Abstract
A large-scale, whole-genome comparison of Canadian Neisseria gonorrhoeae isolates with high-level cephalosporin MICs was used to demonstrate a genomic epidemiology approach to investigate strain relatedness and dynamics. Although current typing methods have been very successful in tracing short-chain transmission of gonorrheal disease, investigating the temporal evolutionary relationships and geographical dissemination of highly clonal lineages requires enhanced resolution only available through whole-genome sequencing (WGS). Phylogenomic cluster analysis grouped 169 Canadian strains into 12 distinct clades. While some N. gonorrhoeae multiantigen sequence types (NG-MAST) agreed with specific phylogenomic clades or subclades, other sequence types (ST) and closely related groups of ST were widely distributed among clades. Decreased susceptibility to extended-spectrum cephalosporins (ESC-DS) emerged among a group of diverse strains in Canada during the 1990s with a variety of nonmosaic penA alleles, followed in 2000/2001 with the penA mosaic X allele and then in 2007 with ST1407 strains with the penA mosaic XXXIV allele. Five genetically distinct ESC-DS lineages were associated with penA mosaic X, XXXV, and XXXIV alleles and nonmosaic XII and XIII alleles. ESC-DS with coresistance to azithromycin was observed in 5 strains with 23S rRNA C2599T or A2143G mutations. As the costs associated with WGS decline and analysis tools are streamlined, WGS can provide a more thorough understanding of strain dynamics, facilitate epidemiological studies to better resolve social networks, and improve surveillance to optimize treatment for gonorrheal infections.
INTRODUCTION
Neisseria gonorrhoeae is a Gram-negative diplococcus bacterium that causes gonorrhea infections. Gonorrhea is the second most reported bacterial sexually transmitted infection (STI) in Canada, with reported cases increasing from 15.5 per 100,000 in 1997 to 36.2 per 100,000 in 2012 (1), and approximately 106 million cases are estimated annually worldwide (2). N. gonorrhoeae bacteria have developed resistance against sulfonamides, penicillins, tetracyclines, and fluoroquinolones (3, 4), and current treatment options now include third-generation extended-spectrum cephalosporins (ESC), namely, cefixime (CFM) and ceftriaxone (CRO) (5). MIC creep has seen the modal MIC values rise between 2001 and 2010 in Canada from 0.016 μg/ml to 0.125 μg/ml and 0.063 μg/ml for CFM and CRO, respectively (6). These results coincide with recent clinical reports of treatment failures to primarily CFM monotherapy in Canada (7, 8) and additional global reports of high-level CRO MICs in isolates from Japan and Europe (9–13). Furthermore, isolates with decreased susceptibility to cephalosporins and coresistance to azithromycin (AZM), a recommended cotherapy (5), have recently been identified in Canada (14).
Decreased susceptibility to extended-spectrum cephalosporins (ESC-DS) and resistance to AZM have been attributed to several molecular mechanisms. The primary mechanism for ESC-DS is modification of the penA gene (penicillin binding protein 2 [PBP2]) including various mutations of the wild-type gene and a recombinant mosaic allele containing portions of genetic sequences from commensal Neisseria (3, 4, 9, 15–18). Mutations in the promoter and/or the coding region of the repressor gene mtrR cause overexpression of the MtrCDE efflux pump (3, 4, 15, 19, 20) and porB1b gene mutations (penB G120 and/or A121 mutation) have been shown to cause decreased membrane permeability and contribute to decreased susceptibility toward cephalosporins (3, 4, 15, 19–22). Other genes associated with penicillin resistance include pilQ and ponA; however, the role in ESC-DS remains unclear (3, 9, 10, 20, 23–27). The currently identified resistance determinants do not fully account for the observed ESC MICs, and it is hypothesized that other factors may be involved (4, 9, 10, 15, 20, 24, 25). AZM resistance has been associated with mutations in 23S rRNA alleles (4, 28) as well as mtrR (29–31).
Monitoring the spread of resistant N. gonorrhoeae involves phenotypic and molecular biology-based typing methodologies (32, 33) that have been used in combination with antimicrobial susceptibility data for epidemiological studies. The internationally recognized N. gonorrhoeae multiantigen sequence typing (NG-MAST) system has been effective in tracking short-chain transmission of infections (33) (www.ng-mast.net). However, application of whole-genome sequencing (WGS) for comparative studies provides higher resolution through a genomic epidemiology approach to investigate strain relatedness and dynamics (34–36).
In this report, we have applied the enhanced discriminatory power of WGS to describe the dissemination, relatedness, and emergence of Canadian N. gonorrhoeae isolates with elevated ESC MICs.
MATERIALS AND METHODS
N. gonorrhoeae isolates.
This study included 169 N. gonorrhoeae isolates collected between 1989 and 2013 from across Canada and 10 international reference strains. Canadian isolates were primarily selected for decreased susceptibility to CRO (n = 65) consisting of 19 CRO-DS isolates that were submitted to the National Microbiology Laboratory (NML), Public Health Agency of Canada, between 1989 and 2006 (83%), 6/18 from 2007, 5/83 from 2008, 7/96 from 2009, 12/217 from 2010, 5/207 from 2011, and 11/168 from 2012. Additional isolates (n = 105) were included to provide a broad range of NG-MAST sequence types (ST), geographical distributions, and antimicrobial susceptibilities.
Isolate characterization and antimicrobial susceptibility testing.
Antimicrobial susceptibilities of N. gonorrhoeae to spectinomycin, ceftriaxone, erythromycin, penicillin, tetracycline, azithromycin, cefixime (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada), and ciprofloxacin (generously provided by Bayer, Etobicoke, Ontario, Canada) were determined using the agar dilution method as previously described (37). Interpretation of the MIC was based on established criteria (38, 39); cefixime decreased susceptibility MIC of ≥0.25 μg/ml, ceftriaxone decreased susceptibility MIC of ≥0.125 μg/ml (2), and azithromycin resistance MIC of ≥2.0 μg/ml (40). N. gonorrhoeae ATCC 49226, WHO-B, WHO-C, WHO-D, WHO-F, WHO-G, WHO-K, and WHO-P reference cultures were used as controls (41). Susceptibility categories were assigned as very susceptible, susceptible, moderate susceptibility, decreased susceptibility or resistant, and high-level resistance for drugs as follows: for CRO, 0.00025 to 0.004 μg/ml, 0.008 to 0.016 μg/ml, 0.032 to 0.063 μg/ml, 0.125 to 0.5 μg/ml, and 1 to 2 μg/ml, respectively; for CFM, 0.00025 to 0.004 μg/ml, 0.008 to 0.016 μg/ml, 0.032 to 0.125 μg/ml, 0.25 to 0.5 μg/ml, and 1 to 4 μg/ml, respectively; and for AZM, 0.032 to 0.063 μg/ml, 0.125 to 0.25 μg/ml, 0.5 to 1 μg/ml, 2 to 16 μg/ml, and 32 to 256 μg/ml, respectively.
Statistical comparisons.
To determine the magnitude of the contributions of the known antimicrobial resistance markers to ESC-DS, isolates were divided into four categories based on CRO MIC values and the presence or absence of the molecular marker of interest (including penA genotype, mtrR −35 A deletion, the G-to-D change at position 39 encoded by mtrR [mtrR A39T], mtrR G45D, ponA L421P, porB G120, and porB A121): true positive (TP) having moderate to very high MICs (0.032 to 2 μg/ml) and the presence of the genetic marker of interest; false positive (FP) having very low to low MICs (≤0.016 μg/ml) and the marker; false negative (FN) having moderate to very high MICs and no marker; and true negative (TN) having very low to low MICs and no marker. Calculations were performed as follows: sensitivity (SENS) = TP/(FN + TP) × 100 and specificity (SPEC) = TN/(FP + TN) × 100 (42).
The measure of association was determined using χ2 or Fisher exact test using OpenEpi version 3.01 (43). Two-tailed differences of P < 0.05 at 95% confidence were considered statistically significant.
Whole-genome sequencing and assembly.
DNA samples were extracted from cultures following standard protocol with Epicentre Masterpure complete DNA and RNA extraction kit (Mandel Scientific, Guelph, Ontario, Canada). Multiplexed libraries were created with Nextera XT sample preparation kits (Illumina, San Diego, CA). Paired-end, 250-bp indexed reads were generated on the Illumina MiSeq platform (Illumina, San Diego, CA) yielding an average of 1,167,540 reads/genome and an average genome coverage of 127.
De novo assembly.
The quality of the reads was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), merged using FLASH (44), and assembled with SPAdes (45) and annotation was accomplished with Prokka (46). The average number of contigs was 147, the average contig length was 16,506, and the average N50 contig length was 62,730.
Phylogenomic analysis based on core single nucleotide polymorphisms.
FASTQ files for forward and reverse reads were concatenated into one fastq FASTQ file per isolate and were used for further analysis. Read ends were trimmed, and poor-quality reads were filtered to improve assembly quality using the script run_assembly_trimClean.pl from CG-Pipeline (47) with the following options: “–min_quality 25 –bases_to_trim 10 –min_avg_quality 25 –min_length 36 -p 1”, and read qualities were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The high-quality reads were then mapped to the publically available reference genome, N. gonorrhoeae NCCP11945 (NCBI accession no. NC_011035) (G. T. Chung et al. PubMed identifier [PMID] 18586945) with SMALT version 0.7.4 (http://www.sanger.ac.uk/resources/software/smalt/) with the following options: smalt_index “-k 13 -s 6” and smalt_map “-f samsoft -r -1”. Single nucleotide variants were called using FreeBayes (Erik Garrison, Garbor Marth [2012] arXiv:1207.3907[q-bio.GN]) using the following parameters: “–pvar 0 –ploidy 1 –left-align-indels –min-mapping-quality 30 –min-base-quality 30 –min-alternate-fraction 0.75 –min-coverage 15” with additional variant confirmation using SAMtools mpileup (48). The following parameters were used to run SAMtools: “samtools mpileup -BQ0 -d100000000” and “bcftools view –cg”. Positions where variant calls were not in agreement between both variant callers were excluded. Variant calls within potential problematic regions including repetitive regions (MUMmer v.3.23), predicted phages (PHAST), genomic islands (IslandViewer), and 10 suspected highly recombinant regions were excluded from the analysis containing >90 single nucleotide polymorphisms (SNPs) per 10,000 bp (see Fig. S1 and Table S1 in the supplemental material). All remaining variant calls were merged into a single meta-alignment file. There were a total of 6,509 core SNP positions for the population (Table S2). The meta-alignment of informative core SNP positions was used to create a maximum likelihood phylogenetic tree using PhyML with generalized time reversible model (49) using parameters “–quiet -b -4 -m GTR -s BEST.” Phylogenomic clades were assigned by cluster analysis using PhyloPart (http://sourceforge.net/projects/phylopart/files) with a percentile distance threshold value of 0.10 for all clades except clade F that clustered at a threshold value of 0.105.
N. gonorrhoeae multiantigen sequence types (NG-MAST) and antimicrobial resistance molecular markers.
NG-MAST types were determined by the NG-MAST PCR method as previously described (33) and in silico using WGS data. The sequences were submitted to the NG-MAST website (http://www.ng-mast.net/) to determine the sequence type. NG-MAST groups were determined by using the trimmed concatenated porB and tbpB NG-MAST gene sequences. Sequences were aligned, and a neighbor-joining phylogenetic tree was generated, using ClustalX (50) and visualized radially using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). NG-MAST ST types were grouped according to their relative distances on the phylogenomic tree guided by PhyloPart cluster analysis software set to a percentile distance threshold value of 0.05.
NG-MAST gene sequences and antimicrobial resistance markers including penA genotype, mtrR −35 A deletion, mtrR A39T, mtrR G45D, ponA L421P, porB G120, porB A121, 23S rRNA C2599T and A2143G mutations were identified in silico from the WGS data.
RESULTS
Strain distribution.
Core SNP phylogenetic analysis was performed on 169 Canadian isolates of N. gonorrhoeae isolated between 1989 and 2013 from Ontario (n = 82), British Columbia (n = 23), Quebec (n = 30), Saskatchewan (n = 13), Nova Scotia (n = 12), Alberta (n = 4), New Brunswick (n = 4), and Manitoba (n = 1) and 10 international reference isolates including France F89 (sample 34842) (10), ATCC 49226, WHO-F, WHO-L, WHO-O, WHO-K, WHO-G, WHO-N, WHO-M, and WHO-P (41). The Canadian isolates included 41 (24%) from female patients, 126 (75%) from male patients, and 2 (1%) with no gender provided. Patient ages ranged from 2 months to 64 years with a median age of 27 years for the 137 isolates for which an age was available (see Table S3 in the supplemental material). Enhanced epidemiological data such as patient sexual orientation, clinical isolation site, and treatment history were not available.
Phylogenomic tree.
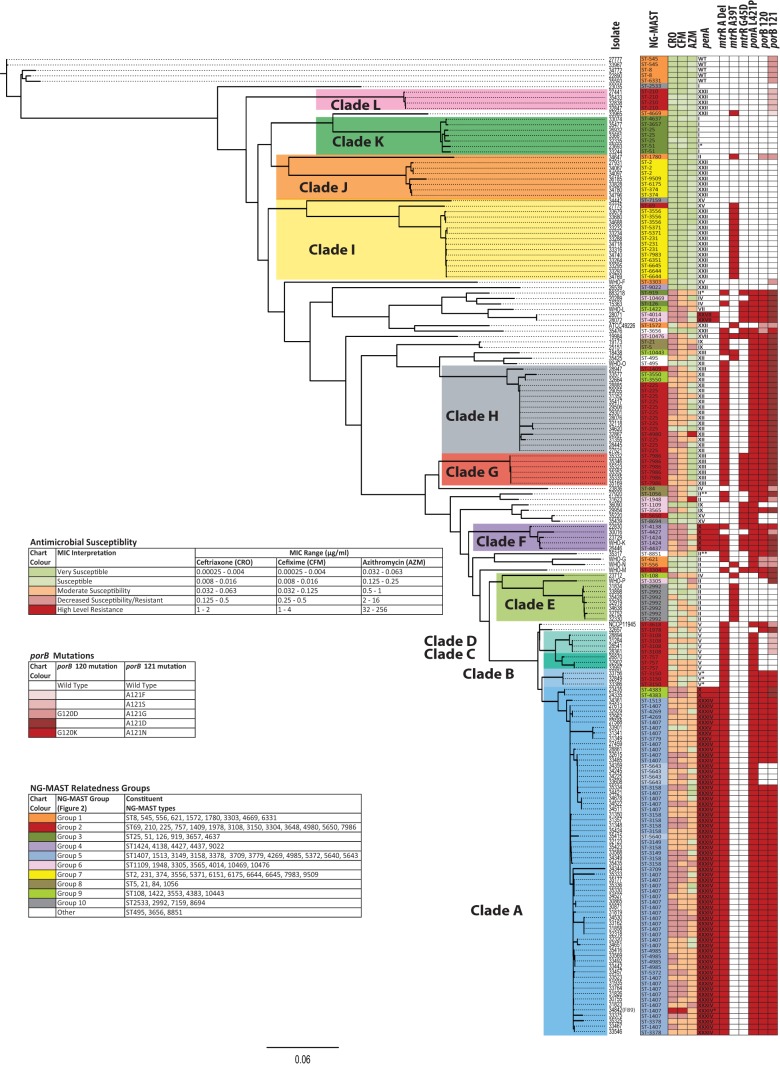
Maximum likelihood phylogeny using core SNPs and cluster analysis grouped 143 of the 179 isolates into 12 major clades (Fig. 1). The reference strains WHO-G, WHO-N, and WHO-M appeared independently between clades E and F, and reference strain NCCP11945 appeared between clades D and E. Reference strains ATCC 49226, WHO-F, WHO-L, and WHO-O appeared between clades H and I, while ESC-resistant France F89 reference strain (strain 34842) clustered in clade A, reference strain WHO-K (Japan) clustered in clade F; and reference strain WHO-P (United States) clustered in clade E.
FIG 1.
Whole-genome-based core SNP phylogenomic tree of N. gonorrhoeae strains based on maximum likelihood. NG-MAST types, antimicrobial susceptibility, and antimicrobial resistance molecular markers are indicated. Antimicrobial susceptibility to ceftriaxone (CRO), cefixime (CFM), and azithromycin (AZM) is shown. Red in the molecular marker columns (rightmost columns) represents presence of the marker. For penA, red represents a mosaic allele. In the penA column, WT, wild type; I*, penA nonmosaic I allele with a P413S substitution; II*, nonmosaic II allele with A505V; II**, nonmosaic II allele with A502V; V*, nonmosaic V allele with Y201H, G202A, E203G, D204E, and Q214E; XXXIV*, mosaic XXXIV allele with A502P.
Isolates 35593, 34772, 33967, 27777, and 22890 were the most genetically distant averaging 1,039, 1,022, 1,017, 1,013, and 1,004 SNPs from the rest of the 175 strains, respectively. These isolates were also members of the most distantly related NG-MAST group 1 (Fig. 2) and were the only strains in the data set with the wild-type penA allele (GenBank accession no. M32091 [Table S4 in the supplemental material]) (9). The greatest pairwise distances observed were 1,183 SNPs between isolates 35593 and 23836 and 1,181 SNPs between isolate 35593 and clade E isolates (Table S2). Clade G (n = 6) had the most closely related isolates with a maximum SNP difference of 3 and an average of 1.6 SNPs between each isolate, whereas clade J (n = 8) had the highest strain diversity with a maximum of 865 SNPs and an average of 245 SNPs between isolates (Table 1).
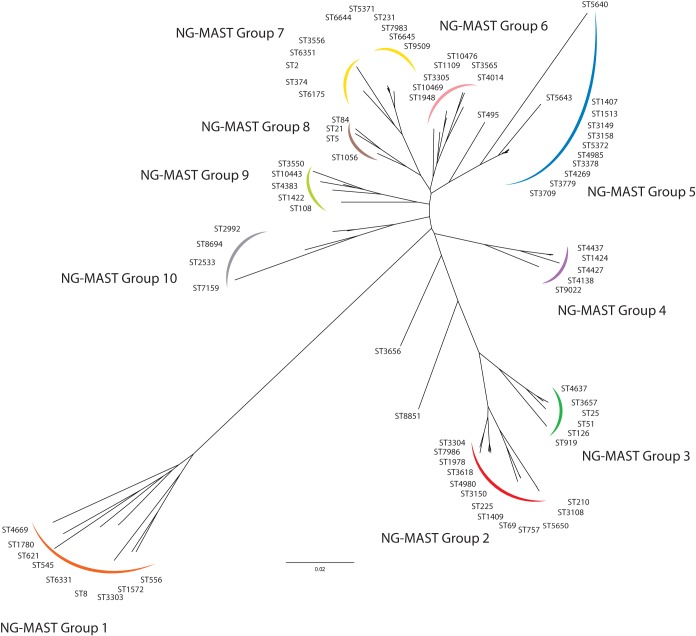
FIG 2.
Genetic relatedness of NG-MAST sequence types. Genetic relationships were inferred using a neighbor-joining method of trimmed, aligned, and concatenated NG-MAST internal porB and tbpB gene sequences with ClustalX and visualized using FigTree software. NG-MAST types clustering on each branch were assigned to the same group.
TABLE 1.
Number of single nucleotide polymorphisms in the core genome within and between the major phylogenomic clades of N. gonorrhoeae strains
| Clade | No. of isolates | Avg no. of SNPs within clade | Maximum no. of SNPs within clade | Avg no. of SNPs from previous cladea | Minimum no. of SNPs from previous cladea | Maximum no. of SNPs from previous cladea |
|---|---|---|---|---|---|---|
| A | 64 | 48.3 | 214 | |||
| B | 3 | 3.0 | 3 | 181.7 | 154 | 281 |
| C | 3 | 34.0 | 51 | 206.0 | 181 | 219 |
| D | 4 | 9.0 | 16 | 114.3 | 85 | 128 |
| E | 9 | 158.4 | 463 | 530.0 | 467 | 545 |
| F | 5 | 50.0 | 91 | 535.0 | 451 | 575 |
| G | 6 | 1.6 | 3 | 510.1 | 483 | 531 |
| H | 16 | 45.2 | 159 | 695.3 | 667 | 741 |
| I | 15 | 155.4 | 816 | 852.7 | 819 | 949 |
| J | 8 | 245.3 | 865 | 889.7 | 845 | 927 |
| K | 7 | 50.2 | 63 | 862.6 | 819 | 893 |
| L | 4 | 5.2 | 9 | 878.8 | 868 | 887 |
Number of single nucleotide polymorphisms (SNPs) compared to the previous closest common ancestal clade.
In our data set, 75% of the isolates were from male patients. Clades A, C, D, E, F, G, H, and L had a higher proportion of isolates from males (88% male [93/106]) compared to the isolates that did not cluster into clades and those of clade I (63% male [27/43]; P = 0.002). In clades J, K, and B, isolates from females predominated (33% male [6/18]; P < 0.001).
Antimicrobial resistance.
Of the 169 Canadian isolates analyzed, 67 (40%) exhibited decreased susceptibility to extended-spectrum cephalosporins (ESC-DS), 29 exhibited CRO-DS/CFM-DS, 36 exhibited CRO-DS/CFM-S, and 2 exhibited CRO-S/CFM-DS. ESC-DS strains were present in 5 genetically distinct lineages corresponding to clades A (35 ESC-DS/63 isolates in clade), E (1/8), F (4/4), G (4/6), and H (10/16) and some that did not cluster into distinct clades (13 ESC-DS/28 nonclade isolates) (Fig. 1). All isolates of clades B, C, D, I, J, K, and L were ESC-S.
ESC-DS was observed in 60.4% (55/91) of the isolates that clustered into clades A, F, G, and H, significantly greater than isolates of clades B, C, D, E, I, J, K, and L where only 1.9% (1/53) exhibited ESC-DS (P < 0.001). Among the isolates that did not group into clades, ESC-DS (40% [14/35]) was significantly greater than those of clades B, C, D, E, I, J, K, and L (P < 0.001) isolates, but not significantly different from the proportion of ESC-DS seen in the clade A, F, G, and H isolates (P = 0.06).
The earliest ESC-DS isolates were observed among miscellaneous isolates located between clades I and H on the phylogenomic tree (Fig. 1). The first CRO-DS strain isolated in Canada in 1989 from Saskatchewan (isolate 883218) was ST919 (NG-MAST group 3 in Fig. 2) and possessed a novel penA nonmosaic allele most closely resembling type II with an additional A550V substitution (see Table S4 in the supplemental material). Other early CRO-DS strains clustering early in the phylogeny included strains from 1994 to 1998 isolated in Quebec, Alberta, and Ontario in Canada with a variety of ST types, NG-MAST groups, and penA alleles (Fig. 1 and 2; see Table S3 in the supplemental material). During 2000 to 2001, ESC-DS strains with the penA mosaic X allele were observed in clade F from Quebec (NG-MAST group 4) as well as in clade A from Ontario (NG-MAST group 9). Then in 2006, ESC-DS strains were observed to form another lineage, clade H, with the penA nonmosaic XII allele and NG-MAST groups 2 and 9.
The first ST1407 strains of clade A (NG-MAST group 5) were isolated in 2005 from Quebec, Canada, and had moderate cephalosporin susceptibilities (CRO MIC of 0.032 μg/ml and CFM MICs of 0.63 and 0.125 μg/ml) and a penA mosaic XXXIV allele, and in 2007 the first ESC-DS ST1407 strains were identified in Ontario, Canada. A subclade of isolates in clade A with penA mosaic XXXV allele included two isolates from 2008, with ST1407 and ST3779, each having moderate ESC susceptibilities (MICs of 0.032 μg/ml), and one ST1407 isolate from 2010 that was highly susceptible to ESC (MIC of 0.008 μg/ml). The ESC-resistant European strain F89 (isolate number 34842) was also part of clade A and had penA mosaic XXXIV allele with an A501P mutation (see Table S4 in the supplemental material). Most recently in 2012, ESC-DS was seen in a distinct lineage of clade G consisting of isolates that were ST7989 (NG-MAST group 2) and had penA nonmosaic XIII allele.
Five ESC-DS strains were also resistant to AZM. These five strains included one isolate clustering between clades H and I with an MIC of 2 μg/ml and no 23S rRNA mutations but with the mtrR −35 A deletion present, two isolates of clade A with MICs of 2 and 8 μg/ml and C2599T mutations in 2/4 and 4/4 of the 23S rRNA alleles, and two isolates with extremely high MICs of ≥64 μg/ml and ≥256 μg/ml, both with A2143G mutations in all alleles, clustering between clades F and G, and clustering in clade H, respectively.
N. gonorrhoeae multiantigen sequence typing (NG-MAST).
All 79 NG-MAST types determined by the NG-MAST PCR method (including 3 novel types, ST10443, ST10469, and ST10476) matched those determined in silico from the WGS data and clustered into 10 closely related NG-MAST groups (Fig. 2). Clade A (Fig. 1) consisted almost entirely of ST1407-like strains (NG-MAST group 5; blue in Fig. 2) with a single non-ST1407-like NG-MAST type ST4383 (NG-MAST group 9; light green in Fig. 2) present. The 35 ST1407 strains were distributed throughout the clade, and other closely related NG-MAST types such as ST4985, ST3158, and ST5643 formed subclades. Isolates of clades B, C, D, G, H, and L were mainly of NG-MAST group 2 (red in Fig. 2), with specific NG-MAST types correlating into individual clades, with other NG-MAST group 2 isolates falling into clade I and independently between clades. Similarly, clade E consisted largely of ST2992 isolates (NG-MAST group 10; gray in Fig. 2), with other NG-MAST group 10 strains clustered independently and in clade I. Clade F was composed of a variety of NG-MAST types, all of which were members of NG-MAST group 4 (purple in Fig. 2); however, NG-MAST group 4 strains also clustered distantly throughout the phylogenomic tree.
Antimicrobial resistance molecular markers.
The known ESC-DS molecular determinants that have been reported to influence cephalosporin susceptibility, including penA, mtrR, and porB mutations, do not fully account for the ESC susceptibilities observed in this study (Table 2). Of the 114 isolates that had CRO MICs of 0.032 to 2 μg/ml, the factors having the greatest influence on susceptibility were the penA mosaic allele (SENS/SPEC = 61%/98%), the porB G120K mutation (SENS/SPEC = 93%/94%), and the mtrR −35 A deletion (SENS/SPEC = 91%/86%). A combination of markers such as the penA mosaic allele and/or porB G120K mutation (SENS/SPEC = 96%/94%), penA mosaic allele and/or mtrR −35 A deletion (SENS/SPEC = 94%/85%); or porB G120K mutation and/or mtrR −35 A deletion (SENS/SPEC = 99%/85%) slightly improved the correlation with lower CRO susceptibility.
TABLE 2.
Molecular profiles associated with decreased susceptibility to ceftriaxone
| Molecular marker associated with resistancea |
No. of isolates in the ceftriaxone MIC range groupb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| penA mosaic |
porB |
mtrR |
ponAL421P | |||||||||
| G120K | A121c | −35A deletion | A39T | G45D | Very low | Low | Moderate | High | Very high | Total | ||
| + | + | + | + | − | − | + | 1 | 25 | 33 | 1 | 60 | |
| + | + | + | + | − | + | + | 2 | 2 | 4 | |||
| + | + | + | − | − | + | + | 2 | 2 | ||||
| + | + | + | − | − | − | + | 1 | 1 | ||||
| + | − | − | + | − | − | + | 3 | 1 | 4 | |||
| − | + | + | + | − | − | + | 2 | 6 | 13 | 21 | ||
| − | + | + | + | − | + | + | 1 | 2 | 6 | 9 | ||
| − | + | + | + | + | + | + | 1 | 1 | ||||
| − | + | + | + | + | − | + | 1 | 1 | ||||
| − | + | + | + | − | − | + | 1 | 1 | ||||
| − | + | + | − | − | + | + | 2 | 2 | ||||
| − | + | + | − | + | − | − | 1 | 1 | ||||
| − | + | + | − | − | + | + | 1 | 1 | ||||
| − | + | + | − | − | − | + | 1 | 1 | ||||
| − | + | + | + | − | − | + | 3 | 3 | ||||
| − | + | + | + | − | − | − | 1 | 1 | ||||
| − | + | + | − | + | − | + | 1 | 1 | ||||
| − | − | + | + | − | − | + | 3 | 2 | 5 | |||
| − | − | − | + | − | − | + | 2 | 1 | 3 | |||
| − | − | + | + | − | + | + | 1 | 1 | ||||
| − | − | + | − | + | − | + | 1 | 1 | ||||
| − | − | + | − | + | − | − | 2 | 1 | 3 | |||
| − | − | − | − | + | − | − | 14 | 6 | 1 | 21 | ||
| − | − | − | − | − | + | + | 3 | 3 | ||||
| − | − | + | − | − | − | − | 9 | 2 | 11 | |||
| − | − | − | − | − | − | − | 16 | 1 | 17 | |||
| Total | 46 | 19 | 47 | 66 | 1 | 179 | ||||||
Symbols: +, present; −, absent.
MIC range categories for ceftriaxone include the following: very low, 0.00025 to 0.004 μg/ml; low, 0.008 to 0.016 μg/ml; moderate, 0.032 to 0.063 μg/ml; high, 0.125 to 0.5 μg/ml; very high, 1 to 2 μg/ml.
porB A121 substitutions include A121D, A121F, A121G, A121N, and A121G.
Isolates that were ESC-DS generally also had the mtrR −35A deletion and the ponA L421P and porB G120K and A121 (includes A121D, -F, -G, -N, and -S) mutations but had a variety of mosaic and nonmosaic penA alleles (Fig. 1). Clade A strains predominantly had the penA mosaic XXXIV allele (n = 59), with a subclade of mosaic type XXXV allele (n = 3), and one strain with a mosaic type X (n = 1) allele, whereas clade F was associated with the penA mosaic type X allele, clade G was associated with the nonmosaic type XIII allele, and clade H was associated with the nonmosaic type XII alleles. Novel penA genotypes were identified in 2 CRO-DS nonclade strains corresponding to a nonmosaic type II allele with an A502V substitution, and the third moderately CFM-susceptible strain had an A550V substitution (see Table S4 in the supplemental material). Other novel penA genotypes observed included the following: a penA nonmosaic I allele with an P413S mutation in an ESC-S isolate of clade K; a penA mosaic XXXIV allele with an A502P mutation in strain F89 of clade A; and a penA nonmosaic V allele with Y201H, G202A, E203G, D204E, and Q214E substitutions seen in 3 isolates of clade B with CRO MICs of 0.032 to 0.063 μg/ml.
Temporal and regional epidemiological tracking.
Clade C consisted of three ST757 strains (Fig. 1), with two isolates having no SNP differences (isolates 33997 and 32902) collected from male patients in central Canada in March and November of 2010. The third isolate (26870) collected 5 years earlier differed from the other two by 53 SNPs indicating a temporal separation of infections. Similarly, clade D consisted of four ST3108 strains, three of which differed by a maximum of 2 SNPs (strains 18361, 31284, and 28541) and were collected between June and September 2006 in central Canada, and the fourth was collected during January 2006 from western Canada and differed by 16 SNPs, demonstrating regional separation within the cluster. A third cluster of three ST3150 strains (32849, 33386, and 33756) formed clade B, differing by a maximum of three SNPs, were collected in January and February of 2010 from female patients in central Canada suggesting temporal, regional, and a possible social network commonality that could be investigated.
DISCUSSION
N. gonorrhoeae isolates are genetically diverse in Canada with the majority of isolates distributed through 12 phylogenomic clades and 28 miscellaneous clinical isolates falling outside these groups. Along with the recent clonal spread of ST1407, isolates of N. gonorrhoeae with decreased ESC susceptibility have also been observed sporadically in Canada since at least 1989 among several distinct lineages of strains, indicating that the spread of ESC resistance in gonorrhea in Canada is not entirely due to clonal expansion nor restricted solely to the presence of a penA mosaic allele in the genome.
The ESC resistance mechanism is a complex, multifaceted system consisting of numerous components contributing to overall levels of susceptibility. The molecular markers previously described as contributing to cephalosporin resistance, such as penA, mtrR, porB, ponA, and pilQ mutations, do not adequately account for the observed phenotypes (4, 9, 10, 15, 20, 24–26, 51). Of these markers identified in silico using the WGS data, the presence of a penA mosaic allele, mtrR −35A deletion and/or a porB G120K mutation were the best indicators of decreased susceptibility to cephalosporins (Table 2).
ESC-DS has been associated with the presence of a penA mosaic allele that evolved through recombination events with other commensal Neisseria species; the penA mosaic allele was first seen in 1998 in Japan as a penA mosaic X allele (see Table S4 in the supplemental material) (9, 52). The penA mosaic XXXIV allele was first reported in the United States in 2008 (52–55) and has spread worldwide. In 2009, an isolate was identified in Japan (H041) with a novel penA mosaic allele, NG-MAST type ST4220, and CRO and CFM MICs of 2 and 8 μg/ml, respectively (9). A second highly ESC-resistant isolate was identified in France in 2010 (F89) with a novel penA mosaic mutation that resembled the penA mosaic XXXIV allele with an A502P substitution, a NG-MAST type ST1407, and CRO and CFM MICs of 2 and 4 μg/ml, respectively (10). Despite being highly ESC-DS, F89 was genetically similar to Canadian clade A strains in the phylogenomic tree (Fig. 1).
In the United States, two lineages of strains with ESC-DS were observed, and they had CRO and CFM MICs of 0.125 and 0.50 μg/ml (35). Strains with ESC-DS in Canada had MIC values similar to those reported in the United States; however, Canadian strains tend to be more broadly distributed and genetically heterogeneous, grouping into 5 lineages with other isolates grouping outside the defined phylogenomic clades. The emergence of CRO-DS in Canada occurred during the early 1990s among a group of sporadic isolates with nonmosaic penA alleles, located between clades H and I within the phylogenomic tree (Fig. 1), followed by the penA mosaic X allele during 2000 to 2004 among strains of clade F and clade A. Clade A was the largest group of isolates associated with ESC-DS consisting of NG-MAST types closely related to ST1407. The penA mosaic XXXIV allele was predominant in this group, with the first isolates having MIC values of 0.032 μg/ml, which was observed in 2005, and then with decreased susceptibility (MIC = 0.125 μg/ml) in 2007.
Despite a recent report of clonal expansion of strains with penA nonmosaic XXII and IX alleles in Saskatchewan, Canada (56), no regional trends could be deduced among the Canadian isolates. Although only general temporal relationships could be established with the emergence of various lineages of ESC-DS strains, recent isolations indicate that these lineages arose independently—most likely from imported sources (57–59). Despite a paucity of social network information available for this study, certain lineages tended to have a greater proportion of isolates isolated from women; these lineages included clades B, J, K and to a lesser extent the nonclade miscellaneous isolates and clade I, suggesting that these lineages may be more likely to be circulating within the heterosexual population.
In the current study, using core SNP comparisons, we identified several isolate pairs exhibiting close relatedness that represent a promising avenue to determine potential targets for novel antimicrobial resistance mechanisms, including isolates of the ST1407-like clade A that had identical resistance marker profiles (penA mosaic XXXIV allele, mtrR −35A deletion, and porB G121K) with variable ESC MIC values.
AZM resistance is of concern, as it is the recommended cotherapy for gonorrhea (5), and AZM resistance with elevated ESC MICs has recently been described in Ontario (14), Quebec, and British Columbia in Canada (see Table S3 in the supplemental material). In contrast to Grad et al. (35) who reported that AZM resistance was clonal in the United States, we observed five geographically separated AZM-resistant strains distributed widely through the phylogenomic tree, indicating that this phenotype is not clonal but arose spontaneously from independent accumulation of mtrR −35 A deletion and 23S rRNA mutations.
Of interest is the international epidemic clone ST1407 (and related NG-MAST types such as ST3150, ST3158, and ST4985) that has elevated ESC MICs and increased frequency of AZM-resistant strains (6, 52, 60) that threaten the efficacy of currently recommended treatments. The ST1407 clone is thought to have originated in Japan and spread globally, with the first report of a ST1407 strain harboring a penA mosaic XXXIV allele identified in the United States in 2008 (10). Although a precise date for the actual emergence of ST1407 is not known, our study indicates emergence of the strain in North America prior to 2005.
Core SNP phylogenomic analysis provides greater resolution when grouping strains; however, the NG-MAST typing scheme is generally concordant with the phylogenomic tree (Fig. 1). Although some NG-MAST groups were distributed among several clades, individual NG-MAST types tended to correlate with specific clades and subclades. The miscellaneous strains that did not cluster into distinct phylogenomic clades similarly were associated with a variety of NG-MAST groups and types. Although this study lacks detailed epidemiological information, the differentiation of clade B, C, and D strains beyond the level of the NG-MAST type demonstrates the utility of WGS to identify and characterize social networks and transmission of strains.
The selection of isolates for WGS was based primarily on ESC-DS, resulting in a data set that was enriched for isolates from larger population jurisdictions, recent collection dates, and to the ST1407-like strain groups that are currently the most prevalent strain circulating within Canada and most likely to be associated with antimicrobial resistance (6, 61). Although 83% of Canadian ESC-DS strains isolated between 1989 and 2006 were included in this study, it is possible that some recent lineages and clonal groups have not been detected because of fewer isolates analyzed from subsequent years. Similarly, although isolates with a variety of NG-MAST types, geographical jurisdictions, and antibiotic susceptibilities were selected, clusters of isolates within these smaller groups may also be underrepresented in the phylogeny.
WGS was used to catalog the distribution and relatedness of ESC-DS strains collected in Canada between 1989 and 2013. Analysis of the data suggested a heterogeneous landscape which is dominated by the global epidemic clone ST1407, yet ESC-DS has arisen independently in several lineages. While there is a high correlation of NG-MAST sequence types to phylogenomic lineages, the latter provides greater resolution and discrimination of isolates. As WGS becomes more widely adopted to identify the molecular relationships among isolates, only the addition of epidemiologic contact tracing can fully leverage the great resolving power of WGS to more effectively inform public health interventions, ultimately reducing the burden of disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary Liu, Pam Sawatzky, and Ravinder Singh from the Streptococcus and Sexually Transmitted Diseases Unit for their laboratory technical assistance; Shaun Tyler, Adam Olson, and the NML Genomics Core and Heather Kent, Franklin Bristow, Aaron Petkau, Josh Adam, Tom Matthews, Philip Mabon, Shane Thiessen, and Natalie Knox of the NML Bioinformatics Core for their next-generation sequencing and analytical expertise. We also thank Patrice Sednaoui of the Institut Alfred Fournier, Centre National de Référence des Gonocoques, Paris, France, for graciously providing N. gonorrhoeae strain F89.
This work was supported by funding from the Public Health Agency of Canada.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02589-14.
REFERENCES
- 1.Public Health Agency of Canada. 2014. Notifiable diseases on-line. Public Health Agency of Canada, Ottawa, Ontario, Canada: Accessed 4 September 2014 http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/charts.php?c=pl. [Google Scholar]
- 2.World Health Organization. 2012. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. World Health Organization, Geneva, Switzerland: http://www.who.int/reproductivehealth/publications/rtis/9789241503501/en/ Accessed 12 June 2012. [Google Scholar]
- 3.Barry PM, Klausner JD. 2009. The use of cephalosporins for gonorrhea: the impending problem of resistance. Expert Opin Pharmacother 10:555–557. doi: 10.1517/14656560902731993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Public Health Agency of Canada. 2013. Canadian guidelines on sexually transmitted infections, gonococcal infections. Revised July 2013. Community Acquired Infections Division, Centre for Communicable Diseases and Infection Control, Public Health Agency of Canada, Ottawa, Ontario, Canada: Accessed 22 July 2014 http://www.phac-aspc.gc.ca/std-mts/sti-its/index-eng.php. [Google Scholar]
- 6.Martin I, Sawatzky P, Allen V, Hoang L, Lefebvre B, Mina N, Wong T, Gilmour M. 2012. Emergence and characterization of Neisseria gonorrhoeae isolates with decreased susceptibilities to ceftriaxone and cefixime in Canada: 2001-2010. Sex Transm Dis 39:316–323. doi: 10.1097/OLQ.0b013e3182401b69. [DOI] [PubMed] [Google Scholar]
- 7.Allen VG, Mitterni L, Seah C, Rebbapragada A, Martin IE, Lee C, Siebert H, Towns L, Melano RG, Lowe DE. 2013. Neisseria gonorrhoeae treatment failure and susceptibility to cefixime in Toronto, Canada. JAMA 309:163–170. doi: 10.1001/jama.2012.176575. [DOI] [PubMed] [Google Scholar]
- 8.Gratrix J, Bergman J, Egan C, Drews SJ, Read R, Singh AE. 2013. Retrospective review of pharyngeal gonorrhea treatment failures in Alberta, Canada. Sex Transm Dis 40:877–879. doi: 10.1097/OLQ.0000000000000033. [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, Nakayama S, Kitawaki J, Unemo M. 2011. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob. Agents Chemother 55:3538–3545. doi: 10.1128/AAC.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unemo M, Golparian D, Nicholas R, Ohnishi M, Gallay A, Sednaouie P. 2012. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Chemother 56:1273–1280. doi: 10.1128/AAC.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unemo M, Golparian D, Syversen G, Vestrheim DF, Moi H. 2010. Two cases of verified clinical failures using internationally recommended first-line cefixime for gonorrhoea treatment, Norway, 2010. Euro Surveill 15(47):pii=19721 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19721. [DOI] [PubMed] [Google Scholar]
- 12.Ison CA, Hussey J, Sankar KN, Evans J, Alexander S. 2011. Gonorrhoea treatment failures to cefixime and azithromycin in England, 2010. Euro Surveill 16(14):pii=19833 http://www.eurosurveillance.org/ViewArticle.aspx?Articled=19833. [PubMed] [Google Scholar]
- 13.Unemo M, Golparian D, Stary A, Eigentler A. 2011. First Neisseria gonorrhoeae strain with resistance to cefixime causing gonorrhoea treatment failure in Austria, 2011. Euro Surveill 16:pii=19998 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19998. [PubMed] [Google Scholar]
- 14.Allen VG, Seah C, Martin I, Melano RG. 2014. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob. Agents Chemother 58:2528–2534. doi: 10.1128/AAC.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg R, Fredlund H, Nicholas R, Unemo M. 2007. Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime and ceftriaxone: association with genetic polymorphisms in penA, mtrR, porB1b, and ponA. Antimicrob. Agents Chemother 51:2117–2122. doi: 10.1128/AAC.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SG, Lee H, Jeong SH, Yong D, Chung GT, Lee YS, Chong Y, Lee K. 2010. Various penA mutations together with mtrR, porB and ponA mutations in Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime or ceftriaxone. J Antimicrob Chemother 65:669–675. doi: 10.1093/jac/dkp505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osaka K, Takakura T, Narukawa K, Takahata M, Endo K, Kiyota H, Onodera S. 2008. Analysis of amino acid sequences of penicillin-binding protein 2 in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime and ceftriaxone. J Infect Chemother 14:195–203. doi: 10.1007/s10156-008-0610-7. [DOI] [PubMed] [Google Scholar]
- 18.Whiley DM, Limnios EA, Ray S, Sloots TP, Tapsall JW. 2007. Diversity of penA alterations and subtypes in Neisseria gonorrhoeae strains from Sydney, Australia, that are less susceptible to ceftriaxone. Antimicrob Agents Chemother 51:3111–3116. doi: 10.1128/AAC.00306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapsall JW, Ndowa F, Lewis DA, Unemo M. 2009. Meeting the public health challenge of multidrug- and extensively drug-resistant Neisseria gonorrhoeae. Expert Rev Anti Infect Ther 7:821–834. doi: 10.1586/eri.09.63. [DOI] [PubMed] [Google Scholar]
- 20.Zhao S, Duncan M, Tomberg J, Davies C, Unemo M, Nicholas RA. 2009. Genetics of chromosomally mediated intermediate resistance to ceftriaxone and cefixime in Neisseria gonorrhoeae. Antimicrob Agents Chemother 53:3744–3751. doi: 10.1128/AAC.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olesky M, Hobbs M, Nicholas RA. 2002. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46:2811–2820. doi: 10.1128/AAC.46.9.2811-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olesky M, Zhao S, Rosenberg RL, Nicholas RA. 2006. Porin-mediated antibiotic resistance in Neisseria gonorrhoeae: ion, solute, and antibiotic permeation through PIB proteins with penB mutations. J Bacteriol 188:2300–2308. doi: 10.1128/JB.188.7.2300-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balashov S, Mordechai E, Adelson ME, Gygax SE. 2013. Multiplex bead suspension array for screening Neisseria gonorrhoeae antibiotic resistance genetic determinants in noncultured clinical samples. J Mol Diagn 15:116–129. doi: 10.1016/j.jmoldx.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Golparian D, Hellmark B, Fredlund H, Unemo M. 2010. Emergence, spread and characteristics of Neisseria gonorrhoeae isolates with in vitro decreased susceptibility and resistance to extended-spectrum cephalosporins in Sweden. Sex Transm Infect 86:454–460. doi: 10.1136/sti.2010.045377. [DOI] [PubMed] [Google Scholar]
- 25.Ropp PA, Hu M, Olesky M, Nicholas RA. 2002. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46:769–777. doi: 10.1128/AAC.46.3.769-777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiley DM, Jacobsson S, Tapsall JW, Nissen MD, Sloots TP, Unemo M. 2010. Alterations of the pilQ gene in Neisseria gonorrhoeae are unlikely contributors to decreased susceptibility to ceftriaxone and cefixime in clinical gonococcal strains. J Antimicrob Chemother 65:2543–2547. doi: 10.1093/jac/dkq377. [DOI] [PubMed] [Google Scholar]
- 27.Zhao S, Tobiason DM, Hu M, Seifert HS, Nicholas RA. 2005. The penC mutation conferring antibiotic resistance in Neisseria gonorrhoeae arises from a mutation in the PilQ secretin that interferes with multimer stability. Mol Microbiol 57:1238–1251. doi: 10.1111/j.1365-2958.2005.04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng LK, Martin I, Liu G, Bryden L. 2002. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46:3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarantonelli L, Borthagaray G, Lee EH, Shafer WM. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob Agents Chemother 43:2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarantonelli L, Borthagaray G, Lee EH, Veal W, Shafer WM. 2001. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtr(R) promoter mutation in Neisseria gonorrhoeae. J Antimicrob Chemother 47:651–654. doi: 10.1093/jac/47.5.651. [DOI] [PubMed] [Google Scholar]
- 31.Vester B, Douthwaite S. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother 45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unemo M, Dillon JR. 2011. Review and international recommendation of methods for typing Neisseria gonorrhoeae isolates and their implications for improved knowledge of gonococcal epidemiology, treatment, and biology. Clin Microbiol Rev 24:447–458. doi: 10.1128/CMR.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin IMC, Ison CA, Aanensen DM, Fenton KA, Spratt BG. 2004. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J Infect Dis 189:1497–1505. doi: 10.1086/383047. [DOI] [PubMed] [Google Scholar]
- 34.Gilmour MW, Graham M, Reimer A, Van Domselaar G. 2013. Public health genomics and the new molecular epidemiology of bacterial pathogens. Public Health Genomics 16:25–30. 10.1159/000342709. doi: 10.1159/000342709. [DOI] [PubMed] [Google Scholar]
- 35.Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, Goldstein E, Weinstock H, Parkhill J, Hanage WP, Bentley S, Lipsitch M. 2014. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohnishi M, Unemo M. 2014. Phylogenomics for drug-resistant Neisseria gonorrhoeae. Lancet Infect Dis 14:179–180. doi: 10.1016/S1473-3099(13)70700-X. [DOI] [PubMed] [Google Scholar]
- 37.Martin I, Jayaraman G, Wong T, Liu G, Gilmour M. 2011. Trends in antimicrobial resistance in Neisseria gonorrhoeae isolated in Canada: 2000-2009. Sex Transm Dis 38:892–898. doi: 10.1097/OLQ.0b013e31822c664f. [DOI] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 39.Ehret JM, Nims LJ, Judson FN. 1996. A clinical isolate of Neisseria gonorrhoeae with in vitro resistance to erythromycin and decreased susceptibility to azithromycin. Sex Transm Dis 23:270–272. doi: 10.1097/00007435-199607000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Centers for Disease Control and Prevention. 2009. Sexually transmitted disease surveillance 2007 supplement. Gonococcal Isolate Surveillance Project (GISP) annual report 2007. Division of STD Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, GA: Accessed 30 November 2010 http://www.cdc.gov/std/gisp2007/gispsurvsupp2007short.pdf. [Google Scholar]
- 41.Unemo M, Fasth O, Fredlund H, Limnios A, Tapsall J. 2009. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J Antimicrob. Chemother 63:1142–1151. doi: 10.1093/jac/dkp098. [DOI] [PubMed] [Google Scholar]
- 42.Parikh R, Mathai A, Parikh S, Chandra Sekhar G, Thomas R. 2008. Understanding and using sensitivity, specificity and predictive values. Indian J Ophthalmol 56:45–50. doi: 10.4103/0301-4738.37595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean AG, Sullivan KM, Soe MM. 2013. OpenEpi: open source epidemiologic statistics for public health version 3.01. www.openepi.com Accessed 8 August 2014.
- 44.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 47.Kislyuk AO, Katz LS, Agrawal S, Hagen MS, Conley AB, Jayaraman P, Nelakuditi V, Humphrey JC, Sammons SA, Govil D, Mair RD, Tatti KM, Tondella ML, Harcourt BH, Mayer LW, Jordan IK. 2010. A computational genomics pipeline for prokaryotic sequencing projects. Bioinformatics 26:1819–1826. doi: 10.1093/bioinformatics/btq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 51.Takahata S, Senju N, Osaki Y, Yoshida T, Ida T. 2006. Amino acid substitutions in mosaic penicillin-binding protein 2 associated with reduced susceptibility to cefixime in clinical isolates of Neisseria gonorrhoeae. Antimicrob Agents Chemother 50:3638–3645. doi: 10.1128/AAC.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Unemo M, Nicholas RA. 2012. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol 7:1401–1422. doi: 10.2217/fmb.12.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandori M, Barry PM, Wu A, Ren A, Whittington WLH, Liska S, Klausner JD. 2009. Mosaic penicillin-binding protein 2 in Neisseria gonorrhoeae isolates collected in 2008 in San Francisco, California. Antimicrob Agents Chemother 53:4032–4034. doi: 10.1128/AAC.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimuta K, Unemo M, Nakayama S, Morita-Ishihara T, Dorin M, Kawahata T, Ohnishi M. 2013. Antimicrobial resistance and molecular typing of Neisseria gonorrhoeae isolates in Kyoto and Osaka, Japan, 2010 to 2012: intensified surveillance after identification of the first strain (H041) with high-level ceftriaxone resistance. Antimicrob Agents Chemother 57:5225–5232. doi: 10.1128/AAC.01295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chisholm SA, Unemo M, Quaye N, Johansson E, Cole MJ, Ison CA, Van de Laar MJW. 2013. Molecular epidemiological typing within the European Gonococcal Antimicrobial Resistance Surveillance Programme reveals predominance of a multidrug-resistant clone. Euro Surveill 18(3):pii=20358 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20358. [PubMed] [Google Scholar]
- 56.Vidovic S, Caron C, Taheri A, Thakur SD, Read TD, Kusalik A, Dillon JR. 2014. Using crude whole genome assemblies of Neisseria gonorrhoeae as a platform for strain analysis: clonal spread of gonorrhea infection in Saskatchewan, Canada. J Clin Microbiol 52:3772–3776. doi: 10.1128/JCM.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muratani T, Akasaka S, Kobayashi T, Yamada Y, Inatomi H, Takahashi K, Matsumoto T. 2001. Outbreak of cefozopran (penicillin, oral cephems, and aztreonam)-resistant Neisseria gonorrhoeae in Japan. Antimicrob Agents Chemother 45:3603–3606. doi: 10.1128/AAC.45.12.3603-3606.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ameyama S, Onodera S, Takahata M, Minami S, Maki N, Endo K, Goto H, Suzuki H, Oishi Y. 2002. Mosaic-like structure of penicillin-binding protein 2 gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob Agents Chemother 46:3744–3749. doi: 10.1128/AAC.46.12.3744-3749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito M, Deguchi T, Mizutani KS, Yasuda M, Yokoi S, Ito SI, Takahashi Y, Ishihara S, Kawamura Y, Ezaki T. 2005. Emergence and spread of Neisseria gonorrhoeae clinical isolates harboring mosaic-like structure of penicillin-binding protein 2 in central Japan. Antimicrob Agents Chemother 49:137–143. doi: 10.1128/AAC.49.1.137-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heymans R, Bruisten SM, Golparian D, Unemo M, De Vries HJC, Van Dama AP. 2012. Clonally related Neisseria gonorrhoeae isolates with decreased susceptibility to the extended-spectrum cephalosporin cefotaxime in Amsterdam, the Netherlands. Antimicrob Agents Chemother 56:1516–1522. doi: 10.1128/AAC.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohnishi M, Watanabe Y, Ono E, Takahashi C, Oya H, Kuroki T, Shimuta K, Okazaki N, Nakayama S, Watanabe H. 2010. Spread of a chromosomal cefixime-resistant penA gene among different Neisseria gonorrhoeae lineages. Antimicrob Agents Chemother 54:1060–1067. doi: 10.1128/AAC.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.