Abstract
Since its first identification, the epizootic avian influenza A H7N9 virus has continued to cause infections in China. Two waves were observed during this outbreak. No cases were reported from Guangdong Province during the first wave, but this province became one of the prime outbreak sites during the second wave. In order to identify the transmission potential of this continuously evolving infectious virus, our research group monitored all clusters of H7N9 infections during the second wave of the epidemic in Guangdong Province. Epidemiological, clinical, and virological data on these patients were collected and analyzed. Three family clusters including six cases of H7N9 infection were recorded. The virus caused severe disease in two adult patients but only mild symptoms for all four pediatric patients. All patients reported direct poultry or poultry market exposure history. Relevant environment samples collected according to their reported exposures tested H7N9 positive. Virus isolates from patients in the same cluster shared high sequence similarities. In conclusion, although continually evolving, the currently circulating H7N9 viruses in Guangdong Province have not yet demonstrated the capacity for efficient and sustained person-to-person transmission.
INTRODUCTION
Since its initial identification on March 2013 (1), the outbreak of highly pathogenic avian influenza A H7N9 virus (H7N9) infection in China has become an important national public health concern. Two waves were observed during this outbreak (2). The initial wave took place between March and May 2013. Infection was diffused from the Yangtze River Delta region to the surrounding areas and inland (3). Thereafter, human infections with avian influenza H7N9 virus slowed, with only a few sporadic cases reported (4, 5). From December 2013, another outbreak of H7N9 infection was identified. Guangdong Province, a subtropical province in southern China, had no reported cases during the first wave of H7N9 outbreak, but it became one of the major outbreak sites in the second wave. As of 8 April 2014, 100 documented human infections (including 29 deaths) had been reported in Guangdong, representing one-fourth of the cumulative cases, sparking concern about whether this lethal pathogen had adapted to humans (6).
Continuing follow-up of contacts of H7N9 cases, especially identification of case clustering, was conducted, since this can provide signals of genetic and transmission ability changes for the virus. During the first wave of H7N9 infection between March and May 2013, four H7N9 clusters were identified and reported (7). Limited person-to-person transmission was well documented in at least one cluster, which occurred in Wuxi in eastern China. In this family cluster, the patient with the index case became ill 6 days after his last exposure to poultry. The second patient, a daughter who had no known exposure to poultry, developed symptoms 6 days after her last contact with her father (8). Epidemiological investigations indicated that the infection of the daughter probably resulted from contact with her father (the index patient) when she provided unprotected bedside care. This finding suggested that the H7N9 virus in this cluster was able to be transmitted from person to person. To better explore the transmission potential of the H7N9 virus, the H7N9 case clusters in the second wave in Guangdong Province were monitored. Epidemiological, clinical, and virological data on these patients were collected and analyzed.
MATERIALS AND METHODS
Syndromic surveillance in humans.
Enhanced surveillance for influenza A/H7N9 virus was conducted in 28 sentinel hospitals and 23 collaborating laboratories in 21 cities from 16 April 2013 in Guangdong, China. All specimens collected from patients who developed febrile respiratory symptoms were first tested for avian influenza A virus. Positive cases were then tested for influenza A virus subtypes (H1, H5, and H7) using commercial real-time reverse transcription-PCR (RT-PCR) methods in local Center for Disease Control and Prevention (CDC) laboratories and further verified by the Guangdong Provincial CDC. H7-positive specimens were further tested for the presence of N9 gene segments by real-time RT-PCR (1). Through the provincial sentinel surveillance system, all laboratory-confirmed H7N9 infections in Guangdong Province are reported to the Guangdong Center for Disease Control and Prevention.
Definitions.
Laboratory evidence of H7N9 infection was defined as H7N9 viral RNA detection or virus isolation from respiratory specimens. As described previously (7), all household members and health care workers who had close contact (within 1 m) with the confirmed case without effective personal protection at any time from the day before illness onset to when the patient was isolated in the hospital were treated as close contacts and placed under medical observation for 7 days. A cluster of H7N9 infection is defined as at least two close-contact persons having laboratory-confirmed H7N9 infection.
Epidemiological investigation.
Epidemiological data of patients with confirmed H7N9 viral infection and their contacts were obtained through interviews and reviews of medical records. Information such as timelines of progression of the illness and possible exposure history before the onset of illness was collected. Information for H7N9 clusters identified in other provinces during the first wave of outbreak was obtained through public sources.
Sample collection.
Throat swabs and tracheal aspirates or sputum (if available) were collected and tested for potential H7N9 infections. Relevant poultry and environmental samples, including poultry manure, epilator swabs, sewage swabs, and chopping swabs, were collected according to potential exposure history.
For the use of all above-mentioned human samples and personal information, written informed consent from all participants (their parents or legal guardian) involved in the research was obtained. This study was approved by the ethics committee of the Guangdong Provincial Center for Disease Control and Prevention and was in compliance with the Helsinki Declaration.
Laboratory investigation.
Specimens in viral transport medium were tested by real-time reverse transcription PCR (rRT-PCR) analysis in the biosafety level 2 (BSL2) laboratory of the Guangdong Province Center for Disease Control and Prevention (1, 9). H7N9-positive swab materials and sputa were blindly passaged for 2 or 3 generations in 9- or 10-day-old embryonated chicken eggs for virus isolation in the BSL3 laboratory. Hemagglutination-positive allantoic fluids were collected and subtyped by hemagglutination and neuraminidase (NA) inhibition (HAI and NAI) assays using a panel of reference antisera as described by Huang et al. (10). For full-genome sequencing, viral RNA extraction was first performed by using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany). Reverse transcription and PCR amplification of all 8 gene segments were performed by using PathAmp FluA preamplification reagents (Life Technologies, MA). Amplicons were purified and quantitated (Ampure XP purification kit; Beckman, CA). The genomic libraries were prepared with the Ion Xpress Plus fragment library kit (Life Technologies). Enzymatic fragmentation was used for the 200-bp read protocol with a 10-min incubation time. Samples were bar-coded with the Ion Xpress bar code adapter 1–32 kit (Life Technologies). Automated template preparation was performed by using the Ion OneTouch 2 system (Ion PGM Template OT2 200 kit; Life Technologies). Final sequencing was performed with the Ion PGM Sequencing 200 kit v2 (Life Technologies) using the Ion 316 Chip V2 on an Ion PGM system. Full-genome sequences were submitted to the Global Initiative on Sharing All Influenza Data (GISAID). The isolation identifiers and information for each virus isolate are listed in Table S1 in the supplemental material.
Phylogenetic analysis.
Maximum likelihood (ML) trees were estimated for hemagglutinin (HA) and NA gene segments by using MEGA version 6.06 with the General Time Reversible GTR+G model (6, 9, 11). To assess the robustness of individual nodes on phylogenetic trees, a bootstrap resampling process was used with 1,000 bootstrap replicates.
RESULTS
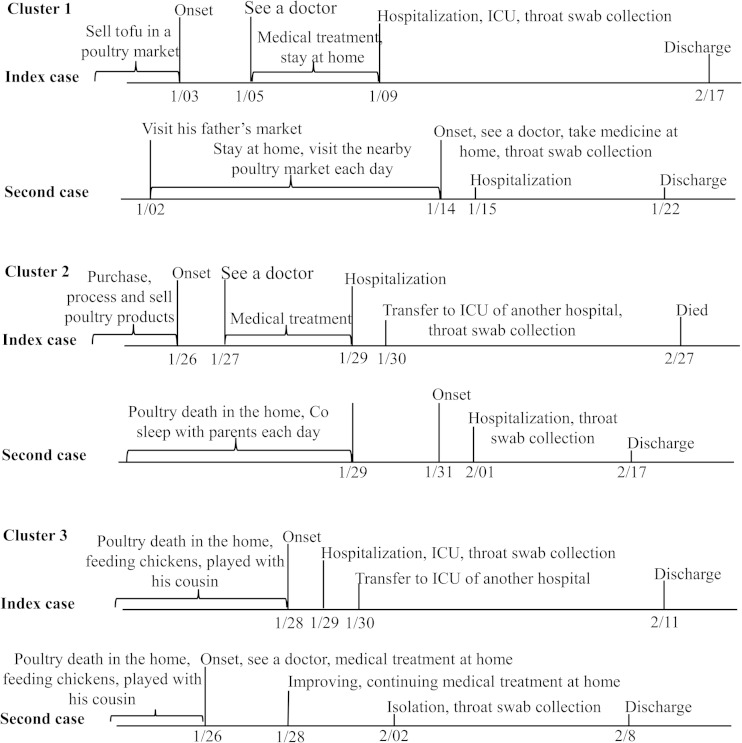
From August 2013 to 8 April 2014, during the second wave of H7N9 infection, 100 cases of human avian influenza A (H7N9) virus infection were officially reported to the Guangdong Center for Disease Control and Prevention. A total of 3,228 close contacts were followed up for a minimum period of 7 days after their last exposure to the cases. Three cluster outbreaks were identified. Each had 2 household members, and no health care workers were involved. Figure 1 summarizes key clinical milestones of each patient, and Table 1 shows their epidemiological and clinical characteristics.
FIG 1.
Time lines for three clusters of H7N9 virus infection in Guangdong Province, China. ICU, intensive care unit.
TABLE 1.
Epidemiological and clinical data for three clusters of H7N9 virus infection in Guangdong Province, Chinaa
| Characteristic | Cluster 1 |
Cluster 2 |
Cluster 3 |
|||
|---|---|---|---|---|---|---|
| Index case | Second case | Index case | Second case | Index case | Second case | |
| Age (yrs)/sex | 29/M | 5/F | 37/M | 2.5/F | 5/M | 4/F |
| Relation | Father | Daughter | Father | Daughter | Cousin | Cousin |
| Onset of illness (mo-day-year) | 1-3-2014 | 1-14-2014 | 1-25-2014 | 1-31-2014 | 1-28-2014 | 1-26-2014 |
| Admission to hospital (mo-day-year) | 1-9-2014 | 1-15-2014 | 1-29-2014 | 2-1-2014 | 1-29-2014 | 2-3-2014 |
| Sign(s) of illness | Fatigue, myalgia | Fever | Chills, fever, sore throat, and dizziness | Fever, rhinorrhea, and cough | Fever | Fever, cough, sneezing, and rhinorrhea |
| Temp (°C) | 40.1 | 38.4 | 39.5 | 39.5 | 38.0 | 39.3 |
| White cell count (×109/liter) | 2.92 | 7.90 | 2.27 | 12.98 | 9.69 | N/A |
| Chest radiography | Bilateral pneumonia | N/A | Bilateral pneumonia | N/A | Bronchopneumonia | N/A |
| Mechanical ventilation | Yes | No | Yes | No | No | No |
| Oseltamivir treatment | Yes | Yes | Yes | Yes | Yes | Yes |
| Type of exposure | Worked in live poultry market | Visited live poultry market | Handled poultry products; poultry death at home | Poultry death at home | Poultry death at home | Poultry death at home |
| Outcome | Recovery | Recovery | Death | Recovery | Recovery | Recovery |
M, male; F, female; N/A, not applicable.
Cluster 1.
The index patient was a 29-year-old man who lived with his wife and two daughters in Guangzhou, Guangdong. He felt fatigue and myalgia on 3 January and presented to a clinic with fever (40.1°C) and sore throat on 5 January. Diagnosed with an upper respiratory tract infection, this man received 4 days (5 to 8 January) of intravenous antibiotic therapy. His condition did not improve during treatment. On 9 January, due to continued fever, more severe cough, and shortness of breath, he attended an outpatient department of Guangzhou South Hospital. Chest radiography revealed bilateral pneumonia. He was transferred to the intensive care unit on the day of admission because of progressive respiratory distress and hypoxemia. Mechanical ventilation was required. Oseltamivir (75 mg twice a day), doxycycline, ceftriaxone sodium, and other symptomatic treatments were used. His condition improved, and he was discharged on 17 February. A throat swab collected on 9 January tested positive for H7N9 by RT-PCR. H7N9 virus was successfully isolated from this specimen.
The second case was the index patient's 5-year-old daughter. She developed fever (38.4°C) and went to the community hospital on 14 January. Although routine blood testing identified no abnormality, she was given oseltamivir (75 mg twice a day) as a precaution. This girl was admitted to the Guangzhou South Hospital on 15 January. Her condition improved gradually, and on 22 January she was discharged in good health. H7N9 infection was confirmed by RT-PCR from a throat swab obtained on the day she attended the hospital (14 January), followed by successful viral isolation.
As a self-employed businessman, the index patient sold tofu in a live poultry market from which H7N9-positive environmental samples have since been collected (see Table S1 in the supplemental material). This market was not close to his home, and his daughter seldom visited this market. The last time she recalled going to the market was 2 January (12 days before she attended the hospital with fever). The girl did go to a live market nearby with her grandmother to purchase food; however, environmental samples collected there were negative for H7N9. From 4 to 8 January during his treatment, the index patient stayed at home every day and slept with his daughter at noon. No other ill persons were identified out of the 38 contacts of these two patients (6 household members and neighbors, 3 friends who visited the hospital, 24 health care workers, and 5 ward mates).
Cluster 2.
Cluster 2 included two of six family members living together in a rural village of Zhongshan, Guangdong Province. The index patient was a 37-year-old man who ran a cooked-food stall in the town. On 26 January, he developed chills, fever, sore throat, and dizziness. He went to the outpatient department of a community hospital on the second day of his illness. Examination revealed a maximum body temperature of 39.7°C, white blood cell (WBC) count of 4.8 × 109/liter, and lymphocyte count of 1.3 × 109/liter. The patient was diagnosed with an upper respiratory infection and treated with compound aminophenazone and barbital for 3 days (26 to 28 January). He did not respond well to treatment, and cough and swelling of the tonsils developed. He was referred to a county-level hospital on 29 January and began to take oseltamivir (75 mg twice a day). Because of progressive respiratory distress and persistent hyperpyrexia, the patient was transferred again to a tertiary hospital in Zhongshan on 30 January. With a diagnosis of severe pneumonitis and type I acute respiratory distress syndrome, he was treated in the hospital's intensive care unit. Mechanical ventilation was required. This patient died on 27 February. A throat swab collected on 30 January tested H7N9 positive by RT-PCR. H7N9 virus was also isolated from the same specimen.
The second case was the 2-year-old daughter of the index patient. She developed fever, rhinorrhea, and cough on 31 January, 6 days after the onset of illness in her father. As a close contact, on 1 February the girl was directly admitted to the tertiary hospital in Zhongshan. Initial routine testing revealed a maximum body temperature of 39.5°C, a WBC count of 12.98 × 109/liter, a neutrophilic granulocyte level of 80.44%, and a lymphocyte level equal to 9.64%. She was treated in isolation. Oseltamivir (75 mg twice a day) was administered. Her condition improved gradually, and on 17 February she was discharged in good health. H7N9 infection was confirmed by RT-PCR from a throat swab obtained on the second day of illness (1 February), followed by successful viral isolation.
The index patient was married and lived with his wife, two daughters, mother, and sister. He and his wife ran a cooked-food stall supplying take-out food in the town. His daily work was to purchase process and sell roast chicken, duck, and goose. All the poultry was processed in his home. There were free-range household chickens kept in the yard of their house, and some had died in the last month before he got sick. The second patient, the daughter of the index patient, spent most of her time at home or in a day care center. She coslept with her parents until her father was hospitalized. Sixteen close contacts of these cases were identified (8 health care workers, 4 relatives, and 4 household members). All remained well during the 7-day surveillance period. Environmental samples collected in the market where the index patient purchased stock and in their house tested H7N9 positive (see Table S1 in the supplemental material).
Cluster 3.
Cluster 3 included 2 of 8 family members living together in a private building in the suburb of Zhaoqing, Guangdong Province, China. The first identified patient was a 5-year-old boy who developed fever (38.0°C) on 28 January. Accompanied by his mother, the boy attended Huaiji County People's Hospital on the second day of illness (29 January). Routine blood testing revealed a normal but high value WBC count (9.69 × 109/liter). He was hospitalized with a diagnosis of bronchopneumonia. A positive reaction was observed in a rapid influenza diagnostic test. This boy was suspected as having avian influenza virus infection and treated with oseltamivir (75 mg twice a day). On the third day of illness (30 January), he was transferred to a tertiary hospital in Zhaoqing and treated in isolation. His condition gradually improved, and the boy was discharged on the 11 February. H7N9 virus was confirmed by RT-PCR, and virus was isolated from a throat swab collected on the second day of his illness (29 January).
The second patient in the cluster was a 4-year-old girl who was recognized coincidentally during the medical observation of close contacts of her cousin (the index patient). According to her medical records, the girl became sick and had sought medical treatment in Huaiji County People's Hospital on 26 January, 3 days before the onset of illness in her cousin. Fever (a maximum body temperature of 39.3°C), cough, sneezing, and rhinorrhea were reported at the onset of illness. Diagnosed with tonsillitis and upper respiratory infection, the girl was treated by intravenous injection of ceftizoxime, Xuebijing, and ribavirin. Paracetamol was administered for 3 days (26 to 28 January). On day 3, her body temperature dropped to normal but a severe cough remained. Oral liquid containing Scutellaria baicalin was used. Her condition gradually improved. No clear and obvious clinical symptom or sign was observed on 2 February, the day the girl was isolated as a close contact of her cousin. The presence of H7N9 virus was confirmed by RT-PCR, and virus was isolated from a throat swab.
The two cases in this cluster involve cousins in an extended family. Aside from their own parents, the index and second patient lived with their grandmother and aunt in separate apartments within an independent house. Their grandma fed 20 chickens on the top floor, 2 of which died on 17 January (9 days before the onset of symptoms in the 5-year-old girl). In their daily life, feeding chickens was reported to be one of the children's favorite pastimes. These two patients also commonly played with each other even during their illness. Direct contact between them lasted until the boy (index patient) was admitted to the hospital. Eighteen close contacts of these patients were identified (8 health care workers, 4 relatives, and 6 household members). All remained well during a 7-day surveillance period. Environmental samples collected around where the chickens were raised were H7N9 positive (see Table S1 in the supplemental material).
Clinical findings.
All three clusters were family clusters that occurred in the first month of 2014, of which 2 were father-daughter and 1 cousin-cousin (Table 1). Of the six patients identified, 4 were pediatric cases. The most common early symptom of illness was fever (5 of 6). For pediatric cases, only mild symptoms developed, and all pediatric patients recovered. In addition to fever, cough and rhinorrhea were also common symptoms in pediatric cases (2 of 4). For the two adult patients, serious symptoms, including bilateral pneumonia and respiratory distress, developed, and mechanical ventilation was required.
H7N9 testing.
All six patients in the three clusters reported a history of direct contact with poultry or a poultry market before symptom onset. To test the presence of influenza A (H7N9) viral RNA in the environment, throat and anal swabs of poultry were taken along with swabs from feces, waste, and sewage at associated live poultry markets or yards. Of the 122 samples collected, 17 samples were H7N9 positive (see Table S1).
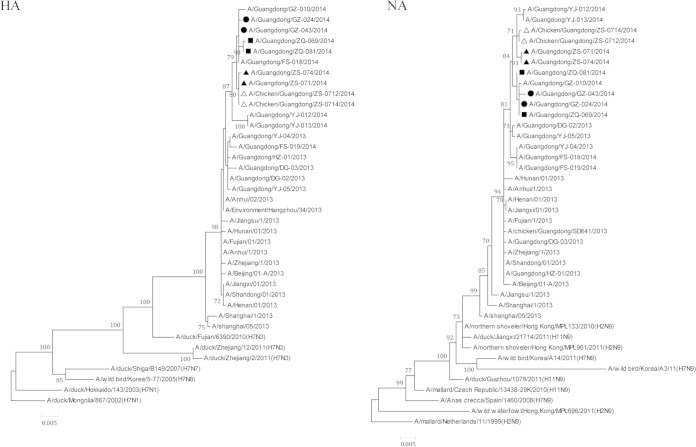
H7N9 viral strains were successfully isolated from all six patients, and whole genomes were sequenced based on next-generation sequencing (NGS) (see Table S1). Phylogenetic analyses showed that strains isolated from patients in the same cluster shared high sequence similarities (see Table S2), and all of the eight internal genes in viral strains isolated from same cluster were clustered together in the phylogenetic trees (Fig. 2; see also Fig. S2 in the supplemental material). Two viral strains [A/Ck/Guangdong/ZS-0712/2014(Mix) and A/Ck/Guangdong/ZS-0714/2014(Mix)] were also successfully isolated from poultry samples. Both were associated with the second cluster (from live poultry market) and isolated as the viral mixtures including H7N9 and H9N2 viruses. Therefore, for these two strains, only HA and NA segments were sequenced. High sequence similarities were also observed between these two viruses and viral strains from index patient and the second patient of cluster 2 (Fig. 2).
FIG 2.
Phylogenetic tree of the HA gene and NA gene of the H7N9 virus isolated from three clusters of H7N9 virus infection in Guangdong Province, China. Nucleotide sequences were analyzed using the maximum likelihood method. Supporting bootstrap values greater than 60 are shown. Scale bar indicates nucleotide substitutions per site. H7N9 viruses isolated from the first, second, and third clusters are marked with circles, solid triangles, and squares, respectively. The viral strains isolated from chicken samples associated with the second cluster are indicated by open triangles.
Several key functional amino acid sites in viral proteins associated with interspecies transmission or drug resistance were closely analyzed. Both human and chicken isolates had substitutions of Gly186Val and Gln226Leu (H3 numbering) at the receptor binding site of HA, which has been reported to contribute to the high-affinity binding of H5 and H7 viruses to the human receptor (12). Ser31Asn mutation in M protein (associated with adamantane resistance) was noted in all human isolates, but Arg292Lys substitution in the NA gene associated with oseltamivir and zanamivir resistance was not recorded, suggesting that oseltamivir, zanamivir, and peramivir should still be active. Interestingly, Glu627Lys substitution on PB2 resulting in mammalian host adaptation was observed in all human isolates except A/Guangdong/ZQ-069/2014(H7N9), which preserves glutamic acid at position 627 of PB2.
DISCUSSION
Since its first identification, the unprecedented epizootic H7N9 virus infection in China has attracted international public health concern. Virology analyses indicate that the currently circulating H7N9 viruses have in fact evolved over time (2, 13), and this evolution raises questions as to whether this lethal pathogen has the capacity for efficient and sustained person-to-person transmission. Since a cluster of cases may be the first suggestion of a viral or epidemiological change (14, 15), our group monitored all close contacts of patients with H7N9 and documented clusters of H7N9 infections in the second wave of the H7N9 outbreak in Guangdong Province.
Studies of four H7N9 clusters that occurred between March and May 2013 (7) found limited person-to-person transmission. For the cluster in Shandong Province, epidemiological information was insufficient to determine whether person-to-person transmission had occurred, although the environment or other sources of infection could not be completely ruled out (16). No additional H7N9 clusters during the first wave of the H7N9 outbreak have been identified in China. This research focused on H7N9 clusters that occurred in Guangdong Province during the second H7N9 outbreak. All cases in these three clusters had a confirmed history of exposure to poultry or poultry markets. Relevant environmental samples collected according to their exposures tested H7N9 positive. In clusters 2 and 3, the durations between the onset of the index and the second case were within the documented average incubation period of ≤6 days (17, 18), suggesting that simultaneous and/or common sources of infection may have been involved. For cluster 1, we were not able to determine the source of infection for the second case. We believe that the most likely source of infection for this case was the poultry market the index patient worked in, although the last time the second patient visited there was 12 days before her illness. Significantly longer incubation times have been reported for cases with exposures on multiple days (18). However, the possibility that the daughter was infected by her father during unprotected exposure could not be completely ruled out since highly identical viruses were isolated from these two patients. Considering that a large number of traced contacts associated with confirmed cases have not been diagnosed as infected with the H7N9 virus, and both the index and secondary cases in these clusters have direct poultry or poultry market exposure history, we believe that the three newly occurring clusters in Guangdong Province do not provide sufficient evidence to indicate human-to-human transmission and that transmission resulted from exposure to risk sources, confirming previous research indicating that visiting live poultry markets or having direct contact with poultry is a primary risk factor for H7N9 infection (19).
Clusters of viral infection are also of interest because they can provide information about the likelihood of genetic susceptibility (20). Studies for other avian influenza infections such as H5N1 clusters revealed that H5N1 infection is influenced by host genetic susceptibility (21). Household contacts who were first-degree relatives of the index patient were more likely to become secondary cases. This finding was also obtained in two of the clusters reported in this study. Of the seven H7N9 clusters, including three in this study and four reported previously (7), all were familial clusters and no health care workers were involved. Patients within each cluster were genetically linked except for 1 cluster, in which a wife and her husband were identified. One possible explanation is that cases within the same cluster are more likely to have common exposure history. However, the preponderance of genetically related cases in clustering outbreaks, with 6 of 7 clusters involving cases genetically linked, suggests genetic susceptibility as an important contributor to H7N9 infection. Further studies are urgently needed to identify the genetic mechanism for H7N9 infection.
Our three clusters included four pediatric patients with clinically mild disease. This finding is consistent with the case series of H7N9 infections detected through China's national sentinel surveillance system for influenza-like illness (22) which demonstrated that all younger patients described mild symptoms. Cowling et al. also suggested a preliminary inference on age-specific seriousness of human disease caused by H7N9 infections (23). It remained obscure why almost all H7N9 mortality has occurred in older individuals. Worobey et al. hypothesized that anamnestic recall of immune responses to the HA stalk antigens of initial childhood exposure may underlie this pattern by predisposing patients to severe disease when they encounter H5N1 or H7N9 viruses with group-mismatched HA but strongly protecting them when these viruses have group-matched HA proteins (24). Our findings reinforced the need to extend the recent H7N9 surveillance, which focused mainly on individuals who were hospitalized with severe symptoms or pneumonia. The potentially large number of unidentified cases is of great importance, as they may serve as a reservoir for continued viral spread in the population, and should be considered when the government develops and implements public health interventions. Clinical features of H7N9 clustering infections were generally similar to those in sporadic infectious patients. Fever and cough are the most common symptoms (25). Previous clinical findings in 111 cases of H7N9 infections showed that like patients with other avian influenza infections, those with H7N9 virus infections were not reported to have a sore throat or rhinorrhea (26). However, rhinorrhea was observed in two of the four pediatric patients in this study. More comprehensive assessment of the clinical features of pediatric patients requires additional case data.
Our study analyzed the clustering of cases during the second wave of H7N9 infection in Guangdong Province in 2014. Epidemiological information suggests common exposures to sources of infection and that host genetic susceptibility may contribute to the H7N9 infection, consistent with other avian influenza virus infections. Importantly, this analysis of the H7N9 virus circulation in 3 clusters identified in Guangdong Province in 2014 provides no evidence that the virus identified in the outbreak period has the capacity for efficient and sustained person-to-person transmission.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by 12th 5-year-major-projects of China's Ministry of Public Health, grant no. 2012zx10004-213, and by the PREDICT Surveillance Animal Human Interface Project of GVF, grant no. 06-09-057-02.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02322-14.
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 2.Meng Z, Han R, Hu Y, Yuan Z, Jiang S, Zhang X, Xu J. 2014. Possible pandemic threat from new reassortment of influenza A(H7N9) virus in China. Euro Surveill 19:20699. [DOI] [PubMed] [Google Scholar]
- 3.Tang RB, Chen HL. 2013. An overview of the recent outbreaks of the avian-origin influenza A (H7N9) virus in the human. J Chin Med Assoc 76:245–248. doi: 10.1016/j.jcma.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Tu C, Fu L, Tang R, He T, Chen J, Fang Y, Wang J, Huang Z. 2014. The first case of avian influenza A (H7N9) virus occurring in the autumn season, China. Am J Infect Control 42:212–213. doi: 10.1016/j.ajic.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Yang P, Wang Q, Pang X, Chen L, Tian L, Deng Y. 2013. A case of avian influenza A (H7N9) virus occurring in the summer season, China. J Infect 67:624–625. doi: 10.1016/j.jinf.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Wu J, Guan D, Yi L, Zeng X, Zou L, Liang L, Ni H, Zhang X, Lin J, Ke C. 2014. Genetic changes of reemerged influenza A(H7N9) viruses, China. Emerg Infect Dis 20:1582–1583. doi: 10.3201/eid2009.140250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, Meng L, Hong Z, Tu W, Cao Y, Li L, Ding F, Liu B, Wang M, Xie R, Gao R, Li X, Bai T, Zou S, He J, Hu J, Xu Y, Chai C, Wang S, Gao Y, Jin L, Zhang Y, Luo H, Yu H, Wang X, Gao L, Pang X, Liu G, Yan Y, Yuan H, Shu Y, Yang W, Wang Y, Wu F, Uyeki TM, Feng Z. 2014. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 370:520–532. doi: 10.1056/NEJMoa1304617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. 2013. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ 347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Wu J, Zeng X, Guan D, Zou L, Yi L, Liang L, Ni H, Kang M, Zhang X, Zhong H, He X, Monagin C, Lin J, Ke C. 2014. Continuing reassortment leads to the genetic diversity of influenza virus H7N9 in Guangdong, China. J Virol 88:8297–8306. doi: 10.1128/JVI.00630-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang K, Zhu H, Fan X, Wang J, Cheung CL, Duan L, Hong W, Liu Y, Li L, Smith DK, Chen H, Webster RG, Webby RJ, Peiris M, Guan Y. 2012. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J Virol 86:6075–6083. doi: 10.1128/JVI.06389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 13.Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5:3142. doi: 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]
- 14.The Health Protection Agency (HPA) UK Novel Coronavirus Investigation Team. 2013. Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill 18:20427. [DOI] [PubMed] [Google Scholar]
- 15.Kandun IN, Wibisono H, Sedyaningsih ER, Yusharmen Hadisoedarsuno W, Purba W, Santoso H, Septiawati C, Tresnaningsih E, Heriyanto B, Yuwono D, Harun S, Soeroso S, Giriputra S, Blair PJ, Jeremijenko A, Kosasih H, Putnam SD, Samaan G, Silitonga M, Chan KH, Poon LL, Lim W, Klimov A, Lindstrom S, Guan Y, Donis R, Katz J, Cox N, Peiris M, Uyeki TM. 2006. Three Indonesian clusters of H5N1 virus infection in 2005. N Engl J Med 355:2186–2194. doi: 10.1056/NEJMoa060930. [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Bi Z, Wang X, Li Z, Ding S, Wang L, Pei Y, Song S, Zhang S, Wang J, Sun D, Pang B, Sun L, Jiang X, Lei J, Yuan Q, Kou Z, Yang B, Shu Y, Yang L, Li X, Lu K, Liu J, Zhang T, Xu A. 2014. One family cluster of avian influenza A(H7N9) virus infection in Shandong, China. BMC Infect Dis 14:98. doi: 10.1186/1471-2334-14-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowling BJ, Jin L, Lau EH, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, Ni MY, Zhang Q, Ip DK, Yu J, Li Y, Wang L, Tu W, Meng L, Wu JT, Luo H, Li Q, Shu Y, Li Z, Feng Z, Yang W, Wang Y, Leung GM, Yu H. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382:129–137. doi: 10.1016/S0140-6736(13)61171-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Xu K, Ren DF, Ai J, Ji H, Ge AH, Bao CJ, Shi GQ, Shen T, Tang FY, Zhu YF, Zhou MH, Wang H. 24 February 2014. Probable longer incubation period for human infection with avian influenza A(H7N9) virus in Jiangsu Province, China, 2013. Epidemiol Infect doi: 10.1017/S0950268814000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Wu JT, Cowling BJ, Liao Q, Fang VJ, Zhou S, Wu P, Zhou H, Lau EH, Guo D, Ni MY, Peng Z, Feng L, Jiang H, Luo H, Li Q, Feng Z, Wang Y, Yang W, Leung GM. 2014. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet 383:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horby P, Sudoyo H, Viprakasit V, Fox A, Thai PQ, Yu H, Davila S, Hibberd M, Dunstan SJ, Monteerarat Y, Farrar JJ, Marzuki S, Hien NT. 2010. What is the evidence of a role for host genetics in susceptibility to influenza A/H5N1? Epidemiol Infect 138:1550–1558. doi: 10.1017/S0950268810000518. [DOI] [PubMed] [Google Scholar]
- 21.Aditama TY, Samaan G, Kusriastuti R, Purba WH, Misriyah Santoso H, Bratasena A, Maruf A, Sariwati E, Setiawaty V, Cook AR, Clements MS, Lokuge K, Kelly PM, Kandun IN. 2011. Risk factors for cluster outbreaks of avian influenza A H5N1 infection, Indonesia. Clin Infect Dis 53:1237–1244. doi: 10.1093/cid/cir740. [DOI] [PubMed] [Google Scholar]
- 22.Ip DK, Liao Q, Wu P, Gao Z, Cao B, Feng L, Xu X, Jiang H, Li M, Bao J, Zheng J, Zhang Q, Chang Z, Li Y, Yu J, Liu F, Ni MY, Wu JT, Cowling BJ, Yang W, Leung GM, Yu H. 2013. Detection of mild to moderate influenza A/H7N9 infection by China's national sentinel surveillance system for influenza-like illness: case series. BMJ 346:f3693. doi: 10.1136/bmj.f3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowling BJ, Freeman G, Wong JY, Wu P, Liao Q, Lau EH, Wu JT, Fielding R, Leung GM. 2013. Preliminary inferences on the age-specific seriousness of human disease caused by avian influenza A(H7N9) infections in China, March to April 2013. Euro Surveill 18:20475. [PMC free article] [PubMed] [Google Scholar]
- 24.Worobey M, Han GZ, Rambaut A. 2014. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A 111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Yu H, Horby PW, Cao B, Wu P, Yang S, Gao H, Li H, Tsang TK, Liao Q, Gao Z, Ip DK, Jia H, Jiang H, Liu B, Ni MY, Dai X, Liu F, Van Kinh N, Liem NT, Hien TT, Li Y, Yang J, Wu JT, Zheng Y, Leung GM, Farrar JJ, Cowling BJ, Uyeki TM, Li L. 2014. Comparison of patients hospitalized with influenza A subtypes H7N9, H5N1, and 2009 pandemic H1N1. Clin Infect Dis 58:1095–1103. doi: 10.1093/cid/ciu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.