Abstract
The evolution of pulmonary disease in cystic fibrosis (CF) usually begins when bacteria get trapped in mucus in the lungs and become established as a chronic infection. While most CF patients experience periods of stability, pulmonary exacerbations (PEs) can occur multiple times per year and result in permanent damage to the lungs. Little is known of the shift from a period of stability to a PE, but this shift is likely to be attributed to changes in the bacterial community. Here, we identified changes in the lung microbiota to determine if they reflect patient health, indicate the onset of exacerbations, or are related to antibiotic treatment. In contrast to most bacterial studies on CF, we collected weekly samples from an adult CF patient over a period of 3 years and performed quantitative PCR (qPCR) and Illumina sequencing on those samples. While many DNA-based studies have shown the CF microbiota to be relatively stable, we observed an increase in the total bacterial abundance over time (P < 0.001), while the number of different taxa (bacterial richness) and the number of different taxa and their abundances (diversity) significantly decreased over time (P < 0.03), which was likely due to repeated antibiotic exposure. Using genus-specific primers with qPCR, we observed an increase in the abundance of Burkholderia multivorans, a CF-associated pathogen, prior to the occurrence of a PE (P = 0.006). Combining these DNA-based techniques with frequent sampling identified a potential initiator for exacerbations and described a response of the CF microbiota to time and antibiotic treatment not observed in previous CF microbiota studies.
INTRODUCTION
Bacterial infections with consequent progressive lung disease are the leading cause of death in persons with cystic fibrosis (CF), a disease that affects an estimated 30,000 people in the United States and 70,000 people worldwide (1). Prior to the past 2 decades, it was assumed that the CF lungs were colonized with only a few different bacteria, including Pseudomonas aeruginosa, Haemophilus influenzae, Staphylococcus aureus, and members of the Burkholderia cepacia complex (BCC) (2). It has been shown in CF patients that chronic infection with these CF-related bacteria (CFRB) is linked to an increase in mortality (3, 4).
Pulmonary exacerbations (PEs), which may develop multiple times per year in CF patients, are often caused by a disturbance to a stable chronic bacterial infection (5, 6). The exact cause of a PE, often identified by an increase in pulmonary disease symptoms, remains uncertain but is commonly attributed to factors associated with established bacteria, and possibly to viruses or newly acquired bacterial strains (7, 8). In 2004, Rogers et al. used terminal restriction fragment length polymorphism (TRFLP) analysis to target the bacterial 16S rRNA gene in order to analyze DNA extracted from sputum samples from CF patients. This method of analysis revealed a complexity that included 15 species not previously identified in the lungs. The study by Rogers et al. laid the foundation for redefining CF as a polymicrobial disease (9). These culture-independent studies have led to clinical treatment for CF lung infections focused on multiple species instead of a single agent.
The frequency of PEs has been connected to increased mortality and results in a permanent impairment in lung function (6). Early treatment intervention might reduce the length and severity of a PE; however, attempts at developing tools to predict a PE have had limited success (10, 11). A DNA-based study by Stressman et al. (12) used quantitative PCR (qPCR) on bacterial DNA isolated from sputum samples collected weekly over a 12-month period to identify the cause of PEs in 12 CF patients. Using this quantitative analysis, Stressman et al. (12) showed that the bacterial load, including the dominant pathogen, P. aeruginosa, did not change 1 to 3 weeks prior to the onset of a PE.
Because a single sputum sample can provide only a snapshot of the community profile at any given time, the identification of changes in the microbiota that precede PE onset would require frequent longitudinal sampling (13, 14). Our long-term study was designed to determine if combining quantitative and deep-sequencing techniques with intensive sampling from a single patient over a time period of years would reveal population changes, undetected in previous studies, prior to the onset of antibiotic treatment or a PE. If bacteria are involved in eliciting a PE, bacterial population changes could be used as an early indicator of a PE (11).
We used Illumina sequencing and qPCR to examine changes in the microbial community diversity and abundance of the total bacteria and of two taxa, Pseudomonas and Burkholderia, in sputum samples collected from a 30-year-old CF patient at least once a week over a 3-year period. The samples collected from this patient were sufficient to generate conclusions based on statistically significant changes in the microbiota. We identified a correlation between the time period shortly before the onset of a PE and an increase in the abundance of Burkholderia, a major CF pathogen within a microbial community dominated by Pseudomonas. Monitoring these changes to the microbiota allowed us to determine how individual taxa, including specific pathogens, change over time, change prior to a PE, and respond to antibiotic therapy.
MATERIALS AND METHODS
Patient characteristics.
The patient is a 30-year-old adult male who was diagnosed with CF at 2 weeks of age and voluntarily participated in this study. His treatment regimen during the study included oral enzymes for CF-related malabsorption, along with antibiotics administered as prophylactic agents and various antibiotics (Table 1), as prescribed by the patient's pulmonologist. The subject has a heterozygous deltaF508/unknown CF transmembrane conductance regulator (CFTR) genotype and no other CF-associated complications. The forced expiratory volume in 1 s (FEV1) values measured during routine clinic appointments over the course of the study ranged from 33% to 40% of the predicted value, which is consistent with severe obstruction. He experienced nine PEs that required antibiotic intervention during the course of this study. Each PE was diagnosed by an experienced CF pulmonologist on the basis of a change in sputum production, an increase in cough, a decrease in lung function, an increase in dyspnea, and/or the occurrence of hemoptysis.
TABLE 1.
Antibiotic treatment regimen for nine pulmonary exacerbations
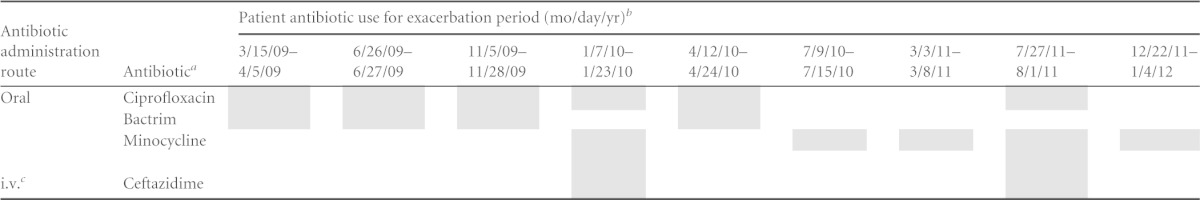
Prophylactic antibiotics, including inhaled tobramycin, inhaled aztreonam, and azithromycin were not included in the analysis due to their intermittent use, which was independent of the occurrence of a PE.
The shaded areas represent periods of patient antibiotic use.
i.v., intravenous.
Samples.
Serial expectorated sputum samples were obtained with informed consent, according to an institutional review board (IRB)-approved protocol, twice weekly for a period of almost 3 years. The samples were collected in the morning by the patient expectorating sputum into a sterile 15-ml Falcon tube, which was placed on ice during transport to the lab and then stored at −80°C until use. The transportation time was no more than 45 min. DNA was extracted within 35 months of collection. The samples chosen for analysis were selected to represent both periods of stability and exacerbation.
Sputum homogenization, viable cell selection, and DNA extraction.
The sputum samples were mixed in a 1:3 ratio of sputum and 0.1% dithiothreitol solution and incubated at 37°C for 1 h, followed by mechanical homogenization for 1 min using a microblender. To amplify DNA from living cells only, propidium monoazide (Biotium, Hayward, CA) was then added to a final concentration of 50 μmol/ml, and DNA cross-linking was induced using a 400-W halogen light source (15). DNA was extracted using the IT 1-2-3 VIBE sample purification kit (BioFire Diagnostics, Inc., Salt Lake City, UT) and its concentration determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). All extracted DNA was immediately stored at −20°C until it was subjected to qPCR.
Abundances of total bacteria, Pseudomonas, and Burkholderia in sputum samples.
The qPCR mixture contained 10 μl of PerfeCTa SYBR green FastMix reagent low ROX (Quanta Biosciences, Gaithersburg, MD), 0.5 μl of 100 pmol/μl each primer, 5 μl of sample DNA, and 4 μl of nuclease-free water to a final volume of 20 μl. Universal primers (16) were used to amplify a conserved 466-bp 16S rRNA gene fragment to measure the abundance of the total bacteria in the sample. B. cepacia complex-specific primers (17) and Pseudomonas-specific primers (18) were used to selectively amplify genomic DNA sequences from each genus, yielding 333-bp and 93-bp fragments, respectively. qPCR was performed using the ABI 7500 Fast real-time PCR system (Applied Biosystems, Carlsbad, CA) with an initial step of 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. Melting curves were determined following qPCR by 1 cycle of 15 s at 95°C, 1 min at 60°C, 30 s at 95°C, and 15 s at 60°C. Standard curves were created for each primer pair using 10-fold dilutions of amplicons generated using an Escherichia coli strain as the DNA template for the 16S rRNA gene sequence primers, P. aeruginosa for the Pseudomonas-specific primers, and Burkholderia multivorans for the BCC-specific primers. The DNA copy number per g of sputum was calculated for each sample based on a standard curve with a 1 × 105-fold linear range in threshold cycle (CT) values. To ensure precision in the qPCR data, three technical replicate reactions were performed for each sample.
Illumina sequencing library preparation.
The samples were prepared for 16S rRNA gene Illumina sequencing targeting the V6 hypervariable region with a two-stage PCR strategy. The samples were PCR amplified in the first stage using primers that included barcodes in both the forward and reverse oligonucleotides for sample identification in a multiplex fashion (see Table S1 in the supplemental material). A secondary stage of PCR utilized a set of primers that overlapped the 5′ ends of the first set of primers and added bases complementary to the Illumina flow cell adapters for sequencing (see Table S1).
The thermal cycling conditions were as follows: an initial denaturation step at 94°C for 3 min, followed by a touchdown protocol beginning at 94°C for 45 s, 61°C for 45 s, with a 1°C drop each cycle for a total of 5 cycles, an additional 15 cycles at 51°C for 45 s, 72°C for 45 s, and a final elongation at 72°C for 2 min. Fifteen microliters of the first PCR products was utilized in the second stage of PCR. The second PCR consisted of one denaturation step of 94°C for 3 min, 15 cycles at 94°C for 45 s, 65°C for 45 s, 72°C for 45 s, and a final extension step at 72°C for 2 min.
The PCR fragments were visualized on a gel, quantitated on a NanoDrop ND-3300 fluorospectrometer (Thermo Scientific, Wilmington, DE) using PicoGreen to determine the concentration of double-stranded DNA (dsDNA), and pooled in equimolar amounts for sequencing.
Sequence analysis.
Illumina HiSeq 2000 technology was used to sequence the 72 samples used for this study. Raw paired-end sequences were processed as described previously (19–21). Briefly, a minimum of 70 continuous matching nucleotides across the length of the ungapped alignment were required to produce each merged sequence. A total of 70,550,318 sequences, with an average length of ∼77 bases, met our merging and extending criteria, which were fed into the AbundantOTU+ version 0.93b program (http://omics.informatics.indiana.edu/AbundantOTU/otu+.php) with the -abundantonly option. AbundantOTU+ clustered those sequences into 127 operational taxonomic units (OTUs), incorporating 70,310,410 (99.66%) of all the merged sequences. The sequences that were not incorporated into an OTU were excluded from further analyses. For the purpose of detecting chimeric OTUs, we used UCHIME (22) and UPARSE (23) in conjunction with the GOLD reference database (24); neither reported any chimeras in our 127 OTUs.
Taxonomic classification was achieved by first aligning the OTU sequences to the Silva database (release 108 [http://www.arb-silva.de/]) using BLASTn version 2.2.26+ with an expectation value of E-5. Next, the standalone version of the Ribosomal Database Project (RDP) classifier version 2.5 (25) was used to classify the full-length Silva sequences with the best BLASTn match to the OTU sequence requiring an RDP confidence score of ≥80%. This was done to compensate for the short read length of the generated OTUs. The raw counts for each OTU were normalized and log transformed according to the equation log10[(OTU raw count/no. of sequences in sample × average no. of sequences per sample) + 1].
Principal coordinate analysis (PCoA) was done using mothur version 1.25.0 (26) and a Bray-Curtis dissimilarity matrix generated from the log10-normalized counts.
Alpha diversity measures (richness for observed species and Shannon diversity) were calculated on the raw counts after rarefying to 198,500 sequences per sample (where 198,500 is number of sequences in the sample with the smallest number of sequences).
RESULTS
qPCR measurements reveal a steady increase in bacterial load.
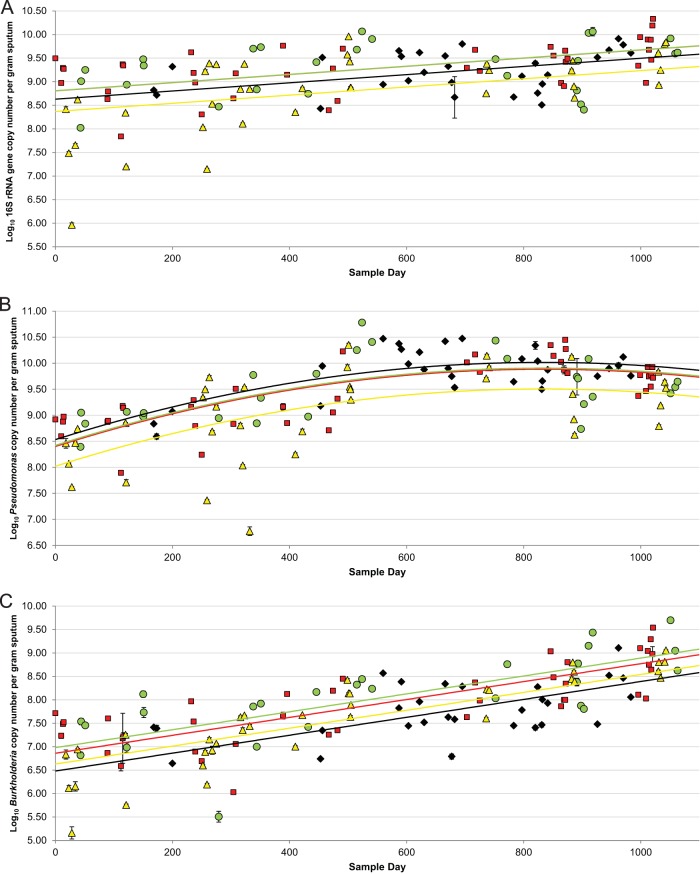
To characterize microbial variation over time in a single CF patient, we collected 130 sputum samples over a time period of 1,063 days. We used qPCR to independently amplify the 16S rRNA gene sequences common to all bacteria (Fig. 1A), Pseudomonas (Fig. 1B), and Burkholderia (Fig. 1C), and we measured the changes in bacterial load over time. Both the total bacterial load (Fig. 1A) and Burkholderia abundance (Fig. 1C) increased steadily over the entire sampling period, while Pseudomonas abundance steadily increased over the first 2 years of our study, followed by an apparent plateau (Fig. 1B).
FIG 1.
Variation in abundances of total bacteria (A), Pseudomonas (B), and Burkholderia (C) in sputum samples collected over a 35-month period during which nine exacerbations occurred. Treatment was considered to be during or within 48 h of termination of antibiotics given to treat an exacerbation. The time points are plotted as red squares, before treatment (≤30 days before an exacerbation); yellow triangles, treatment (samples collected during and within 48 h of termination of treatment); green circles, recovery (≤30 days after an exacerbation); and black diamonds, stable (>30 days prior to or after an exacerbation). Universal primers targeting the 16S rRNA gene, Pseudomonas-specific primers, and Burkholderia-specific primers were used to measure abundance, which is expressed as log10 DNA copy number per g of sputum (y axis) over the collection period (x axis). The best fit line for before treatment in panels A and B is superimposed with the line for recovery, due to their similarity. The error bars represent the standard deviation of three technical replicate reactions; most error bars are smaller than the symbols for each sample. The best fit lines are red for before treatment, yellow for treatment, green for recovery, and black for stable.
Our qPCR measurements have a high variance; it is not uncommon for measurements taken close in time to differ by as much as a log10 unit. Despite this, if we model the relationship between qPCR signal and time, using a simple first-order linear model for Burkholderia and 16S rRNA (see Table S2, model 1 in the supplemental material) or a second-order linear model for Pseudomonas that allows us to capture the saturation relationship (see Table S2, model 2), we observed a highly significant correlation between PCR abundance and time (see Table S2). Despite the short-term variability inherent in any individual qPCR measurement, there is a clearly significant increase in bacterial load over the multiyear time course of our study.
Antibiotic treatment temporarily depresses total bacterial and pathogen loads.
We next asked whether we could observe a difference in bacterial load based on treatment status. We defined four different treatment conditions as (i) before treatment, defined as samples collected ≤30 days prior to antibiotic treatment for a PE, (ii) treatment, defined as any sample collected during the administration of antibiotics used to treat a PE, (iii) recovery, defined as samples collected ≤30 days after antibiotic treatment for a PE, and (iv) stable, defined as any sample not meeting any of the above criteria. We used a series of linear models to evaluate whether these treatment categories were associated with differences in qPCR signals. Our linear models had terms for time (for Burkholderia or total bacteria) or time-squared (second-order model) for Pseudomonas, as well as terms for treatment category and interactions between time and treatment variables (see Table S2, models 3 and 4 in the supplemental material). An analysis of variance (ANOVA) (utilizing an F-test comparing the full model to a reduced model with interactions terms set to zero) found that P values associated with interactions terms were all >0.05. To simplify the interpretation of our models, we dropped the interaction terms, thus removing them as complicating conditional effects, to yield models 5 and 6 in Table S2. The fit from these models is shown in Fig. 1. An ANOVA (utilizing an F-test comparing the full model to a reduced model with terms for treatment set to zero) evaluating the null hypothesis that treatment has no effect was rejected for Burkholderia (P < 0.003), Pseudomonas (P < 0.0006), and 16S rRNA (P < 0.002), as measured by qPCR. In terms of our model, these P values can be interpreted as testing the hypothesis that separate lines for each treatment category have identical intercepts given a constant slope (for 16S rRNA and Burkholderia) or shape (for Pseudomonas). That is, we tested the null hypothesis that a single line (the reduced model) can fit the qPCR data as well as the four separate lines (the full model) shown in Fig. 1. Our model's rejection of this null hypothesis suggests that we are able to detect antibiotic treatment temporarily depressing bacterial load, despite the high variability of each individual measurement.
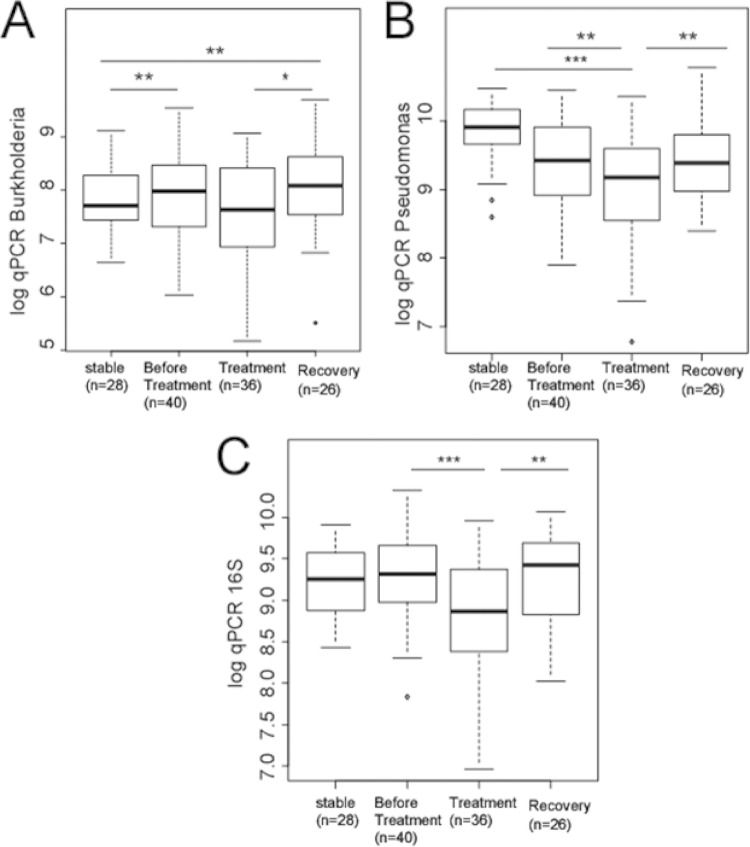
To further explore the idea that antibiotic treatment decreases bacterial load, we visualized our 130 data points by treatment category independent of time (Fig. 2). Viewed this way, antibiotic treatment clearly temporarily depresses Pseudomonas (Fig. 2B) and 16S rRNA (Fig. 2C) but not Burkholderia (Fig. 2A). We could not use simple t tests to evaluate the statistical significances of these relationships because the data viewed in this way contain multiple samples from the same individual binned across time points, which would violate the assumption of independence in a test statistic that did not explicitly consider time. We can, however, use models 5 and 6 in Table S2 in the supplemental material to test the hypothesis that the intercepts (in Fig. 1) from the line of best fit for the data from any pair of treatment conditions are identical. Under this procedure, we find that the higher bacterial loads observed during the postantibiotic recovery time points, compared to those during treatment for Burkholderia (Fig. 2A), Pseudomonas (Fig. 2B), and 16S rRNA (Fig. 2C), are all statistically significant (Table 2). We also find that bacterial load is significantly lower in treatment than in before treatment for Pseudomonas (Fig. 2B) and 16S rRNA (Fig. 2C), with a nonsignificant trend for Burkholderia (Fig. 2A and Table 2). Our data are consistent with the conclusion that antibiotics relieve exacerbating symptoms by temporarily depressing the bacterial load.
FIG 2.
Comparison of stable, before treatment, treatment, and recovery qPCR values for Burkholderia (A), Pseudomonas (B), and the 16S rRNA gene (C). The box plots show the same data as in Fig. 1 collapsed across time points. The asterisks indicate significant differences between the conditions, with significance determined by pairwise comparisons from model 5 (for Burkholderia and 16S rRNA) and model 6 (for Pseudomonas) from Table S2 in the supplemental material, which contains a term for time and is therefore not in violation of the assumption of independence (see the text). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TABLE 2.
Data for Fig. 2 by treatment category
| Treatment category | Bacterial group | Treatment category |
|||
|---|---|---|---|---|---|
| Stable | Before treatment | Treatment | Recovery | ||
| Stable | Total bacteria | 0.178↑a | 0.052↓ | 0.213↑ | |
| Pseudomonas | 0.318↓ | 2.00e-04↓b | 0.440↓ | ||
| Burkholderia | 0.006↑c | 0.307↓ | 0.001↑c | ||
| Before treatment | Total bacteria | 0.291↓d | 3.87E-04↓b | 0.978↑ | |
| Pseudomonas | 0.409↑d | 0.002↓c | 0.867↑ | ||
| Burkholderia | 0.015↓d,e | 0.063↓ | 0.367↑ | ||
| Treatment | Total bacteria | 0.104↑d | 0.003↑c,d | 0.001↑c | |
| Pseudomonas | 0.004↑c,d | 0.007↑c,d | 0.003↑c | ||
| Burkholderia | 0.425↑d | 0.114↑d | 0.012↑e | ||
| Recovery | Total bacteria | 0.319↓d | 0.978↓d | 0.006↓c,d | |
| Pseudomonas | 0.495↑d | 0.918↓d | 0.009↓c,d | ||
| Burkholderia | 0.006↓c,d | 0.440↓d | 0.027↓d,e | ||
Arrows represent an increase (↑) or decrease (↓) in abundance relative to the treatment category in the 1st column.
Significant at a P value of <0.001.
Significant at a P value of <0.01.
Data adjusted for multiple hypothesis testing with the Benjamini-Hochberg method.
Significant at a P value of <0.05.
An increase in Burkholderia abundance is associated with onset of a pulmonary exacerbation.
If any one taxon is an indicator for onset of a PE, we would expect to observe a change in the abundance of that taxon before antibiotic treatment is administered. The abundance of Burkholderia is significantly higher (P = 0.006) in the before treatment time points, which indicate the beginning of a PE, than in the stable time points (Fig. 2A). Pseudomonas is less abundant before treatment than in the stable time period (Fig. 2B), but our model (model 6 of Table S2 in the supplemental material) does not assign significance to this difference (P = 0.32). No significant change was observed in the total bacterial load between the stable and before treatment time points. Interestingly, the change in abundance of Burkholderia exhibits a pattern consistent with a posttreatment rebound, even while the patient showed improved symptoms. Our data are therefore consistent with an increase in Burkholderia being associated with the onset of a PE but not as an indicator of overall patient health.
Relative abundance of microbiota remains stable over time.
qPCR measures absolute abundance but is not an efficient method to generate information about all members of the microbiota found within sputum samples. In order to more fully characterize the microbial community, we utilized 16S rRNA gene Illumina sequencing on some of the sputum samples. Funding limited our analysis to 72 of the 130 samples, chosen based on their time of collection under the four treatment conditions (see above). The sequences were clustered with AbundantOTU+, and the consensus sequences for each OTU were classified to the family level using the RDP classifier (see Materials and Methods). Comparisons between the CF-associated taxa at the family and genus levels within our pipeline and within the QIIME pipeline showed a similar response to antibiotics and time (data not shown). We chose the family level because the RDP calls to the family level were generally consistent between our pipeline and a parallel analysis using the QIIME pipeline, but there was considerable divergence at the genus level (data not shown). Given the short sequence (<100 bp) reads of the Illumina sequencing technology we used, such divergence at more narrow taxonomic levels is not surprising between different analysis methods.
Out of the >70.3 million sequences, Pseudomonadaceae was the dominant family, representing >89% of all sequences (see Table S3 in the supplemental material). Two other typical CF pathogens from the families Burkholderiaceae and Streptococcaceae made up approximately 8% of all sequences (see Table S3). The remaining sequences were mostly nontypical CF-associated bacteria (27) and were classified as either Micrococcaceae, Veillonellaceae, each of which consisted of <1% of all sequences, or less prevalent families, representing <2% of all other sequences (see Table S3).
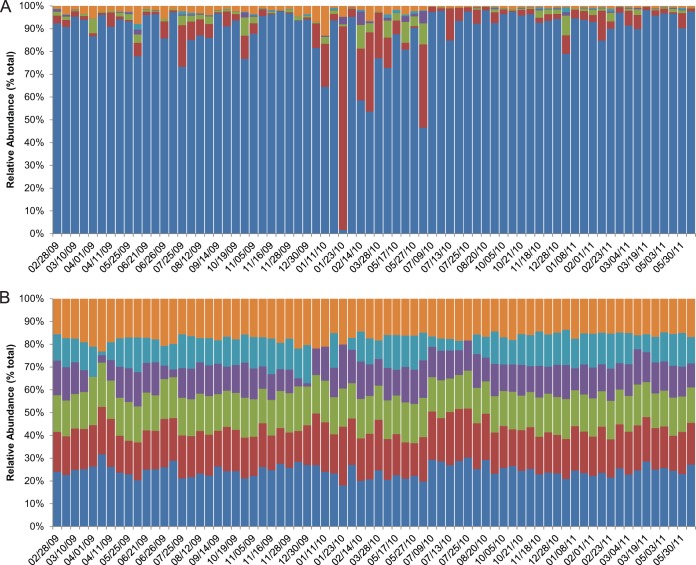
The microbial profiles of the most abundant individual taxa within the sputum samples showed little variability across the study period (Fig. 3). No pattern of change in the relative abundances of the top five families was observed surrounding the occurrence of a PE. Pseudomonadaceae was the most dominant family and showed little change in its relative abundance in the majority of the sputum samples. An increase in the relative abundance of Burkholderiaceae or Streptococcaceae was observed only in those samples in which the relative abundance of Pseudomonadaceae showed short-term decreases (Fig. 3A); this, however, did not correspond to the occurrence of a PE and likely reflects the compositional nature of relative abundance measures (28, 29). Little variation in relative abundance was observed over time when the other less abundant families were combined into the “other” category (Fig. 3B). Using model 10 (see Table S2 in the supplemental material), we were able to test the hypothesis that the relative abundance of any individual family significantly changed over time. We did not achieve significant results for this model for the abundant taxa Pseudomonadaceae, Burkholderiaceae, and Streptococcaceae. The slope of the fit for 14 of the less abundant families, however, did not exhibit a significant decrease, while the slope of the fit for one family significantly increased over time (see Table S3 in the supplemental material). The relative abundance of each of these less abundant families consisted of <0.5% of the total sequences. We conclude that for all but a few of the least abundant taxa, our sequencing data are consistent with overall stability over time.
FIG 3.
Change in relative abundance of taxa classified to the family level during the sampling period. The families in the top five of relative abundance are shown (dark blue, Pseudomonadaceae; red, Burkholderiaceae; purple, Streptococcaceae; purple, Micrococcaceae; light blue, Veillonellaceae). The relative abundances of all other families, each representing <0.5% of the community, are grouped in the “other” (orange) category. The colored bars represent the proportion of (unlogged [A], log10 [B]) reads mapped to each family. The dates on the x axis are in month/day/year format.
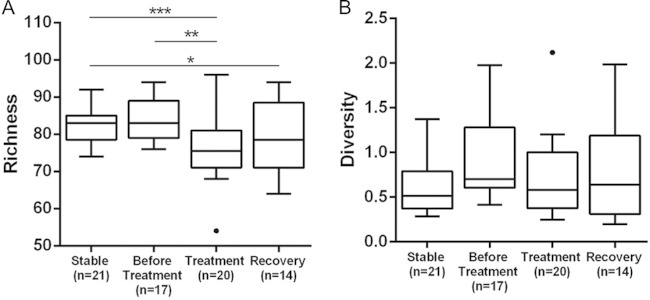
Illumina sequencing reveals a decrease in richness over time.
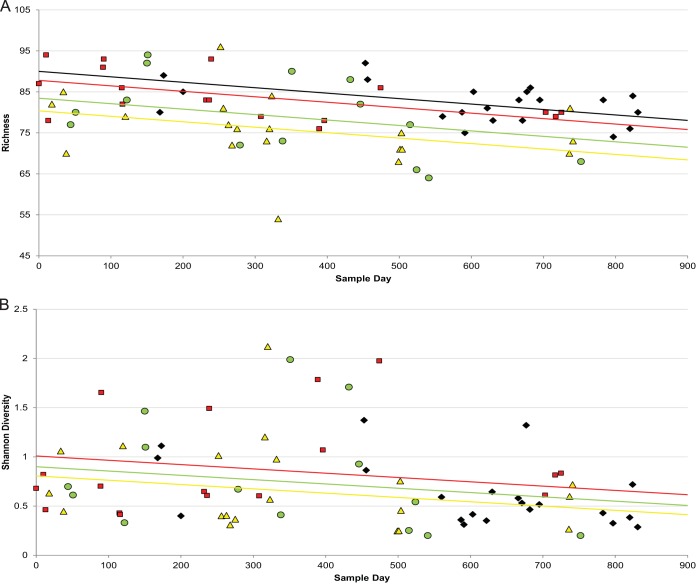
To determine if long-term exposure to antibiotics in our subject decreased microbial diversity, we used a simple first-order linear model to evaluate the relationship between the diversity or the richness of the sequencing data and time (see Table S2, model 7 in the supplemental material). We observed a significant inverse correlation between time and richness (P < 0.02) and between time and diversity (P < 0.03) (see Table S2 and Fig. 4).
FIG 4.
The changes in bacterial richness (A) and bacterial diversity (B) from 72 sputum samples are shown over time. Richness (P = 0.018) and diversity (P < 0.050) decreased significantly over the sampling period. Richness but not diversity showed a significant (P = 6.59e−05) decrease during treatment for a PE compared to those variables in the stable time period. The time points are plotted as before treatment (red squares) (≤30 days before an exacerbation), treatment (yellow triangles) (samples collected during and within 48 h of termination of treatment), recovery (green circles) (≤30 days after an exacerbation), and stable (black diamonds) (>30 days prior to or after an exacerbation). (B) The best fit line for the stable time period is superimposed with the line for recovery due to their similarity. The best fit lines are red for before treatment, yellow for treatment, green for recovery, and black for stable.
To determine if our treatment categories were associated with differences in diversity and richness, we used a series of linear models similar to those used for the qPCR analysis. These models had terms for time, as well as terms for treatment category and interactions between time and treatment variables (see Table S2, model 8 in the supplemental material). ANOVA found that the P values associated with the interaction terms were all >0.05, so the interaction terms were dropped to yield model 9 in Table S2. An ANOVA evaluating the null hypothesis that treatment has no effect was rejected for richness (P < 0.001) but not for diversity (P = 0.688).
Similar to our absolute abundance described above (Fig. 2), we visualized diversity and richness for each of the 72 samples according to treatment status and independent of time (Fig. 5). We once again derived P values for pairwise comparisons for this visualization from the models in Table S2 in the supplemental material). For diversity, no significant change was observed over time or in samples collected during any one of the treatment status categories (Fig. 5B). The decrease in richness, however, was highly significant when the samples collected during treatment (P = 3.67e−05) or recovery (P = 5.49e−03) were compared to those samples collected during the stable time period (Fig. 5A). These data are consistent with repeated antibiotic treatment leading to a temporary decrease in the number of taxa present.
FIG 5.
Comparison of Shannon diversity and richness of sequences in four treatment categories. The asterisks indicate significant differences between the categories of treatment status, with significance determined by pairwise comparisons from model 9 from Table S2 in the supplemental material, which contain a term for time and are therefore not in violation of the assumption of independence (see the text). A highly significant (P = 6.59e−05) decrease in richness was observed in samples collected during antibiotic treatment compared to that in samples collected during periods of stability. Richness also decreased in a comparison of the samples collected before treatment to treatment (P = 0.0013) and stable to recovery (P = 0.0104). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
This study represents the most intensive sampling of a single CF patient to date. Our goals were to determine if changes in bacterial abundance may predict an oncoming PE and to identify changes in the microbiota associated with antibiotic use or time. While other studies have attempted to identify biomarkers that aid in the prediction of a PE and the progression of lung disease in CF (11, 30), ours is the first to obtain samples so frequently from a CF patient over a long-term period. Our study was based upon the assumption that a detectable shift in the bacterial community precedes a PE. Testing our assumption required a longitudinal study to reveal the relationship between disease progression, the occurrence of a PE, and various components of a bacterial community, such as the change in diversity, richness, or the abundances of specific members of the microbiota (31). Our goal was to determine if a bacteria-derived biomarker can be identified and result in antibiotic treatment being initiated early in the course of a PE and reduce the symptoms that would otherwise cause damage to the lungs. Identifying a change in microbiota that precedes a PE requires frequent patient sampling, since the length of time between the changes in the bacterial community and PE onset is unknown (14). Previous cross-sectional studies examining the diversity of the microbiota in the lungs of CF patients collected samples at a frequency or for a length of time that did not reveal potential short-term changes associated with a PE (32, 33).
qPCR reveals unstable microbiota over time.
A great deal of sample-to-sample variation was observed in our qPCR data over the sampling period. This change in absolute bacterial abundance between adjacent samples may be subject to misinterpretation if a sputum sample containing a low number of bacteria was compared to a sequential sample that originated from a region of the lungs containing a large number of bacterial cells. Recent culture-independent studies have revealed a spatial distribution of distinct microbial communities in the CF lung. Goddard et al. (34) and Willner et al. (35) both observed distinct clustering of microbial communities in various areas of the lung by examining explant lung samples. These studies suggest that since the origin of expectorated sputum cannot be identified, any two sputum samples collected from an individual patient are expected to vary. The degree to which the microbiota may vary between samples may be difficult to resolve using 16S rRNA gene sequencing due to, as Willner et al. (35) stated, “spatial heterogeneities.” Based on these studies and our small standard deviation in the qPCR data between at least three technical replicates for each sample (see Fig. 1), it is likely that the variation in our qPCR data may have occurred due to spatial distribution of dense bacterial clusters within the lungs. As we demonstrated across a 3-year period, a high variation between any two samples can be compensated for by frequent collection of samples.
Our qPCR data show a highly significant increase over time for both Pseudomonas and Burkholderia. In contrast, our sequencing data show a much less significant increase in these taxa over time (see Table S3 in the supplemental material). While we recognize that our observations are from a single patient, we would expect that any cross-sectional analysis of sputum samples collected from a CF patient would be at risk for misinterpretation due to there being little association in the change of bacterial abundance in an individual sputum sample and a change in patient health.
An increase in the abundance of a CF pathogen occurs prior to PE onset.
A 2011 study by Stressman et al. (12) found no evidence of changes in bacterial density in sputum samples obtained from 12 patients at 21, 14, and 7 days prior to the occurrence of a PE (12). Unlike the 1-year sampling period of the Stressman et al. study, we sampled at least weekly for 3 years, which spanned nine PEs, allowing us to group samples into categories relative to the occurrence of a PE. Our strategy revealed moderate changes in bacterial abundance that occurred before, during, and after a PE, which likely would have been missed with less frequent sampling over a shorter time period.
The antibiotic treatment regimen for each PE (see Table S2 in the supplemental material) was prescribed to target the two major pathogens in this patient, Pseudomonas and Burkholderia, and a different pattern of bacterial abundance change was observed for each. Interestingly, we observed a possible saturation phase of the most abundant pathogen, Pseudomonas, toward the second half of our study period. This saturation may reflect the effectiveness of the antibiotic regimen that was tailored to Pseudomonas. Consistent with this idea is the finding that Pseudomonas was significantly lower in abundance in the treatment category than in any of the other treatment status categories (Table 2). The second most abundant pathogen measured in our samples, Burkholderia, decreased in abundance during treatment compared to that before treatment, but this decrease was not significant (Table 2). The lack of a significant decrease in the abundance of Burkholderia in samples collected before treatment compared to those collected during treatment suggests that the antibiotic regimen was less effective against this taxon, which may be due to increased antibiotic resistance or other unidentified factors.
One anomaly in our data is that the total bacterial abundances are less than those for Pseudomonas in several of the sputum samples, as determined by qPCR. Possible explanations for this include a difference in the efficiencies of these two reactions due to using a different amplicon size or to stochastic effects generated by primers targeting different regions of genomic DNA. Even with this anomaly, we were able to use both data sets in this study because we generated separate standards for each primer set, used the same genomic DNA for each reaction, and compared each target separately.
The age and disease status of our patient may be reasons why we observed so few changes over time. Adult CF patients typically exhibit low diversity, with few established pathogens dominating the microbial community (31, 36). Even with these factors, we saw consistent trends in which (i) there was a spike in Burkholderia abundance before most, but not all PEs, (ii) antibiotics temporarily depressed bacterial load, and (iii) over a time frame of years, bacterial load increased. These data are consistent with the observation that antibiotics provide a temporary relief of symptoms, but they are in apparent contradiction with studies that indicate that a spike in bacterial load is not the cause of PEs (12, 14, 37, 38).
Antibiotics reduce richness but not diversity.
We used a barcoded strategy for Illumina sequencing to determine the changes in bacterial diversity and richness. Next-generation sequencing has shown that little change in diversity occurs over time in sputum samples collected from CF patients (31). A decade-long bacterial study by Zhao et al. (31) examined the diversity in sputum samples, which were collected and grouped according to treatment status, from six adult CF patients, three with stable lung disease and three with progressive lung disease. Bacterial diversity remained stable over the course of the study in the stable patients but showed a significant decrease over time in the progressive patients. The authors did detect a significant decrease in diversity during treatment for all six patients (31). Similar to the Zhao et al. study, we observed a significant decrease in diversity over time in our patient, an adult with CF who was considered to have progressive lung disease, but unlike in their study, diversity remained stable across changes in treatment status. A possible explanation for the difference in the change in diversity during treatment between these two studies may have been due to a difference in sampling frequency. Another possibility arises from our having used propidium monoazide (PMA) pretreatment. Rogers et al. (39) reported that diversity can be under- or overestimated without prior PMA treatment.
The fact that we saw no significant change in diversity between the samples collected during our defined treatment categories indicates that changes in diversity were not involved in the occurrence of a PE in our patient. Our observations again demonstrate the importance of frequent sampling. Since changes in patient health may occur between sample collections, there is a risk of choosing a sample that provides data that do not properly represent or may underestimate associated responses in the microbial community. Because the duration of the response may be short-lived, infrequent sampling may not provide an accurate baseline against which to compare changes in the microbial community that correlate with antibiotic treatment.
Change in richness was also not an indicator of PE in our patient. We observed no significant change in richness in the samples collected prior to antibiotic treatment compared to that in the samples collected during periods of stability, although a highly significant decrease in richness was seen during antibiotic treatment. A similar decreased richness during antibiotic treatment has been reported in other studies that examined serially collected sputum samples (14). For example, Fodor et al. (37) observed an impact of antibiotic treatment on both the relative sequence abundances of seven OTUs and on taxon richness from sputum samples collected from 23 patients at the onset of a PE and at the end of treatment. Additionally, Daniels et al. (14) used TRFLP to show that mean taxon richness decreases in sputum samples as a response to antibiotic treatment for a PE. That richness is decreased during treatment in the Fodor and Daniels studies and significantly reduced during treatment in our study suggests there is a concomitant effect of antibiotics on bacteria other than the major pathogens (see Table S3 in the supplemental material).
Relative abundance of microbiota appears to be stable over time.
Our sequencing results suggest an overall stability in the microbial community over a long-term period. These results are consistent with those of previous studies that used pyrosequencing to characterize the microbial community and found high overall levels of stability (34, 37). In contrast, our qPCR results suggest a steady increase in bacterial load over time. We are investigating the possibility that the discrepancy between the two methods is caused by saturation effects and the compositional nature of the sequencing data. In sequencing reactions in which Pseudomonas represents the vast majority of sequencing reads, as it does here, increases in absolute Pseudomonas abundance may not be reflected by a corresponding increase in relative abundance. The discrepancy between our Burkholderia qPCR and sequencing results is harder to understand but may involve the saturation of nonquantitative PCR steps that are a necessary precursor to 16S rRNA sequencing, as well as complex compositional effects that can skew sequencing results in low-diversity environments (28, 29). Taken together, our qPCR results suggest that our sequencing results, as well as the sequencing results from other studies, may exaggerate the stability of the dominant pathogens within the low-diversity CF microbial community.
Conclusions.
Ours is the first study to use Illumina sequencing and qPCR to examine the diversity and abundance of bacteria in frequently collected sputum samples from a single CF patient over a multiyear period. Based on our sequencing data alone, time and antibiotic use appeared to decrease the number of different taxa in the lungs but only of those taxa that are the least abundant (see Table S3 in the supplemental material) in the sputum of our CF patient. That antibiotics usually alleviate PE symptoms while affecting the less abundant taxa suggests that identifying the role of taxa other than the major pathogens may be useful in understanding the progression of lung disease and the occurrence of exacerbations.
Our study also demonstrates the advantage of combining genus-specific qPCR with statistical linear models to identify changes in abundance over time and in treatment status. For example, we observed a reduced load of Pseudomonas but not of Burkholderia during times of antibiotic use over our study period, throughout which there was no decline in patient lung function. The results suggest that antibiotics helped to maintain a relatively stable health condition but did not affect the persistent increase in Burkholderia, a potential driver of PE onset. Based on the qPCR results alone, we can speculate that if the greater recovery of Burkholderia after antibiotic treatment that we observed were to continue to exceed that of Pseudomonas, Burkholderia would eventually become the dominant pathogen.
There are statistical limitations to using a single patient only, such as not being able to generalize our results. However, we were able to identify changes over time that have been only partially documented in other quantitative and deep-sequencing studies of the CF lung microbiota. These changes were identified because of the frequent sampling regimen used. The current sampling methods of a CF patient typically limit the identification of changes in the microbial community to after the PE has been diagnosed in a clinical setting. Frequent sampling increases the likelihood of collecting a pre-PE sample, which would be beneficial in studies aimed at identifying the biological initiators of a PE. The identification of microbiota indicators is the first step toward making this approach technically and financially viable in a clinical setting. We plan to similarly examine additional patients to determine if our observations represent a theme common to CF or if microbiota changes are patient specific.
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Donaldson for his contribution to the editing and reviewing of this paper and Wei Sha for critical comments on the manuscript.
This work was supported in part by funds from The University of North Carolina at Charlotte (to T.R.S.), and a Lucille P. and Edward C. Giles Dissertation-Year Graduate Fellowship, Cystic Fibrosis Foundation Student Traineeship award STOKEL11H0, and a Charlotte Research Institute–Duke Energy postdoctoral fellowship (to J.R.S.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02555-14.
REFERENCES
- 1.Cystic Fibrosis Foundation. 2011. About cystic fibrosis. Cystic Fibrosis Foundation, Bethesda, MD: http://www.cff.org/AboutCF/. [Google Scholar]
- 2.Sibley CD, Surette MG. 2011. The polymicrobial nature of airway infections in cystic fibrosis: Cangene Gold Medal Lecture. Can J Microbiol 57:69–77. doi: 10.1139/W10-105. [DOI] [PubMed] [Google Scholar]
- 3.Kidd TJ, Douglas JM, Bergh HA, Coulter C, Bell SC. 2008. Burkholderia cepacia complex epidemiology in persons with cystic fibrosis from Australia and New Zealand. Res Microbiol 159:194–199. doi: 10.1016/j.resmic.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Feliziani S, Luján AM, Moyano AJ, Sola C, Bocco JL, Montanaro P, Canigia LF, Argaraña CE, Smania AM. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:e12669. doi: 10.1371/journal.pone.0012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goss CH, Burns JL. 2007. Exacerbations in cystic fibrosis. 1: epidemiology and pathogenesis. Thorax 62:360–367. doi: 10.1136/thx.2006.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amadori A, Antonelli A, Balteri I, Schreiber A, Bugiani M, De Rose V. 2009. Recurrent exacerbations affect FEV(1) decline in adult patients with cystic fibrosis. Respir Med 103:407–413. doi: 10.1016/j.rmed.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld M, Emerson J, Williams-Warren J, Pepe M, Smith A, Montgomery AB, Ramsey B. 2001. Defining a pulmonary exacerbation in cystic fibrosis. J Pediatr 139:359–365. doi: 10.1067/mpd.2001.117288. [DOI] [PubMed] [Google Scholar]
- 8.Aaron SD, Ramotar K, Ferris W, Vandemheen K, Saginur R, Tullis E, Haase D, Kottachchi D, St Denis M, Chan F. 2004. Adult cystic fibrosis exacerbations and new strains of Pseudomonas aeruginosa. Am J Respir Crit Care Med 169:811–815. doi: 10.1164/rccm.200309-1306OC. [DOI] [PubMed] [Google Scholar]
- 9.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. 2004. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall BC. 2004. Pulmonary exacerbations in cystic fibrosis: it's time to be explicit! Am J Respir Crit Care Med 169:781–782. doi: 10.1164/rccm.2401009. [DOI] [PubMed] [Google Scholar]
- 11.Rogers GB, Hoffman LR, Johnson MW, Mayer-Hamblett N, Schwarze J, Carroll MP, Bruce KD. 2011. Using bacterial biomarkers to identify early indicators of cystic fibrosis pulmonary exacerbation onset. Expert Rev Mol Diagn 11:197–206. doi: 10.1586/erm.10.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stressmann FA, Rogers GB, Marsh P, Lilley AK, Daniels TW, Carroll MP, Hoffman LR, Jones G, Allen CE, Patel N, Forbes B, Tuck A, Bruce KD. 2011. Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J Cyst Fibros 10:357–365. doi: 10.1016/j.jcf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 13.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, Carroll MP, Parkhill J, Bruce KD. 2011. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels TW, Rogers GB, Stressmann FA, van der Gast CJ, Bruce KD, Jones GR, Connett GJ, Legg JP, Carroll MP. 2013. Impact of antibiotic treatment for pulmonary exacerbations on bacterial diversity in cystic fibrosis. J Cyst Fibros 12:22–28. doi: 10.1016/j.jcf.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. 2007. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol 73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266. [DOI] [PubMed] [Google Scholar]
- 17.Ho CC, Lau CC, Martelli P, Chan SY, Tse CW, Wu AK, Yuen KY, Lau SK, Woo PC. 2011. Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: a proof-of-concept study. J Clin Microbiol 49:814–821. doi: 10.1128/JCM.01702-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volkmann H, Schwartz T, Kirchen S, Stofer C, Obst U. 2007. Evaluation of inhibition and cross-reaction effects on real-time PCR applied to the total DNA of wastewater samples for the quantification of bacterial antibiotic resistance genes and taxon-specific targets. Mol Cell Probes 21:125–133. doi: 10.1016/j.mcp.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCafferty J, Mühlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. 2013. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J 7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arthur JC, Gharaibeh RZ, Uronis JM, Perez-Chanona E, Sha W, Tomkovich S, Mühlbauer M, Fodor AA, Jobin C. 2013. VSL#3 probiotic modifies mucosal microbial composition but does not reduce colitis-associated colorectal cancer. Sci Rep 3:2868. doi: 10.1038/srep02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 24.Pagani I, Liolios K, Jansson J, Chen I-MA, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. 2012. The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bittar F, Rolain JM. 2010. Detection and accurate identification of new or emerging bacteria in cystic fibrosis patients. Clin Microbiol Infect 16:809–820. doi: 10.1111/j.1469-0691.2010.03236.x. [DOI] [PubMed] [Google Scholar]
- 28.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLoS Comput Biol 8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8:e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eickmeier O, Huebner M, Herrmann E, Zissler U, Rosewich M, Baer PC, Buhl R, Schmitt-Grohé S, Zielen S, Schubert R. 2010. Sputum biomarker profiles in cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD) and association between pulmonary function. Cytokine 50:152–157. doi: 10.1016/j.cyto.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guss AM, Roeselers G, Newton IL, Young CR, Klepac-Ceraj V, Lory S, Cavanaugh CM. 2011. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J 5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price KE, Hampton TH, Gifford AH, Dolben EL, Hogan DA, Morrison HG, Sogin ML, O'Toole GA. 2013. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome 1:27. doi: 10.1186/2049-2618-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. 2012. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A 109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. 2012. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J 6:471–474. doi: 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, Wu B, Tran D, Koff J, Kleinhenz ME, Nielson D, Brodie EL, Lynch SV. 2010. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tunney MM, Klem ER, Fodor AA, Gilpin DF, Moriarty TF, McGrath SJ, Muhlebach MS, Boucher RC, Cardwell C, Doering G, Elborn JS, Wolfgang MC. 2011. Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax 66:579–584. doi: 10.1136/thx.2010.137281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogers GB, Cuthbertson L, Hoffman LR, Wing PA, Pope C, Hooftman DA, Lilley AK, Oliver A, Carroll MP, Bruce KD, van der Gast CJ. 2013. Reducing bias in bacterial community analysis of lower respiratory infections. ISME J 7:697–706. doi: 10.1038/ismej.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.