showing serologically false-positive reactions for Brucella melitensis. Infect Ecol Epidemiol doi: 10.3402/iee.v2i0.19027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godfroid J, Nielsen K, Saegerman C. 2010. Diagnosis of brucellosis in livestock and wildlife. Croat Med J 51:296–305. doi: 10.3325/cmj.2010.51.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nesbakken T, Eckner K, Hoidal HK, Rotterud OJ. 2003. Occurrence of Y. enterocolitica in slaughter pigs and consequences for meat inspection, slaughtering and dressing procedures. Adv Exp Med Biol 529:303–308. doi: 10.1007/0-306-48416-1_57. [DOI] [PubMed] [Google Scholar]
- 51.Fredriksson-Ahomaa M, Stolle A, Siitonen A, Korkeala H. 2006. Sporadic human Yersinia enterocolitica infections caused by bioserotype 4/O: 3 originate mainly from pigs. J Med Microbiol 55:747–749. doi: 10.1099/jmm.0.46523-0. [DOI] [PubMed] [Google Scholar]
- 52.Quintard B, Petit T, Ruvoen N, Carniel E, Demeure CE. 2010. Efficacy of an oral live vaccine for veterinary use against pseudotuberculosis. Comp Immunol Microbiol Infect Dis 33:e59–e65. doi: 10.1016/j.cimid.2009.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.