Abstract
The genus Yersinia is a large and diverse bacterial genus consisting of human-pathogenic species, a fish-pathogenic species, and a large number of environmental species. Recently, the phylogenetic and population structure of the entire genus was elucidated through the genome sequence data of 241 strains encompassing every known species in the genus. Here we report the mining of this enormous data set to create a multilocus sequence typing-based scheme that can identify Yersinia strains to the species level to a level of resolution equal to that for whole-genome sequencing. Our assay is designed to be able to accurately subtype the important human-pathogenic species Yersinia enterocolitica to whole-genome resolution levels. We also report the validation of the scheme on 386 strains from reference laboratory collections across Europe. We propose that the scheme is an important molecular typing system to allow accurate and reproducible identification of Yersinia isolates to the species level, a process often inconsistent in nonspecialist laboratories. Additionally, our assay is the most phylogenetically informative typing scheme available for Y. enterocolitica.
INTRODUCTION
The Gram-negative Yersinia is one of the most important and well-studied bacterial genera, consisting of three human pathogens. Y. pestis is the causative agent of bubonic and pneumonic plague and is a recently diverged clone of Yersinia pseudotuberculosis (1), which alongside Y. enterocolitica is a zoonotic gastrointestinal pathogen (2). The remaining species are not associated with human disease and are considered to be environmental organisms, with the exception of the common fish pathogen Y. ruckeri (2) and the insecticidal species Y. entomophaga. Of the human-pathogenic species, Y. enterocolitica is the most common etiological agent of human disease, and in Germany and Scandinavia, the numbers of cases of human intestinal yersiniosis caused by Y. enterocolitica rival those caused by Salmonella (3). Y. enterocolitica is in itself a very diverse species that is classically subdivided into nonpathogenic, low-pathogenic, and high-pathogenic biotypes based on virulence in a mouse infection model (4). Biotype 1A isolates are considered nonpathogenic, which is concordant with a lack of the major Y. enterocolitica virulence factors such as pYV, invasin, YadA, and Ail (5), although there are numerous reports of biotype 1A human carriage (6, 7). Biotype 1B isolates are high pathogenic, which is concordant with carriage of the high-pathogenicity island, but isolation from human disease cases is very rare with the exception of notable outbreaks such as the recent emergence in Poland (8). Biotype 2 to 4 isolates are low pathogenic and are globally the most common causes of human gastrointestinal yersiniosis (4). Biotype 5 isolates are also considered low pathogenic but have only been isolated from wild hare populations and are very rare in nature (5).
From a clinical perspective, the isolation and subsequent identification of Yersinia and in particular Y. enterocolitica to the species and subspecies levels can be challenging, with recent publications striving to improve the efficacy of selective culturing of Yersinia from clinical and environmental samples (9). Once isolated, strains are most commonly identified to the species level by comparing the differential utilization of a panel of 17 biochemical substrates (4, 10). Further subdivision of Y. enterocolitica into biotypes is also performed based on utilization of a further 12 substrates. In both cases, the interpretation of such biochemical typing may often be subjective and affected by environmental factors such as temperature of incubation (4, 10). There is also further subdivision based on classical serotyping. As such, the identification of Yersinia to the species and subspecies levels can be very problematic for nonspecialist laboratories with misidentification at the species level and subtyping level not an uncommon occurrence, as exemplified by recent assignment of new species by molecular methods following inconclusive species determination by biochemical methods (11, 12).
Recent work by our group definitively characterized the phylogeny of the entire Yersinia genus using genome sequences of 241 strains encompassing the full diversity within the genus (13). Particular attention was given to Y. enterocolitica, of which 94 strains encompassing all biotypes and serotypes were sequenced. The whole-genus phylogeny was constructed using 84 housekeeping genes that are located on 23 syntenic blocks, regions of DNA-containing genes conserved across the genus, and showed the presence of 14 species clusters as determined by Bayesian analysis of population structure (BAPS) software (13). The resulting phylogeny also accurately distinguished Y. pestis as a distinct clone of Y. pseudotuberculosis and phylogenetically split Y. enterocolitica on the basis of high-pathogenic, low-pathogenic, and nonpathogenic biotypes (13). A core genome single nucleotide polymorphism (SNP)-based phylogeny provided greater resolution for Y. enterocolitica and showed that the phylogenetic separation within the low-pathogenic strains is concordant with serotype and not biotype, which is almost certainly due to difficulties in interpreting variable reactions discriminating biotypes 2 and 3 (13).
Given our significant findings on the true phylogenetic structure of the entire Yersinia genus and the fact that this phylogeny can be determined from housekeeping genes present on conserved syntenic blocks, we sought to determine if a standard, seven-gene multilocus sequence typing (MLST) scheme could be developed from a subset of those genes. Such a scheme would then be able to rapidly and with complete accuracy identify any member of the Yersinia genus to the species and subspecies levels upon the initial isolation. There is a well-established MLST scheme available for Y. pseudotuberculosis (14) that has been used to delineate the population structure of the species complex (15); however, this scheme has not been designed to be robust across the genus. Similarly, there have been attempts to create MLST schemes for Y. enterocolitica (16–18); however, these have not been informed by genomic data and their suitability for identification to the species and subspecies levels is questionable compared to that of our previous whole-genome phylogeny study (13). Here, we present the design and validation of a new pan-Yersinia MLST scheme that provides identification to the species level that is completely concordant with our previous whole-genome phylogeny (13). Furthermore, it accurately differentiates Y. pestis and Y. similis from Y. pseudotuberculosis and, more significantly, the scheme subtypes low-pathogenic Y. enterocolitica on the basis of serotype in complete concordance with whole-genome phylogeny of the species. We propose that the pan-Yersinia MLST scheme is an invaluable tool in the identification of Yersinia to the species and subspecies levels from clinical samples and that the classification of low-pathogenic Y. enterocolitica on the basis of phylogenetically distinct serotypes be adopted.
MATERIALS AND METHODS
Strains.
The initial design and development of the MLST scheme utilized de novo assembled genome sequences of 171 Yersinia strains that were part of our previously published work (13). This strain collection was made up of the Yersinia species as follows: 1 Y. aldovae, 2 Y. aleksiciae, 3 Y. bercovieri, 58 Y. enterocolitica, 22 Y. frederiksenii, 16 Y. intermedia, 9 Y. kristensenii, 1 Y. massiliensis, 10 Y. mollaretii, 1 Y. pekkanenii, 3 Y. pestis, 33 Y. pseudotuberculosis, 5 Y. rohdei, 3 Y. ruckeri, and 4 Y. similis. The 171 strains are a subset of the 241 sequenced in our previous study and were chosen because their assembled genomes contained no ambiguous base calls or contig breaks in the syntenic blocks our study design focused on.
Selection of phylogenetically informative genes within conserved syntenic blocks.
To establish the level of genetic diversity of each of the common housekeeping genes, GenBank files of each of the 23 syntenic blocks from Y. enterocolitica 8081, Y. pseudotuberculosis IP32953, and Y. pestis D106004 were created using Artemis (19). The sequences of each of the conserved housekeeping genes were then extracted and aligned in MEGA 5.0 (20), as these represent the three human-pathogenic species that are located at diametrically opposite ends of the genus phylogeny. The genes that had a level of SNPs between 10 and 25% were retained for further analysis. The sequences of the remaining genes were used to create individual gene maximum likelihood trees using MEGA 5.0 and compared to the Yersinia phylogeny (13). Seven genes that were able to closely match the branching order and clearly discriminate between the species clusters, with <2% strain displacement, and that were disseminated across the syntenic blocks were chosen. Pan-Yersinia gene primers for the seven selected genes were designed based on the multiple alignments.
PCR and sequence analysis.
The culture was grown overnight in 1.5 ml LB broth at 25°C with shaking, and genomic DNA was extracted using the GenElute bacterial genomic DNA kit (Sigma-Aldrich), following the manufacturer's instructions. A temperature gradient PCR was used to establish the optimum annealing temperatures for the primers. The result was optimized by carrying out the PCR on representative strains of all the species for each primer pair as follows: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 30 s, annealing temperature dependent upon the primer set for 30 s; elongation at 72°C for 30 s; and final elongation at 72°C for 5 min. PCRs were carried out using the GoTaq Flexi DNA polymerase kit (Promega) and deoxynucleoside triphosphates (dNTPs) (Promega) as follows: 5 μl 1.5 mM MgCl2, 5 μl 10× PCR buffer, 2 μl 10 μM dNTPs, 0.3 μl 5 U/μM Taq DNA polymerase, 40 μl sterilized distilled water, 0.5 μl 10 pmol forward and reverse primers, 1 μl ∼10 ng/μl DNA. The amplification product was then cleaned using ExoSAP-IT (Affymetrix) and Sanger sequenced in duplicate to obtain independent forward and reverse reactions.
The sequence data obtained for each gene were aligned and trimmed to a uniform length, using MEGA 5.0. Each unique sequence was identified using the Web tool Non-redundant databases (http://pubmlst.org/analysis/) and allocated a specific allele number. All of the sequence and isolate data were uploaded to the publically available MLST database (http://pubmlst.org/yersinia) using the BIGSdb genomics platform (21).
Phylogenetic and population analysis of MLST data.
The freely available software START (22) was used to calculate the ratio of nonsynonymous (dN) to synonymous (dS) nucleotide substitutions to determine the level of selective pressure acting upon each MLST gene. START was also used to determine that the GC content in the MLST genes was comparable to that of the whole-genome GC content. To detect recombination within the Y. enterocolitica MLST data, SplitsTree 4.2 (23) was used to compute the pairwise homoplasy index (PHI). An MLST database and Web interface were created for the scheme (http://pubmlst.org/yersinia/), and the sequence data for all seven loci from all 171 individual strains were input to assign allele numbers. From these sequences, types were ascribed to each unique allele combination occurring in the data set. The designated allele numbers were visualized by creating minimum spanning trees using the goeBURST Full MLST algorithm in PHYLOViZ (24). Maximum likelihood phylogenies were created by concatenation of the sequence of the seven loci and alignment with ClustalW in MEGA 5.0, before the phylogeny was determined with the GTR gamma model in RAxML 7.2.8–2 (25).
RESULTS
Selection of genes and validation of a pan-Yersinia MLST scheme on in silico genome sequence data.
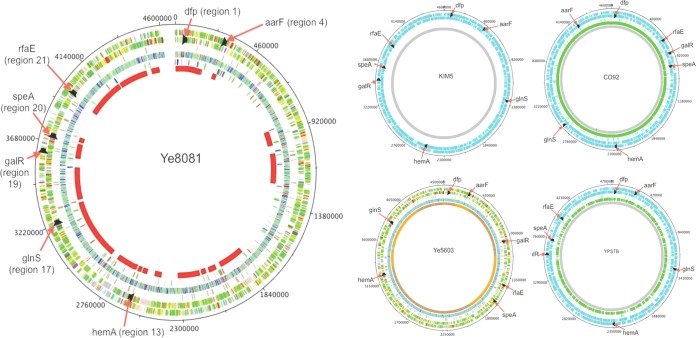
The sequences of 73 genes conserved across the genus (Table 1) from 171 de novo assembled genomes were used to create individual gene phylogenies. Additionally, the alignments were used to identify regions of high similarity in each gene that would permit the design of universal primers capable of amplifying the gene across the genus. From this analysis, seven optimal gene loci were selected based on their ability to mirror the genome-informed phylogeny and the ability to design primers that would work across the genus (Table 2), as well as their separation across the syntenic blocks (Fig. 1).
TABLE 1.
The 73 housekeeping genes selected for investigation for use in the genus MLST scheme
| Syntenic block | Relative location on each syntenic block in relation to the Y. enterocolitica 8081 reference genome: |
Size (bp) | Housekeeping gene(s) in each block | |
|---|---|---|---|---|
| Beginning | End | |||
| 1 | 0 | 107030 | 107,030 | asnA, dfp, tpiA, glnA |
| 2 | 108300 | 191330 | 83,030 | sthA |
| 3 | 191500 | 202555 | 11,055 | rhlB, rho |
| 4 | 282830 | 313630 | 30,800 | udp, aarF, hemB |
| 5 | 879520 | 980830 | 101,310 | pcm, recA |
| 6 | 1039400 | 1251800 | 212,400 | gloB, nadB, guaA, nrdF, nrdE |
| 7 | 1802900 | 1991400 | 188,500 | purB, ptsG, phoQ, phoP, purT, pip, tmk, icdA |
| 8 | 2027865 | 2087750 | 59,885 | kduD1 |
| 9 | 2108500 | 2142600 | 34,100 | NDa |
| 10 | 2154500 | 2325300 | 170,800 | tyrR |
| 11 | 2447240 | 2499700 | 52,460 | topB, ansA, dadA, nhaB, fadR, xthA |
| 12 | 2554700 | 2591553 | 36,853 | minD, zwf, aspS, znuC, znuA, znuB, minC, rnd, msbB |
| 13 | 2602500 | 2630230 | 27,730 | kdsA, prfA, hemA |
| 14 | 2640950 | 2668185 | 27,235 | chaA |
| 15 | 2668285 | 2709585 | 41,300 | ND |
| 16 | 2709700 | 2800900 | 91,200 | ND |
| 17 | 2854263 | 3294700 | 440,437 | folE, nadA, udk, sfcA, glnS |
| 18 | 3313400 | 3544778 | 231,378 | proB, rosA, hemH, adk |
| 19 | 3610900 | 3712864 | 101,964 | thyA, tas, lgt, galR, lysS, prfB |
| 20 | 3726200 | 3761260 | 35,060 | tktA, speA, gshB, endA |
| 21 | 3960000 | 4238800 | 278,800 | rfaE, pyrB, parC, gcp, uxaC |
| 22 | 4245400 | 4464400 | 219,000 | ND |
| 23 | 4504400 | 4561500 | 57,100 | fdoI, fdhE, glnQ |
| Total size of syntenic blocks | 2,639,427 | |||
| Total size of Y. enterocolitica 8081 genome | 4,615,899 | |||
ND, no housekeeping genes present in the syntenic block.
TABLE 2.
Primer sequences, the sizes of the amplified regions, and the annealing temperature for the final seven selected MLST genes
| MLST gene | Primer |
PCR product length (bp) | MLST region length (bp) | Annealing temperature (°C) | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| aarF | 5′-TTCCATGCAGATATGCACC-3′ | 3′-CCACTCACTAATAGTGTAGC-5′ | 650 | 500 | 52 |
| dfp | 5′-GATCCGGTACGCTTTATCAG-3′ | 3′-CATAACGGCTGACAATCTCG-5′ | 547 | 455 | 59 |
| galR | 5′-ATTGGTAACGGTTACCATG-3′ | 3′-GTTGGGCTGAACATATTGGT-5′ | 648 | 500 | 59 |
| glnS | 5′-GAATCATGTATCCGTGATG-3′ | 3′-GCACAGAAATAACCTTCAC-5′ | 557 | 442 | 56.5 |
| hemA | 5′-ATGACTCTGCTCGCATTAGG-3′ | 3′-CGGTTGGCAATAATCATATG-5′ | 602 | 490 | 54 |
| speA | 5′-ATGTCTGATGATAACTTGATT-3′ | 3′-CAGATAAACTTTATGGCCC-5′ | 550 | 452 | 55.5 |
| rfaE | 5′-ATGAAAGTCACTCTGCCTGA-3′ | 3′-ATCACTGCCTTTAGGATC-5′ | 509 | 429 | 55.5 |
FIG 1.
Diagram showing the positioning of the seven selected loci and their native syntenic block on reference genomes across the genus. Ye8081, Y. enterocolitica bioserotype 1B/O:8; KIM5, Y. pestis Medievalis; CO92, Y. pestis Orientalis; Ye5603, Y. enterocolitica bioserotype 4/O:3; YPSTB, Y. pseudotuberculosis YPIII serotype III.
There was a high level of diversity shown across the seven selected MLST regions, averaging around 60 alleles and 40% polymorphic sites for each (Table 3). The dN/dS ratios were far below 1 for each MLST region, suggesting that the nucleotide substitutions are not a result of selective pressure. The average GC content found in the MLST gene regions corresponds to that of the Yersinia chromosomes, which ranges from 46.9% in Y. frederiksenii to 49.0% in Y. mollaretii (data accessible at the xBASE website http://www.xbase.ac.uk/taxon/Yersinia). The PHI test also failed to detect any recombination within the MLST amplicons from the Y. enterocolitica data set.
TABLE 3.
Level diversity across all 171 genome-sequenced strains for each of the MLST regions as determined by START
| Gene | Size of fragment (bp) | % GC content | % polymorphic sites | dN/dS ratio | No. of alleles |
|---|---|---|---|---|---|
| aarF | 500 | 44 | 37.4 | 0.0049 | 58 |
| dfp | 500 | 47.8 | 40.8 | 0.0599 | 61 |
| galR | 500 | 49.7 | 44.8 | 0.028 | 70 |
| glnS | 500 | 48.5 | 38.4 | 0.0221 | 68 |
| hemA | 500 | 51.3 | 39.8 | 0.0222 | 65 |
| rfaE | 429 | 54.1 | 40.3 | 0.019 | 60 |
| speA | 490 | 48.9 | 34.7 | 0.0232 | 50 |
| Mean | 488.4 | 49.2 | 39.5 | 0.0256 | 61.7 |
Pan-Yersinia MLST scheme is phylogenetically informative to genome sequence level.
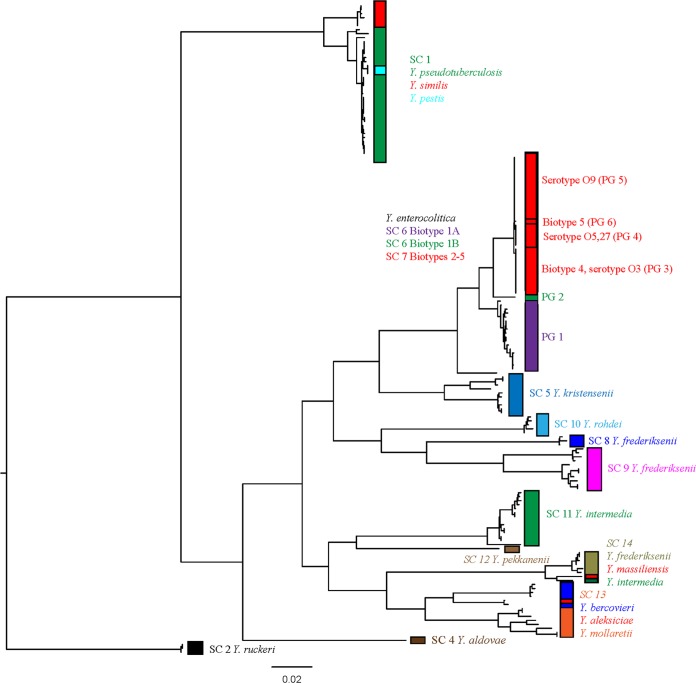
A maximum likelihood phylogeny of the concatenated MLST data obtained from the 171 genome-sequenced strains was constructed. The resulting tree showed accurate phylogenetic separation of all of the species identified by the 84-gene tree approach taken in our previous work (Fig. 2) with 100% concordance between the two phylogenies and identical separation into 14 distinct species clusters as determined by BAPS. The Y. pseudotuberculosis complex was accurately split with distinct clades containing Y. pestis and Y. similis within the larger Y. pseudotuberculosis complex, showing that the scheme is capable of differentiating accurately within this lineage. Closer investigation of the Y. enterocolitica complex showed that the MLST scheme also differentiates on the basis of high-pathogenic, low-pathogenic, and nonpathogenic groups, and within the low-pathogenic group differentiates on the basis of serotype into defined phylogroups as observed when the whole-genome phylogeny is used. As such, the pan-Yersinia MLST scheme provides a completely robust mechanism by which to accurately assign any Yersinia isolate to a defined species cluster and further subtype without any additional growth requirements beyond initial isolation.
FIG 2.
Maximum likelihood phylogeny of concatenated alleles derived from every unique sequence type obtained from 171 genomes from across the genus. The species contained within each sequence type are indicated, and species clusters are labeled as defined in our previous genome study (13), with the MLST tree showing complete concordance with our previous phylogeny.
Validation of the pan-Yersinia MLST scheme on reference laboratory isolate collections.
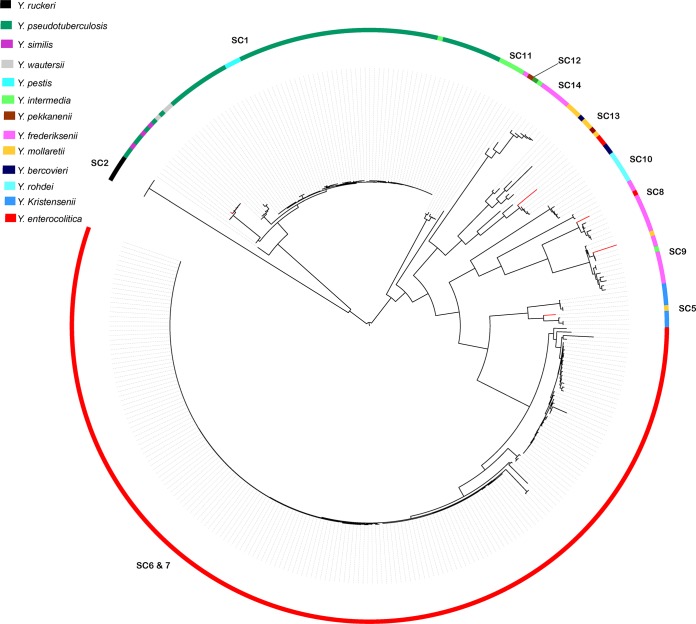
To validate the in silico results for our genus-wide typing scheme, we performed MLST on a further 214 Yersinia strains archived in the national Yersinia reference laboratories of Belgium, Germany, United Kingdom, and France (see Table S1 in the supplemental material). The concatenated MLST sequence data for all 385 strains were then used to construct a maximum likelihood phylogeny and compare the results of the classical biochemical typing and subtyping with those for our phylogenetic approach (Fig. 3). The phylogeny once again shows unambiguous separation of strains into the previously designated species clusters, with 97.83% of strains tested being assigned to the corresponding species cluster based on their biochemical typing. Included here are strains of Y. wautersii, a newly proposed species which is a sublineage of Y. pseudotuberculosis. Two strains biochemically defined as Y. pseudotuberculosis by the reference laboratories with the Y. similis subgroup and a further 6 isolates were assigned to species clusters in disagreement with their classical biochemical typing designation by the reference laboratories.
FIG 3.
Maximum likelihood phylogeny of concatenated alleles derived from 385 reference laboratory strains of Yersinia. The species clusters are designated on each clade as SC, with the biochemically determined species of each strain denoted by the described color coding. Strains whose species cluster differed from biochemical typing results are denoted by red branches on the phylogeny.
To validate the in silico results showing that our MLST scheme was able to successfully subtype Y. enterocolitica, we separately analyzed the MLST data for the 188 Y. enterocolitica isolates contained within the entire data set generated here (Fig. 4). Our phylogeny perfectly assigns every strain to a defined phylogroup on the basis of serotype as previously reported with whole-genome SNP-based phylogeny. There are no ambiguous phylogroup assignations on the basis of serotype, although, as with the whole-genome study, biotype is not phylogenetically robust. To allow an easy comparator for use of the scheme, we assigned which species cluster and/or Y. enterocolitica phylogroup each sequence type belongs to (see Table S2 in the supplemental material).
FIG 4.
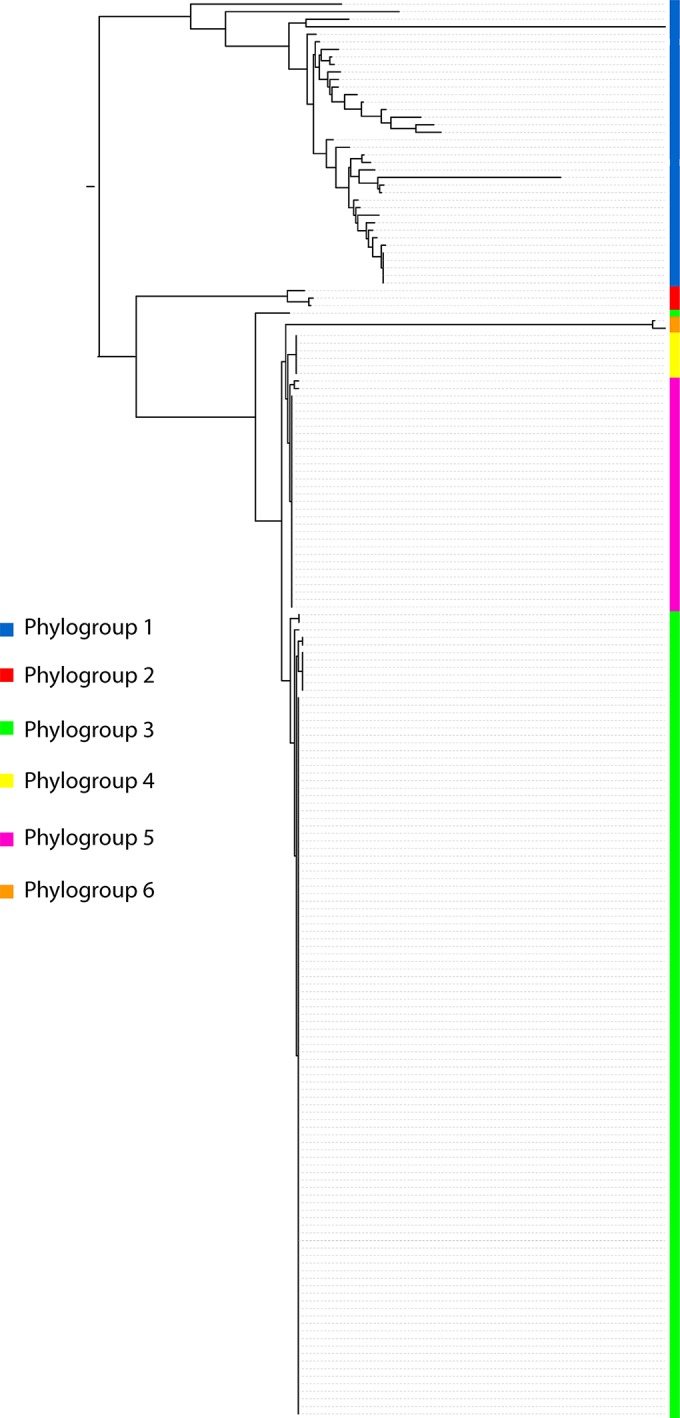
Maximum likelihood phylogeny of concatenated alleles derived from 188 Y. enterocolitica strains. The phylogroups are indicated by the described color coding.
DISCUSSION
The enteropathogenic Yersinia spp. are the third most common cause of bacterial infectious intestinal disease in the developed world (5). Despite this, the isolation and identification of infections with Y. enterocolitica and Y. pseudotuberculosis are still heavily reliant on classical biochemical techniques that may be open to subjective interpretation to provide a definitive identification (4, 10). This subjective biochemical typing is even more problematic when applied to subtyping of isolates, which is of importance in epidemiological tracking, and in the case of Y. enterocolitica may be of clinical importance in distinguishing between the carriage of a nonpathogenic organism, a self-limiting infection with a low-pathogenic strain, or an infection with a more aggressive high-pathogenic strain type. Similarly, nonpathogenic species within the genus may be biochemically typed as atypical Y. enterocolitica, leading to misidentification of clinical episodes, administration of unnecessary treatments, and skewed data in environmental and livestock surveys of enteropathogenic Yersinia prevalence (26, 27).
Despite the proven levels of resolution offered by molecular typing techniques for bacterial pathogens to overcome such problems, there is no such approved and standardized methodology in place for Y. enterocolitica, the most common cause of human gastrointestinal yersiniosis. An MLST scheme does exist for Y. pseudotuberculosis but is designed and validated to be used as an epidemiological and population genetics tool solely for that species (15). In this study, we have utilized the comprehensive genus genome sequence data set previously produced by our group (13) to inform the design of an MLST-based scheme that can rapidly and reproducibly assign any strain to a defined species cluster and any Y. enterocolitica to a defined phylogroup.
Previous attempts have been made to create MLST typing tools for Y. enterocolitica. The first scheme (16) was a 5-locus scheme incorporating 16S that was developed to allow phylogenetic inferences within the genus Yersinia. However, when the phylogeny published in that pregenomics era study is compared to our definitive phylogeny recently published (13), it is clear that the 5-locus phylogeny is inaccurate with Y. enterocolitica deeply embedded within environmental species (16). As such, determining the species using this scheme on an unknown isolate would not offer sufficiently robust resolution for reference laboratory adoption. A conventional 7-locus scheme was developed from a semirandom selection of housekeeping genes to investigate subgrouping within the nonpathogenic biotype 1A Y. enterocolitica isolates (17). While the loci in this scheme are among the 84 genes conserved across the genus, in silico analysis suggests that the primers designed may not be optimal across the genus due to base mismatches at the primer sites and as such would not be suitable for the purposes of identifying Yersinia isolates to the species level. Most recently, a scheme was developed to differentiate the three human-pathogenic species of the Yersinia genus using a 7-locus MLST scheme (18). This scheme accurately subtyped Y. enterocolitica into distinct subtypes, including serotype-specific clades within the low-pathogenic strains, as observed both in our scheme and in our genomic phylogeny (13). However, this scheme also uses primers that in silico analysis suggests would not anneal to sequences from some species. Additionally, neither of the latter two schemes has been set up with a database and protocols to allow its wide-scale adoption for reference typing.
In conclusion, we present a model and novel design strategy for molecular typing tools based on genome sequence data across an entire genus containing human-pathogenic species. By using these data, we can design a simple MLST-based scheme that provides the power of resolution of whole-genome sequencing to quickly and accurately identify isolates to the species level and also subtype strains of Y. enterocolitica. While next-generation sequencing is becoming commonplace in a small number of public health laboratories, there are still many front-line clinical microbiology laboratories that are not yet in a position to employ benchtop sequencing due to the cost or bioinformatics resources. Our scheme provides a blueprint for the efficient design of simple molecular-based tools that provide an equal level of resolution for typing, although obviously not for SNP-based molecular epidemiological investigations. We encourage the public health microbiology community to adopt our scheme and further validate it as a universal typing tool for the entire Yersinia genus and as a subtyping and population genetics tool for the important human pathogen Y. enterocolitica.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by NTU VC studentships awarded to M.H. and S.R. The French Yersinia Reference Laboratory was funded in part by the Institute for Health Surveillance (InVS).
We thank the National Collection of Type Cultures for provision of strains from across the genus for our validation work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02395-14.
REFERENCES
- 1.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A 101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wren BW. 2003. The Yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol 1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- 3.Rosner BM, Stark K, Werber D. 2010. Epidemiology of reported Yersinia enterocolitica infections in Germany, 2001–2008. BMC Public Health 10:337. doi: 10.1186/1471-2458-10-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottone EJ. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect 1:323–333. doi: 10.1016/S1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 5.Bottone EJ. 1997. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev 10:257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tennant SM, Grant TH, Robins-Browne RM. 2003. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol Med Microbiol 38:127–137. doi: 10.1016/S0928-8244(03)00180-9. [DOI] [PubMed] [Google Scholar]
- 7.Singh I, Virdi JS. 2004. Production of Yersinia stable toxin (YST) and distribution of yst genes in biotype 1A strains of Yersinia enterocolitica. J Med Microbiol 53:1065–1068. doi: 10.1099/jmm.0.45527-0. [DOI] [PubMed] [Google Scholar]
- 8.Gierczyñski R, Szych J, Rastawicki W, Wardak S, Jagielski M. 2009. Molecular characterization of human clinical isolates of Yersinia enterocolitica bioserotype 1B/O8 in Poland: emergence and dissemination of three highly related clones. J Clin Microbiol 47:1225–1228. doi: 10.1128/JCM.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savin C, Leclercq A, Carniel E. 2012. Evaluation of a single procedure allowing the isolation of enteropathogenic Yersinia along with other bacterial enteropathogens from human stools. PLoS One 7:e41176. doi: 10.1371/journal.pone.0041176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wauters G, Kandolo K, Janssens M. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol 9:14–21. [PubMed] [Google Scholar]
- 11.Murros-Kontiainen A, Fredriksson-Ahomaa M, Korkeala H, Johansson P, Rahkila R, Björkroth J. 2011. Yersinia nurmii sp. nov. Int J Syst Evol Microbiol 61:2368−2372. doi: 10.1099/ijs.0.024836-0. [DOI] [PubMed] [Google Scholar]
- 12.Murros-Kontiainen A, Johansson P, Niskanen T, Fredriksson-Ahomaa M, Korkeala H, Björkroth J. 2011. Yersinia pekkanenii sp. nov. Int J Syst Evol Microbiol 61:2363−2367. doi: 10.1099/ijs.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 13.Reuter S, Connor T, Barquist L, Walker D, Feltwell T, Harris S, Fookes M, Hall M, Petty N, Fuchs T, Corander J, Dufour M, Ringwood T, Savin C, Bouchier C, Martin L, Miettinen M, Shubin M, Riehm J, Laukkanen-Ninios R, Sihvonen L, Siitonen A, Skurnik M, Falcão J, Fukushima H, Scholz H, Prentice M, Wren B, Parkhill J, Carniel E, Achtman M, McNally A, Thomson N. 2014. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc Natl Acad Sci U S A 111:6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laukkanen-Ninios R, Didelot X, Jolley K, Morelli G, Sangal V, Kristo P, Brehony C, Imori P, Fukushima H, Siitonen A, Tseneva G, Voskressenskaya E, Falcao J, Korkeala H, Maiden M, Mazzoni C, Carniel E, Skurnik M, Achtman M. 2011. Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ Microbiol 13:3114–3127. doi: 10.1111/j.1462-2920.2011.02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotetishvili M, Kreger A, Wauters G, Morris JG Jr, Sulakvelidze A, Stine OC. 2005. Multilocus sequence typing for studying genetic relationships among Yersinia species. J Clin Microbiol 43:2674–2684. doi: 10.1128/JCM.43.6.2674-2684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sihvonen L, Jalkanen K, Huovinen E, Toivonen S, Corander J, Kuusi M, Skurnik M, Siitonen A, Haukka K. 2012. Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties. BMC Microbiol 12:208. doi: 10.1186/1471-2180-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan R, Liang J, Shi G, Cui Z, Hai R, Wang P, Xiao Y, Li K, Qiu H, Gu W, Du X, Jing H, Wang X. 2014. Homology analysis of pathogenic Yersinia species Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis based on multilocus sequence typing. J Clin Microbiol 52:20–29. doi: 10.1128/JCM.02185-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731−2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolley KS, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolley KA, Feil EJ, Chan MS, Maiden MC. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- 23.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 24.Francisco AP, Bugalho M, Ramirez M, Carrico JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 26.Milnes A, Stewart I, Clifton-Hadley F, Davies R, Newell D, Sayers A, Cheasty T, Cassar C, Ridley A, Cook A, Evans S, Teale C, Smith R, McNally A, Toszeghy M, Futter R, Kay A, Paiba G. 2008. Intestinal carriage of verocytotoxigenic Escherichia coli O157, Salmonella, thermophilic Campylobacter and Yersinia enterocolitica, in cattle, sheep and pigs at slaughter in Great Britain during 2003. Epidemiol Infect 136:739–751. doi: 10.1017/S0950268807009223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNally A, Cheasty T, Fearnley C, Dalziel RW, Paiba GA, Manning G, Newell DG. 2004. Comparison of the biotypes of Yersinia enterocolitica isolated from pigs, cattle and sheep at slaughter and from humans with yersiniosis in Great Britain during 1999–2000. Lett Appl Microbiol 39:103–108. doi: 10.1111/j.1472-765X.2004.01548.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.