Abstract
Conventional microscopy is the gold standard for malaria diagnosis. The CellaVision DM96 is a digital hematology analyzer that utilizes neural networks to locate, digitize, and preclassify leukocytes and characterize red blood cell morphology. This study compared the detection rates of Plasmodium and Babesia species on peripheral blood smears utilizing the CellaVision DM96 with the rates for a routine red blood cell morphology scan. A total of 281 slides were analyzed, consisting of 130 slides positive for Plasmodium or Babesia species and 151 negative controls. Slides were blinded, randomized, and analyzed by CellaVision and microscopy for red cell morphology scans. The technologists were blinded to prior identification results. The parasite detection rate was 73% (95/130) for CellaVision and 81% (105/130) for microscopy for positive samples. The interobserver agreement between CellaVision and microscopy was fair, as Cohen's kappa coefficient equaled 0.36. Pathologist review of CellaVision images identified an additional 15 slides with parasites, bringing the total number of detectable positive slides to 110 of 130 (85%). Plasmodium ovale had the lowest rate of detection at 56% (5 of 9); Plasmodium malariae and Babesia spp. had the highest rate of detection at 100% (3/3 and 6/6, respectively). The detection rate by CellaVision was 100% (23/23) when the parasitemia was ≥2.5%. The detection rate for <0.1% parasitemia was 63% (15/24). Technologists appropriately classified all negative specimens. The percentage of positive specimens detectable by CellaVision (73%) approaches results for microscopy on routine scan of peripheral blood smears for red blood cell morphology.
INTRODUCTION
Malaria is a global disease with an estimated 219 million cases and 660,000 deaths reported in 2010 (1). In the United States, 1,691 cases were reported in 2010, up from 767 cases in 2003, mostly from travelers to areas of endemicity (2). The recognition and diagnosis of malaria often depend on clinical signs and symptoms, including fever, headache, fatigue, myalgia, abdominal pain, nausea, vomiting, and splenomegaly. These symptoms are often nonspecific and can be easily missed by physicians unfamiliar with the presentation of malaria (3). In addition, a large proportion of infected individuals in countries in which malaria is endemic are asymptomatic or subclinical. Some studies have reported microscopy-detected malaria carriage in up to 39.2% of asymptomatic individuals (4). The prevalence of asymptomatic carriers has been reported to be highest in children and adolescents due to the fact that children in regions where malaria is endemic often acquire clinical immunity to malaria from repetitive infections, resulting in the ability to tolerate malaria parasites without developing fever (4, 5). These asymptomatic carriers typically do not seek medical treatment and can serve as a reservoir of parasites not only in high-prevalence areas but also potentially in developed countries due to increased international travel (3, 4). Effective routine screening tests for malaria are therefore essential not only in areas where malaria is endemic but also in developed countries.
Optical microscopy has been the diagnostic gold standard for malaria since the early 1900s (6). The reported sensitivity of microscopy ranges from 85% to 93% (7, 8), with experienced microscopists being able to detect <100 to 500 parasites/μl (7, 9). There have been diagnostic advances for malaria, including PCR (10), rapid diagnostic tests (RDTs) (11, 12), and automated hematology analyzers (13–18). PCR has a higher sensitivity than microscopy (100% versus 93%, respectively), with the ability to detect less than 5 parasites/μl (8). The availability of PCR is limited to high-volume reference laboratories, and specimen transport time has limited the use of PCR in the clinical setting. RDTs have a wide range of sensitivity for detecting Plasmodium falciparum (76% to 100%), with decreased sensitivities when the parasite load drops to less than 500 parasites/μl (11). One disadvantage of RDTs is that microscopy is needed to estimate the percent parasitemia and verify the species. There has been an increase in the number of automated hematology analyzers used for detection of malaria (13–18) by flow cytometry methods. The sensitivity has ranged from 48 to 95% compared to microscopy. These tools do not yet allow for species identification, and microscopy is required as an adjunct diagnostic tool.
The CellaVision DM96 (Lund, Sweden) is an FDA-approved digital hematology analyzer that utilizes neural networks to locate, digitize, and preclassify leukocytes; provide a differential count; and characterize red blood cell (RBC) morphology. CellaVision scans a peripheral blood smear slide and finds a well-dispersed area of the monolayer to perform an automated differential count of white blood cells (WBCs). The microscope has a camera attachment which captures individual images of each leukocyte utilized in the count. These images are displayed on the remote viewer interface, where technologists verify that the leukocytes have been categorized appropriately. Images can be accessed from remote locations, including other sites in the hospital where clinicians can access their patients' peripheral smear images. In addition to categorizing leukocytes, the instrument takes an image of one field of red blood cells (RBCs) at ×10 magnification. The technologists can view the image and classify characteristics of RBCs, including anisocytosis, poikilocytosis, polychromasia, microcytosis, macrocytosis, hypochromia, sickle cells, RBC inclusions, and intracellular parasites. Our laboratory's current standard operating procedure is to evaluate red blood cell morphology on every peripheral blood smear that requires a manual differential count. In addition, every complete blood count (CBC) specimen from patients in the emergency department is also evaluated for red blood cell morphology regardless of whether a manual differential count is required. These scans for red blood cell morphology involve examination of at least 10 fields on low magnification and usually take 60 s or less. Our laboratory has on occasion discovered malaria as an incidental finding on these routine red blood cell morphology scans of emergency room patients not clinically suspected of having malaria, prompting the evaluation of CellaVision for detecting malaria.
CellaVision is not FDA approved for detection of intracellular parasites. If a patient is clinically suspected of having malaria and a clinician has requested a formal malaria screen, our current operating procedure for malaria screening includes examination of two thick smears and two thin smears at 100× oil immersion for at least 300 fields. The purpose of this study was to evaluate the sensitivity of detecting intracellular red blood cell parasites, specifically Plasmodium and Babesia species, utilizing CellaVision compared to microscopy for routine red blood cell morphology scans.
MATERIALS AND METHODS
One hundred ninety-six slides positive for intracellular parasites were obtained from proficiency testing surveys from the College of American Pathologists (CAP) from 2001 to 2012 and from patients who were diagnosed by conventional microscopy at the University of Texas (UT) Southwestern Medical Center, Dallas, TX, and Children's Medical Center, Dallas, TX, from 2001 to 2013. Of the slides selected for further study, 130 positive slides were prepared from 88 specimens (two specimens with four slides each, six specimens with three slides each, and 15 specimens with two slides each). The percent parasitemia ranged from <0.1% to >10%. One hundred fifty-one negative-control slides were selected from patients with routine complete blood counts and differentials. The University of Texas Southwestern Medical Center in Dallas, Texas, Institutional Review Board reviewed and approved the study protocol, and a waiver of informed consent was granted to this minimal-risk study.
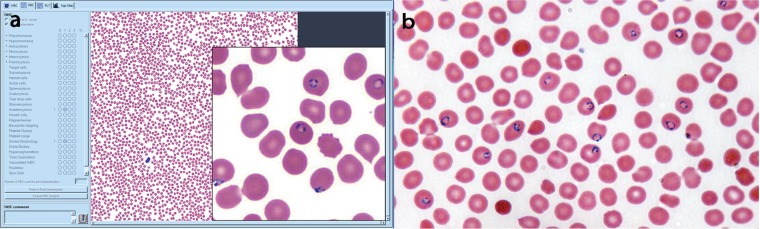
Slides were blinded, randomized, and analyzed on CellaVision utilizing CellaVision Remote Review software version 3.2.1 build 23, as well as by microscopy for red cell morphology scans by laboratory technologists. Medical technologists performed a routine scan for red blood cell morphology as per our laboratory protocol with examination of at least 10 fields at low power (10×), with higher magnification as needed by light microscopy. The analogous examination was performed on CellaVision utilizing the 10× image in the red blood cell morphology window with magnification of areas utilizing software tools (Fig. 1). After technologists evaluated the samples, a pathologist reviewed slides analyzed in CellaVision in the same manner utilizing the red blood cell morphology window. The pathologists were not blinded to the study and were included to determine if parasites could be detected in the single image, even when technologists did not identify them.
FIG 1.
(a) Plasmodium falciparum-infected red blood cells in the RBC view of the CellaVision Remote Review software. (b) Plasmodium falciparum at 1,000× oil immersion with conventional light microscopy.
Sensitivity, specificity, confidence intervals, Cohen's kappa analysis, and regression analyses were performed with SAS 9.2 statistical software (Cary, NC). Sensitivity and specificity were calculated for the technologists using CellaVision as well as for the pathologists reviewing CellaVision results. Chi-square analysis using SAS was performed to calculate sensitivity and specificity, with statistical significance determined using McNemar's test. Regression analysis was performed on RBC morphology characteristics, including polychromasia, hypochromasia, anisocytosis, microcytosis, macrocytosis, poikilocytosis, target cells, schistocytes, helmet cells, sickle cells, spherocytes, ovalocytes, tear drop cells, Howell-Jolly bodies, and acanthocytes to determine any association with malaria.
RESULTS
A total of 130 cases and 151 negative controls were included in the study. One hundred ninety-six slides positive for Plasmodium species were initially selected for analysis. However, CellaVision was unable to read 66 samples due to incompatible smear preparation. These samples were from the CAP proficiency testing surveys, which prepare their peripheral smear starting opposite the labeled end of the slide. CellaVision is programmed to look for the monolayer opposite the label, not near the label, rendering these slides unreadable by CellaVision. This decreased the total number of analyzable positive slides to 130. One hundred fifty-one negative slides were included for a total of 281 peripheral smear slides. The 151 negative-control slides were verified by a pathologist.
Of the 130 positive slides, there were 51 with P. falciparum; 25 with Plasmodium vivax; 9 with Plasmodium ovale; 3 with Plasmodium malariae; 4 with Plasmodium vivax or P. ovale; 25 with Plasmodium species, not otherwise specified; 7 mixed infections, which included 2 with P. falciparum and P. malariae, 2 with P. falciparum and P. ovale, 2 with P. ovale and P. vivax, and 1 with P. vivax and P. malariae; and 6 with Babesia species. Using CellaVision (Fig. 1a), technologists identified intracellular parasites on 95 of the 130 positive samples. The sensitivity and specificity (95% confidence intervals) for CellaVision detection of 95 cases by the technologists were 73% (65% to 80%) and 100% (98 to 100%), respectively. The detection rates by species are compiled in Table 1. Pathologist review of CellaVision images identified an additional 15 positive slides. The sensitivity and specificity (95% confidence intervals) for CellaVision detection of 110 cases by pathologists were 85% (77 to 90%) and 100% (98 to 100%). All negative slides were appropriately classified, yielding a specificity of 100% by CellaVision. Using microscopy (Fig. 1b), technologists identified 105 (81%) of the positive samples with a sensitivity and specificity (95% confidence intervals) of 84% (76 to 90%) and 100% (98 to 100%), respectively. Pathologists confirmed that the remaining false-negative slides had identifiable intracellular parasites.
TABLE 1.
Detection of intracellular parasites by species
| Species | n | CellaVision |
Microscopy |
% parasitemia | ||||
|---|---|---|---|---|---|---|---|---|
| Detection by technologist |
Detection by pathologist |
No. of samples detected by technologist | % sensitivity | |||||
| No. of samples | % sensitivity | No. of samples | % sensitivity | |||||
| P. falciparum | 51 | 37 | 73 | 43 | 84 | 42 | 82 | <0.1–30 |
| P. vivax | 25 | 18 | 72 | 23 | 92 | 21 | 84 | <0.1–2 |
| P. ovale | 9 | 5 | 56 | 5 | 56 | 5 | 56 | <0.1–0.9 |
| P. malariae | 3 | 3 | 100 | 3 | 100 | 3 | 100 | 0.16–1 |
| Plasmodium spp. | 25 | 20 | 80 | 20 | 80 | 19 | 76 | <0.1–23.5 |
| P. ovale or P. vivax | 4 | 0 | 0 | 3 | 75 | 3 | 75 | <1% |
| Mixed infectionsa | 7 | 6 | 86 | 7 | 100 | 7 | 100 | 0.1–0.9 |
| Babesia | 6 | 6 | 100 | 6 | 100 | 5 | 83 | 1–5 |
| Total | 130 | 95 | 73 | 110 | 85 | 105 | 81 | <0.1–30 |
Mixed infections included P. falciparum and P. malariae (two), P. falciparum and P. ovale (two), P. ovale and P. vivax (two), and P. vivax and P. malariae (one).
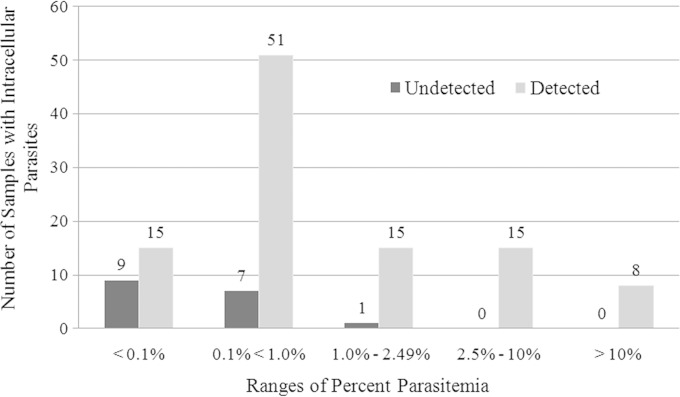
Percent parasitemia was categorized based on the College of American Pathologists proficiency testing categories and is shown in Fig. 2. Intracellular parasites in samples with less than 0.1% parasitemia were detected 63% of the time. Higher parasitemia increased the rate of detection by CellaVision. At a parasitemia of 1% or higher, only one specimen containing P. falciparum was misclassified as negative. Specimens with a parasitemia of 2.5% or higher contained either P. falciparum or Babesia species and were correctly classified 100% of the time utilizing CellaVision.
FIG 2.
Detection of intracellular parasites by percent parasitemia. Parasitemia ranges are those used by the College of American Pathologists in proficiency testing surveys. The original percent parasitemia values were available for 68 out of 130 specimens used in the study.
The average number of red cells counted by CellaVision for all samples was 1,189. The average number of red cells counted on the positive samples was 858. When a higher number of RBCs was counted, the likelihood of detecting Plasmodium increased. No particular RBC count threshold was associated with increased detection of malaria.
Regression analysis on red cell morphology features showed a statistical significance for acanthocytes being associated with Plasmodium species overall (P = 0.03). Polychromasia was associated with the presence of P. vivax (P = 0.03).
DISCUSSION
This is the first study to evaluate the sensitivity of CellaVision for detecting Plasmodium and Babesia species. CellaVision was developed as a tool to differentiate leukocytes and does not currently have software to detect intracellular parasites based upon predictive models in place for the WBC differential. Our goal was to evaluate the ability to detect intracellular parasites of Plasmodium and Babesia species utilizing the images captured by CellaVision for the classification of RBC morphology.
Overall, the detection rate by technologists using CellaVision was 73%. Of the positive specimens missed by technologists' review, it was observed that six samples contained late trophozoites and two had gametocytes, without any ring forms. These forms were present on the image of the red blood cells but missed being recognized by the technologists as malaria. The CellaVision neural network classified these forms as either leukocytes, smudge cells, or artifacts; however, the technologists were not asked to perform a manual differential on these specimens and therefore did not access the WBC classification screen in CellaVision for these samples. Education of core laboratory or hematology technologists on recognizing the different forms of Plasmodium species could improve the detection rate of malaria, bringing the sensitivity up to the same limits of detection as those of conventional microscopy, which have been reported to range between 85% and 93% (8, 19, 20).
A standard method for estimating parasitemia involves counting the number of parasitized red cells per 10,000 RBCs (40 monolayer cell fields using 100× oil immersion) (12). The use of CellaVision for detecting malaria includes the red cell count, which would offer another method for the estimation of parasitemia. Future CellaVision software developments include the ability to sort images of individual red blood cells by morphological features, including intracellular inclusions such as those of malaria and Babesia species, and would in essence provide the ability to automate the estimation of parasitemia.
One of the limitations of CellaVision appears to be the inconsistent detection of malaria species at lower levels of parasitemia, particularly at levels of <1% (Fig. 2). P. ovale, P. vivax, and P. malariae are reported to be associated with lower parasitemia than is P. falciparum (21). The utilization of CellaVision for routine screening may be problematic in asymptomatic patients from areas where these species are endemic.
One limitation of the study was the use of repository slides instead of prospective patient samples. Using fresh patient specimens would have allowed additional slides to be prepared and analyzed when CellaVision could not read the initial slides. In addition, although the technologists were blinded to the study, the supervisor had knowledge of the study protocol. It is unclear if this biased the technologists and contributed to a higher detection rate than would be seen for a routine patient sample. Lastly, in order to definitively rule out malaria, some references recommend screening 300 fields at 100× (21), which exceeds the limitations of CellaVision. Additional microscopy or molecular methods would be needed to definitively rule out malaria. The resolution of CellaVision images may need to be improved in order to be utilized as a tool for identifying Plasmodium to the species level.
While it would be cost-prohibitive to perform a formal malaria screen with evaluation of 300 fields utilizing 100× oil immersion for all emergency room patients, our institutional practice of performing a scan for red blood cell morphology consisting of examination of at least 10 fields at 10× for every emergency room patient with a CBC has yielded the very rare “incidental” malaria finding in patients not clinically recognized as being at risk for malaria infection. While we are not advocating for the red blood cell morphology scan as a replacement for the formal malaria screen or other ancillary studies when clinically indicated, it is recognized at our institution as providing another opportunity to detect malaria in an otherwise subclinical malaria patient.
One promising advantage of CellaVision compared to conventional slides is that all images collected by the slide scanner can be viewed remotely using the CellaVision remote viewer software. The manual differential and analysis of red blood cell morphology could therefore occur in a remote site far from the collection locale. In developed countries, and some developing countries where capital investments may be easier to come by than adequately trained laboratory professionals, this may provide a way to make diagnostic expertise available from anywhere in the world. In addition, pathologists and clinicians can access these images from home through their institution's virtual private network, potentially decreasing delays in diagnosis and improving patient care.
This study demonstrates that CellaVision has the potential to detect intracellular parasites in routine screening of blood smears for RBC morphology. In addition, the software is a valuable resource for storing images to be used for educational purposes, including training technologists in the detection of intracellular parasites.
ACKNOWLEDGMENTS
We thank Patrick Racsa and Rong Huang for the statistical analysis of this study.
The authors declare no conflict of interest.
REFERENCES
- 1.World Health Organization. 2012. World malaria report 2012. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2012. Malaria surveillance—United States, 2010. MMWR Surveill Summ 61(2):1–17. [PubMed] [Google Scholar]
- 3.Lee HK, Kim SI, Chae H, Kim M, Lim J, Oh EJ, Kim Y, Park YJ, Lee W, Han J. 2012. Sensitive detection and accurate monitoring of Plasmodium vivax on routine complete blood count using automatic blood cell analyzer (DxH800). Int J Lab Hematol 34:201–207. doi: 10.1111/j.1751-553X.2011.01383.x. [DOI] [PubMed] [Google Scholar]
- 4.Ogutu B, Tiono AB, Makanga M, Premji Z, Gbadoe AD, Ubben D, Marrast AC, Gaye O. 2010. Treatment of asymptomatic carriers with artemether-lumefantrine: an opportunity to reduce the burden of malaria? Malar J 9:30. doi: 10.1186/1475-2875-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabunda S, Aponte JJ, Tiago A, Alonso P. 2009. A country-wide malaria survey in Mozambique. II. Malaria attributable proportion of fever and establishment of malaria case definition in children across different epidemiological settings. Malar J 8:74. doi: 10.1186/1475-2875-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilles HM, Warrell DA (ed). 1993. Bruce-Chwatt's essential malariology, 3rd ed. Edward Arnold, London, United Kingdom. [Google Scholar]
- 7.Maguire JD, Lederman ER, Barcus MJ, O'Meara WA, Jordon RG, Duong S, Muth S, Sismadi P, Bangs MJ, Prescott WR, Baird JK, Wongsrichanalai C. 2006. Production and validation of durable, high quality standardized malaria microscopy slides for teaching, testing, and quality assurance during an era of declining diagnostic proficiency. Malar J 5:92. doi: 10.1186/1475-2875-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston SP, Pienizaek NJ, Xayavong MV, Slemenda SB, Wilkins PP, da Silva AJ. 2006. PCR as a confirmatory technique for laboratory diagnosis of malaria. J Clin Microbiol 44:1087–1089. doi: 10.1128/JCM.44.3.1087-1089.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. 1988. Malaria diagnosis: memorandum from a WHO meeting. Bull World Health Organ 66:575–594. [PMC free article] [PubMed] [Google Scholar]
- 10.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. 1993. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol 58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 11.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. 2007. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg 77:119–127. [PubMed] [Google Scholar]
- 12.Moody A. 2002. Rapid diagnostic test for malaria parasites. Clin Microbiol Rev 15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendelow BV, Lyons C, Nhlangothi P, Tana M, Munster M, Wypkema E, Liebowitz L, Marshall L, Scott S, Coetzer TL. 1999. Automated malaria detection by depolarization of laser light. Br J Haematol 104:499–503. doi: 10.1046/j.1365-2141.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 14.Hanscheid T, Melo-Cristino J, Pinto B. 2001. Automated detection of malaria pigment in white blood cells for the diagnosis of malaria in Portugal. Am J Trop Med Hyg 64:290–292. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Ezra J, St Louis M, Riley RS. 2001. Automated malarial detection with the Abbott Cell-Dyn 4000 hematology analyzer. Lab Hematol 7:61–64. [Google Scholar]
- 16.Huh HJ, Oh GY, Huh JW, Chae SL. 2008. Malaria detection with the Sysmex XE-2100 hematology analyzer using pseudoeosinophilia and abnormal WBC scattergram. Ann Hematol 87:755–759. doi: 10.1007/s00277-008-0486-8. [DOI] [PubMed] [Google Scholar]
- 17.Campuzano-Zuluaga G, Avarez-Sanchez G, Escobar-Gallo GE, Valencia-Zuluaga LM, Ríos-Orrego AM, Pabón-Vidal A, Miranda-Arboleda AF, Blair-Trujillo S, Campuzano-Maya G. 2010. Design of malaria diagnostic criteria for the Sysmex XE-2100 hematology analyzer. Am J Trop Med Hyg 82:402–411. doi: 10.4269/ajtmh.2010.09-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wever PC, Henskens YMC, Kager PA, Dankert J, van Gool T. 2002. Detection of imported malaria with the Cell-Dyn 4000 hematology analyzer. J Clin Microbiol 40:4729–4731. doi: 10.1128/JCM.40.12.4729-4731.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne LM, Kyi MS, Chiodini PL, Warhurst DC. 1994. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J Clin Pathol 47:740–742. doi: 10.1136/jcp.47.8.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pöschl B, Waneesorn J, Thekisoe O, Chutipongvivate S, Panagiotis K. 2010. Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP in northern Thailand. Am J Trop Med Hyg 83:56–60. doi: 10.4269/ajtmh.2010.09-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia LS. (ed). 2001. Diagnostic medical parasitology, 4th ed. ASM Press, Washington, DC. [Google Scholar]