Abstract
We compared two multistep diagnostic algorithms based on C. Diff Quik Chek Complete and, as confirmatory tests, GenomEra C. difficile and Xpert C. difficile. The sensitivity, specificity, positive predictive value, and negative predictive value were 87.2%, 99.7%, 97.1%, and 98.3%, respectively, for the GenomEra-based algorithm and 89.7%, 99.4%, 95.5%, and 98.6%, respectively, for the Xpert-based algorithm. GenomEra represents an alternative to Xpert as a confirmatory test of a multistep algorithm for Clostridium difficile infection (CDI) diagnosis.
TEXT
Rapid diagnosis of Clostridium difficile infection (CDI) is crucial for optimal disease control (1, 2). For this reason, many microbiology laboratories used sensitive algorithms based on enzyme immunoassay (EIA) for detection of glutamate dehydrogenase (GDH) with EIA detection of toxins A and B, followed by a confirmatory test based on toxin A or B gene amplification (3–6).
C. Diff Quik Chek Complete (QC) (TechLab, Blacksburg, VA, USA) detects by immunochromatography both GDH and toxins A and B as a single procedure device (4). The real-time PCR assay Xpert C. difficile assay (Xpert) (GeneXpert; Cepheid, Sunnyvale, CA, USA) that detects the toxin B gene (tcdB), binary toxin genes, and tcdC 117-nucleotide (nt) deletion (epidemic 027 ribotype) is frequently used as a confirmatory test because of its speed and good internal validity values (7–11).
The new assay, GenomEra C. difficile (GenomEra) (Abacus Diagnostica, Turku, Finland), is a promising amplification system that detects the tcdB gene in approximately 1 h using rapid thermal cycling by means of a multiblock thermal cycler and homogeneous time-resolved fluorescence detection technology using lanthanide chelates which has proved to be resistant to background effects (12). This molecular method has the CE mark but is not cleared at this moment by the FDA.
The purpose of this study was to compare the diagnostic accuracy of two algorithms based on QC as the screening test and Xpert or GenomEra as confirmatory tests for the rapid diagnosis of CDI.
From October 2012 to March 2013, all loose stool specimens sent to the laboratory of the Hospital General Universitario Gregorio Marañón (Madrid, Spain) for CDI diagnosis were tested in parallel with the direct cytotoxicity assay, toxigenic culture, and the two multistep algorithms evaluated. The gold standard was the combination of direct cytotoxicity assay with stool specimens and cytotoxicity assay with isolates as previously described (13). The multistep algorithm consisted of an initial test, QC, performed according to the manufacturer's recommendations. Specimens positive for both GDH and toxins were considered positive, while specimens negative for both antigens were considered negative. Specimens with uncertain (GDH-positive and toxin-negative) results were tested in parallel using Xpert and GenomEra for confirmation. Xpert was performed according to the manufacturer's recommendations. GenomEra was performed by diluting 1 μl of sample in a tube with 1 ml of sample buffer and transferring 400 μl of the mixture into a tube containing glass beads to be vortexed for 5 min. Approximately 35 μl of the mixture was then transferred into a single-use disposable test chip that was introduced in the GenomEra CDX instrument for the automatic amplification procedure. Test results were reported by the GenomEra software in numerical form (−15 to +100), interpreted as “C. difficile tcdB negative” (values < −5), “borderline” (values from −5 to +5), or “positive” (values > +5). When there was a borderline result, the specimen was retested. The retest was considered positive if the value was more than +5 and negative otherwise. In both amplification procedures, the test was repeated for failed results (invalid results plus errors). Samples with two repeatedly failed results were excluded. Proportions were calculated with a 95% confidence interval following a binomial distribution. The sensitivity (Se) and specificity (Sp) were compared using a 2-tailed McNemar test for paired samples. The positive predictive value (PPV) and negative predictive value (NPV) were compared using a 2-tailed Fisher exact test. Data were analyzed using IBM SPSS Statistics, version 19.0 (Armonk, New York, USA).
During the study period, a total of 981 stool specimens from 801 patients (median age, 67.1 years; interquartile range [IQR], 50.4 to 79.3 years) were collected. The results were failed for three specimens using Xpert (0.3% of all specimens and 2.9% of specimens tested using molecular techniques) and two specimens using GenomEra (0.2% and 1.9%, respectively), although the result for only one specimen for each diagnostic system remained failed on retesting (0.1% and 1.0%, respectively, for each molecular test).
Therefore, 979 specimens from 799 patients were analyzed (Fig. 1). Toxigenic C. difficile was detected in 117 (11.9%) specimens from 96 patients using the gold standard procedure. Molecular systems were used as confirmatory tests in 103 specimens (10.5% of specimens) (Fig. 1). Four specimens (all positive for Xpert and toxigenic culture) yielded borderline results with GenomEra (values of −1, 0, 1, and 4). On retesting, three specimens had positive results, and one specimen yielded a borderline result (values of 65, 5, 64, and 45).
FIG 1.
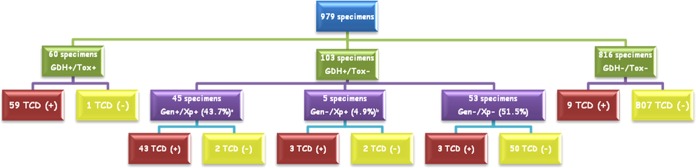
Results obtained in the comparison of diagnostic methods. The results are shown as follows: GDH, glutamate dehydrogenase detection; Tox, toxin A and B detection; TCD, toxigenic Clostridium difficile; Gen, GenomEra C. difficile assay; Xp, Xpert C. difficile assay; +, positive; −, negative. The superscript a after Gen+/Xp+ (43.7%) indicates that three culture-positive specimens had a borderline result using the GenomEra C. difficile assay; all 3 had a positive result on retest. The superscript b after Gen−/Xp+ (4.9%) indicates that one culture-positive specimen had a borderline result using the GenomEra C. difficile assay both on the first test and on retest; therefore, the final molecular result was considered negative.
When validity values of the evaluated procedures were compared, there were no statistically significant differences between both molecular biology-based algorithms although they showed significantly greater Se and NPV than those obtained with QC as a stand-alone test (Table 1).
TABLE 1.
Sensitivities, specificities, and positive and negative predictive values of the evaluated diagnostic procedures
| Test (manufacturer) | Mean (95% CI)a |
|||
|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | |
| Multistep algorithm using the following as confirmatory test | ||||
| GenomEra C. difficile assay | 87.2 (79.7–92.6) | 99.7 (99.0–99.9) | 97.1 (91.9–99.4) | 98.3 (97.2–99.0) |
| Xpert C. difficile assay | 89.7 (82.8–94.6) | 99.4 (98.7–99.8) | 95.5 (89.7–98.5) | 98.6 (97.6–99.3) |
| Quik Chek Complete as a stand-alone test | 50.4 (41.0–59.8) | 99.9 (99.4–99.9) | 98.3 (91.1–99.9) | 93.7 (91.9–95.2) |
The mean (95% confidence interval [95% CI]) values are shown. The P values for the comparison of validity values for the multistep algorithm using GenomEra C. difficile and that using Xpert C. difficile were 0.250 (sensitivity), 0.500 (specificity), 0.722 (positive predictive value), and 0.699 (negative predictive value). The P values for the comparison of validity values for the multistep algorithm using GenomEra C. difficile and Quik Chek Complete as a stand-alone test were <0.001 (sensitivity), 0.500 (specificity), 1 (positive predictive value), and <0.001 (negative predictive value). The P values for the comparison of validity values for the multistep algorithm using Xpert C. difficile and Quik Chek Complete as a stand-alone test were <0.001 (sensitivity), 0.125 (specificity), 0.426 (positive predictive value), and <0.001 (negative predictive value).
Our results show that QC as a stand-alone test, although very specific, had an Se of 50% owing to the limited Se of the toxin detection component, confirming that toxin EIA with or without GDH detection cannot be used as an accurate stand-alone test. For these reasons, most international guidelines recommend multistep algorithms (GDH screening test and molecular confirmation of GDH-positive results) (http://www.asm.org/images/pdf/Clinical/clostridiumdifficile9-21.pdf) (1, 2). Some authors also recommend including an intermediate test (i.e., toxin A and B EIA) to reduce the overall number of molecular tests used and the final diagnostic cost (4, 14).
We compared an algorithm using detection of GDH and toxins A and B in a single device (QC). For confirmation, we used two different molecular procedures, Xpert and a new system, GenomEra. Both molecular assays proved to be very easy to perform because most of the steps were automated. The hands-on time of Xpert was <2 min, whereas that of GenomEra was greater (8 to 10 min, including a vortex step of 5 min). The assay run time was 45 to 50 min for both; therefore, the overall turnaround time for both was <1 h. The proportion of failed results was low for both, decreasing to <1% after retesting, similar to that of another study evaluating Xpert (15).
Our study showed that although the Se of the Xpert-based procedure was slightly greater than that of the GenomEra-based one, the differences were not statistically significant. The Sp of both procedures were very high (close to 100%), resulting in very low number of false-positive results. GenomEra yielded 4 borderline results (3.9% of GenomEra tested specimens), all positive for toxigenic culture. After retesting, all except 1 gave positive results. In the only published evaluation of this assay, Hirvonen et al. found only 1 borderline result from 310 specimens tested with GenomEra (0.3%) (16). After retesting, this specimen, which was positive for toxigenic culture, yielded a negative result. It also showed that GenomEra had an Se, Sp, PPV, and NPV of 98.8%, 99.6%, 98.8%, and 99.6%, respectively. Unfortunately, these values cannot be compared with ours because GenomEra was used as a stand-alone test and not as part of a multistep algorithm.
Although Xpert has been widely evaluated (10, 15, 17–25), we found only 1 study evaluating it with the same algorithm as ours and using toxigenic culture as the gold standard (15). The authors found that the Se, Sp, PPV, and NPV of the algorithm were 86.1%, 97.8%, 88.6%, and 97.2%, respectively. The Se and NPV were similar to ours, while the Sp and PPV were lower, possibly because the large percentage of specimens positive by Xpert but negative by toxigenic culture were, according to the authors, true-positive results, given the positivity of other tests (e.g., toxin EIA and cytotoxicity assay).
In conclusion, detection of GDH and toxins A and B is insufficiently sensitive to be used as a stand-alone test for CDI diagnosis. The incorporation of a molecular test detecting toxin B gene to confirm GDH-positive and toxin-negative results can significantly increase the Se without decreasing the Sp and provides a cost-effective algorithm for rapid CDI diagnosis. In this sense, the multistep algorithm based on GenomEra proved to be a rapid and simple procedure for CDI diagnosis and represents an alternative to the Xpert-based multistep algorithm.
ACKNOWLEDGMENTS
The GenomEra C. difficile tests kits were kindly provided by Abacus Diagnostica (Turku, Finland). We are indebted to Thomas O'Boyle for editorial assistance.
This work was partially supported by Fondo de Investigaciones Sanitarias (FIS), Research Project number PI13/00687. Elena Reigadas holds a grant from the Río Hortega program of the Carlos III Health Institute, Spanish Government.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 2.Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect 15:1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee JH, Park SJ, Lee YM, Lee CH, Cho JH. 2012. Evaluation of the diagnostic algorithm consisting of enzyme immunoassay for toxins and polymerase chain reaction, for the diagnosis of Clostridium difficile-associated diarrhoea. Scand J Infect Dis 44:969–972. doi: 10.3109/00365548.2012.695456. [DOI] [PubMed] [Google Scholar]
- 4.Quinn CD, Sefers SE, Babiker W, He Y, Alcabasa R, Stratton CW, Carroll KC, Tang YW. 2010. C. Diff Quik Chek complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol 48:603–605. doi: 10.1128/JCM.01614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasoo S, Stevens J, Portillo L, Barza R, Schejbal D, Wu MM, Chancey C, Singh K. 2014. Cost-effectiveness of a modified two-step algorithm using a combined glutamate dehydrogenase/toxin enzyme immunoassay and real-time PCR for the diagnosis of Clostridium difficile infection. J Microbiol Immunol Infect 47:75–78. doi: 10.1016/j.jmii.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox MH, Planche T, Fang FC, Gilligan P. 2010. What is the current role of algorithmic approaches for diagnosis of Clostridium difficile infection? J Clin Microbiol 48:4347–4353. doi: 10.1128/JCM.02028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalpke AH, Hofko M, Zorn M, Zimmermann S. 2013. Evaluation of the fully automated BD MAX Cdiff and Xpert C. difficile assays for direct detection of Clostridium difficile in stool specimens. J Clin Microbiol 51:1906–1908. doi: 10.1128/JCM.00344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyorke CE, Wang S, Leslie J L, Cohen SH, Solnick J V, Polage CR. 2013. Evaluation of Clostridium difficile fecal load and limit of detection during a prospective comparison of two molecular tests, the illumigene C. difficile and Xpert C. difficile/Epi tests. J Clin Microbiol 51:278–280. doi: 10.1128/JCM.02120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM. 2012. Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J Clin Microbiol 50:1331–1335. doi: 10.1128/JCM.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin S, Kim M, Lim H, Kim H, Lee K, Chong Y. 2012. Evaluation of the Xpert Clostridium difficile assay for the diagnosis of Clostridium difficile infection. Ann Lab Med 32:355–358. doi: 10.3343/alm.2012.32.5.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover FC, Baron EJ, Peterson LR, Persing DH. 2011. Laboratory diagnosis of Clostridium difficile infection can molecular amplification methods move us out of uncertainty? J Mol Diagn 13:573–582. doi: 10.1016/j.jmoldx.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Lode P, Rosenberg J, Pettersson K, Takalo H. 2003. A europium chelate for quantitative point-of-care immunoassays using direct surface measurement. Anal Chem 75:3193–3201. doi: 10.1021/ac0340051. [DOI] [PubMed] [Google Scholar]
- 13.Alcala L, Sanchez-Cambronero L, Catalan MP, Sanchez-Somolinos M, Pelaez MT, Marin M, Bouza E. 2008. Comparison of three commercial methods for rapid detection of Clostridium difficile toxins A and B from fecal specimens. J Clin Microbiol 46:3833–3835. doi: 10.1128/JCM.01060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp SE, Ruden LO, Pohl J C, Hatcher PA, Jayne LM, Ivie WM. 2010. Evaluation of the C. Diff Quik Chek Complete Assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of Clostridium difficile disease. J Clin Microbiol 48:2082–2086. doi: 10.1128/JCM.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novak-Weekley SM, Marlowe EM, Miller J M, Cumpio J, Nomura J H, Vance PH, Weissfeld A. 2010. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol 48:889–893. doi: 10.1128/JCM.01801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirvonen JJ, Mentula S, Kaukoranta SS. 2013. Evaluation of a new automated homogeneous PCR assay, GenomEra C difficile, for rapid detection of toxigenic Clostridium difficile in fecal specimens. J Clin Microbiol 51:2908–2912. doi: 10.1128/JCM.01083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babady NE, Stiles J, Ruggiero P, Khosa P, Huang D, Shuptar S, Kamboj M, Kiehn TE. 2010. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol 48:4519–4524. doi: 10.1128/JCM.01648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan BW, Mackey TL, Daly J A, Alger G, Denys GA, Peterson LR, Kehl SC, Ledeboer NA. 2012. Multicenter clinical evaluation of the portrait toxigenic C. difficile assay for detection of toxigenic Clostridium difficile strains in clinical stool specimens. J Clin Microbiol 50:3932–3936. doi: 10.1128/JCM.02083-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapin KC, Dickenson RA, Wu F, Andrea SB. 2011. Comparison of five assays for detection of Clostridium difficile toxin. J Mol Diagn 13:395–400. doi: 10.1016/j.jmoldx.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldenberg SD, Dieringer T, French GL. 2010. Detection of toxigenic Clostridium difficile in diarrheal stools by rapid real-time polymerase chain reaction. Diagn Microbiol Infect Dis 67:304–307. doi: 10.1016/j.diagmicrobio.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Goldenberg SD, Gumban M, Hall A, Patel A, French GL. 2011. Lack of effect of strain type on detection of toxigenic Clostridium difficile by glutamate dehydrogenase and polymerase chain reaction. Diagn Microbiol Infect Dis 70:417–419. doi: 10.1016/j.diagmicrobio.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Rocha C, Barra-Carrasco J, Alvarez-Lobos M, Paredes-Sabja D, Guzman-Duran AM. 2013. Prospective comparison of a commercial multiplex real-time polymerase chain reaction and an enzyme immunoassay with toxigenic culture in the diagnosis of Clostridium difficile-associated infections. Diagn Microbiol Infect Dis 75:361–365. doi: 10.1016/j.diagmicrobio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Huang H, Weintraub A, Fang H, Nord CE. 2009. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of Clostridium difficile infections. J Clin Microbiol 47:3729–3731. doi: 10.1128/JCM.01280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong E, Marlowe EM, Whitmore J, Persing DH. 2010. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol 48:3719–3724. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zidaric V, Kevorkijan BK, Oresic N, Janezic S, Rupnik M. 2011. Comparison of two commercial molecular tests for the detection of Clostridium difficile in the routine diagnostic laboratory. J Med Microbiol 60:1131–1136. doi: 10.1099/jmm.0.030163-0. [DOI] [PubMed] [Google Scholar]


