Abstract
We report two cases of infantile diarrhea due to multidrug-resistant, NDM-1 metallo-β-lactamase-producing Salmonella enterica serovar Agona from Pakistan. This study alerts toward possible risk of NDM-1 transmission to enteric fever pathogens and encourages microbiologists to consider active screening of carbapenem resistance in nontyphoidal Salmonella isolates.
CASE REPORT
Case 1 is a 9-month-old child from Nosheroferoz, a small town in Sindh, Pakistan, who presented to Aga Khan Hospital, Karachi, with high-grade fever, vomiting, diarrhea, and abdominal pain. Stool microscopy revealed 10 leukocytes (WBC)/low-power field and mucous threads. The patient was admitted and started on 10 mg intravenous ciprofloxacin/kg of body weight twice a day. The patient became afebrile after 2 days of treatment and was discharged from the hospital on 10 mg oral ciprofloxacin/kg twice a day for 5 days. Stool culture yielded carbapenem-resistant Salmonella sp. (isolate 1a) that was susceptible only to azithromycin, fosfomycin, and colistin. On the follow-up visit, the child was still passing loose stools 5 or 6 times/day. A repeat stool culture yielded a Salmonella sp. isolate (isolate 1b) with the same susceptibility profile as the first. Treatment was changed to 250 mg oral fosfomycin three times daily, and the patient finally improved.
Case 2 is a 1-year-old child from Karachi who was admitted to the Aga Khan Hospital, Karachi, with history of diarrhea and vomiting for 2 days. Stool microscopy revealed >20 WBC/low-power field with mucous threads. The patient was admitted and started on 65 mg intravenous ceftriaxone/kg once a day. Diarrhea subsided within 3 days of admission, and the patient was discharged on 10 mg oral ciprofloxacin/kg twice a day for 5 days. Stool culture grew a carbapenem-resistant Salmonella sp. (isolate 2) susceptible only to azithromycin, fosfomycin, and colistin. This patient had no further follow-up at our hospital.
Gastroenteritis caused by nontyphoidal Salmonella species (NTS) is a major public health problem worldwide. Children less than 2 years old are the main sufferers of enteritis caused by NTS (1). The majority of these infections are self-limiting; therefore, antimicrobial therapy is reserved for treatment of serious infections only. Some patients may develop prolonged enteritis, and septicemia and extraintestinal complications are more commonly seen in immunocompromised and malnourished populations, with an associated case fatality of 20 to 25% (2).
Diarrheal diseases are very common in Pakistan. Laboratory-based surveillance data from our laboratory showed a 13% isolation rate of enteric bacterial pathogens, including Campylobacter spp., Salmonella spp., Shigella spp., and Vibrio cholerae, from clinically suspected cases of diarrhea (3). The frequency of isolation of Salmonella spp. among stool pathogens is 18.4%, and it is the third most commonly isolated enteric pathogen in our setting (4). Reported rates of antimicrobial resistance in NTS vary geographically. Resistance also varies between different serotypes and different antibiotics. Generally, low rates of resistance to ampicillin, chloramphenicol, and ciprofloxacin have been reported in Salmonella enterica serovar Enteritidis. In contrast, a higher rate of resistance has been reported in S. enterica serovar Typhimurium (5). With the emergence globally of ceftriaxone and quinolone resistance in NTS, empirical therapy for severe gastroenteritis and invasive infections has become challenging. Moreover, since the emergence of the NDM metallo-β-lactamase enzyme in enteric organisms, there has been a continuous threat of its spread into S. enterica (6). Two cases of diarrhea caused by NDM-positive S. enterica serovar Agona are presented in this report. A literature search revealed few reports of carbapenem-resistant NTS; however, the genes identified were either blaKPC, blaIMP, or blaVIM (7, 8). In 2011 and 2012, two studies from the United States and Reunion Island reported colonization with NDM-1-positive S. enterica serovars Senftenberg and Westhampton, respectively; however, the clinical significance of the reported isolates was questionable (9). Only one clinically significant infection has been reported, an NDM-1-producing S. enterica serovar Stanley from China (10). Our report is the first report of clinically significant NDM-1-producing S. enterica from the Indian subcontinent and the first report of this enzyme in S. enterica serovar Agona.
Bacterial culture, identification, susceptibility testing, and preliminary serotyping of all three isolates were performed at the clinical laboratory of the Aga Khan University Hospital, while advance serotyping, PCR, sequencing, and pulsed-field gel electrophoresis (PFGE) were performed at Public Health England's Salmonella Reference Section, United Kingdom. Samples were processed by direct plating on MacConkey's and xylose-lysine-deoxycholate agar and inoculating selenite F broth as an enrichment medium, which was subcultured the next day on Salmonella and Shigella agar. Suspected colonies were identified by conventional biochemical methods as Salmonella spp. and confirmed by API 20E (bioMérieux, France). Initial serotyping of the isolates revealed that all three isolates belonged to Salmonella group B. Further serological identification was performed at Public Health England's Salmonella Reference Section, United Kingdom, and isolates were identified as S. enterica serovar Agona (4,12:f,g,s:−) by using the Kauffmann-White scheme (11). Antimicrobial susceptibilities were performed by the disk diffusion method and Vitek 2 Compact system (bioMérieux, France) and interpreted according to Clinical and Laboratory Standards Institute guidelines (12), shown in Table 1. For azithromycin susceptibility, British Society of Antimicrobial and Chemotherapy breakpoints (13) for S. enterica serovar Typhi were used. The multidrug resistance pattern of these isolates is alarming, as the treatment options are extremely limited. Both isolates were tested for other treatment options, like azithromycin, fosfomycin, and colistin. Both children received treatment for gastroenteritis; however, as the majority of diarrheal infections are self-limiting, treatment success could not be evaluated.
TABLE 1.
MIC of Salmonella enterica serovar Agona isolates 1a and 1b (case 1) and isolate 2 (case 2), against various antibiotics determined by the Vitek 2 Compact system
| Antibiotic | MIC (μg/ml) |
||
|---|---|---|---|
| Isolate 1a | Isolate 1b | Isolate 2 | |
| Ampicillin | ≥32 | ≥32 | ≥32 |
| Amoxicillin-clavulanic acid | ≥32 | ≥32 | ≥32 |
| Piperacillin-tazobactam | ≥128 | ≥128 | ≥128 |
| Ceftriaxone | ≥64 | ≥64 | ≥64 |
| Trimethoprim-sulfamethoxazole | ≥320 | ≥320 | ≥320 |
| Gentamicin | ≥16 | ≥16 | ≥16 |
| Ciprofloxacin | 2 | 2 | 2 |
| Imipenem | ≥8 | ≥8 | ≥16 |
| Meropenem | ≥16 | ≥16 | ≥16 |
| Colistin | ≤0.5 | ≤0.5 | ≤0.5 |
| Azithromycin | 4 | 8 | 8 |
All these isolates were tested for carbapenemase production by using a modified Hodge test (14) and were also positive for blaNDM PCR, using primers 5′-GGG CAG TCG CTT CCA ACG GT-3′ and 5′-GTA GTG CTC AGT GTC GGC AT-3′ (15). Further confirmation for the presence of an NDM carbapenemase gene and screening for extended-spectrum-beta-lactamase (ESBL) genes were performed using the commercial Check-MDR CT102 ESBL-carbapenemase microarray (Check-Points Health BV, Wageningen, The Netherlands), which detected NDM carbapenemase, a group 1 CTX-M ESBL, and a non-ESBL TEM gene. Gene sequencing identified the NDM variant as NDM-1 in all three isolates. So far, NDM-1 has also been reported in other diarrheal agents, like Shigella spp. and Vibrio cholerae, from the Indian subcontinent, from the environmental water samples with potential contamination from the sewage system (16). However, no NDM-1-positive Shigella spp. and Vibrio cholerae have been reported from Pakistan. Similarly, this gene has not yet been identified in typhoidal salmonellae. As the NDM-1 gene is already spreading in community Enterobacteriaceae isolates, like Escherichia coli and Klebsiella pneumoniae (4), there is real potential for this gene to spread into enteric fever isolates.
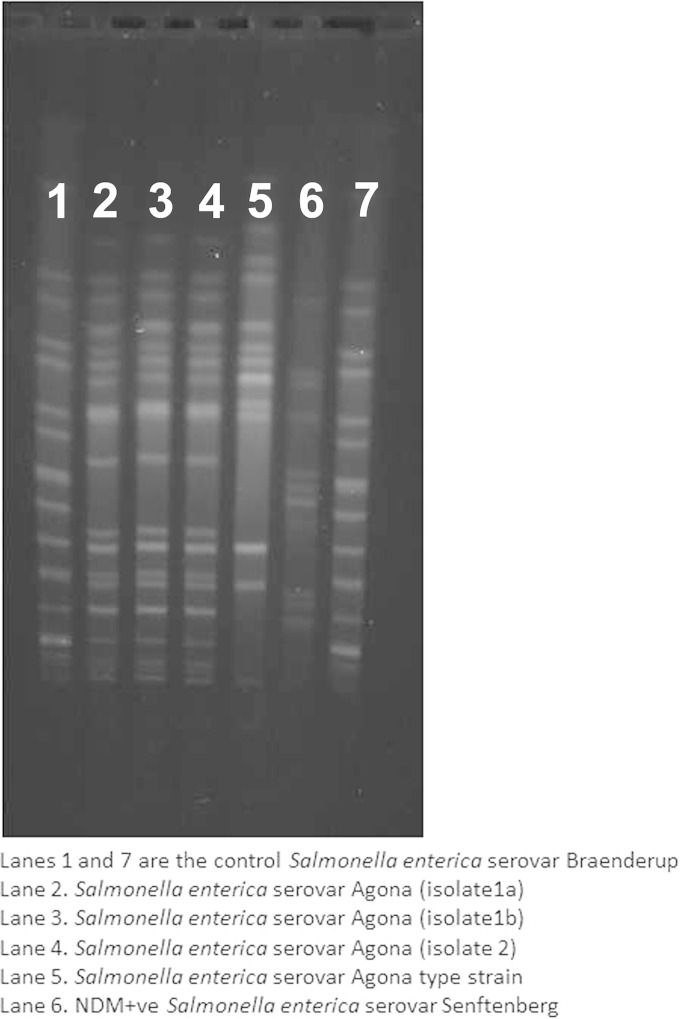
When analyzed by pulsed-field gel electrophoresis, the three isolates shared identical profiles, as shown in Fig. 1. Moreover, S. enterica serovar Agona isolates of these two cases had similar antibiograms, suggesting they originated from a common source. However, because of the lack of laboratory-based data and active surveillance of diarrheal diseases in the community, whether these two isolates were part of a larger outbreak cannot be established.
FIG 1.
PFGE gel showing similarity of all three Salmonella enterica serovar Agona isolates. To digest the DNA, restriction enzyme XbaI (New England BioLabs) was used, and the following PFGE conditions were applied: 2 to 64 s at 200 V for 22 h (17).
This study alerts toward possible risk of NDM-1 transmission to enteric fever pathogens and emphasizes microbiologists to consider active screening of NTS isolates. Any carbapenem-resistant isolates should be further investigated for the presence of NDM enzymes and other carbapenemase genes. In addition, clinicians are alerted to the emergence of these isolates, especially when treating relapsing infections with Salmonella spp.
In conclusion, there is need for active surveillance and screening of resistant Salmonella spp. in the region to avoid emergence of NDM-1-positive typhoidal salmonella strains.
REFERENCES
- 1.Chen HM, Wang Y, Su LH, Chiu CH. 2013. Nontyphoid salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr Neonatol 3:147–152. doi: 10.1016/j.pedneo.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive nontyphoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irfan S, Ahmad A, Guhar D, Khan E, Malik F, Mahmood S, Zafar A. 2010. Fluoroquinolone and macrolide co-resistance in clinical isolates of Campylobacter species: a 15-year study in Karachi, Pakistan. East Mediterr Health J 16:1226–1230. [DOI] [PubMed] [Google Scholar]
- 4.Sultan BA, Khan E, Hussain F, Nasir A, Irfan S. 2013. Effectiveness of modified Hodge test to detect NDM-1 carbapenemases: an experience from Pakistan. J Pak Med Assoc 63:955–960. [Google Scholar]
- 5.Su LH, Chiu CH, Chu C, Ou JT. 2004. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis 39:546–551. doi: 10.1086/422726. [DOI] [PubMed] [Google Scholar]
- 6.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordmann P, Poirel L, Mak JK, White PA, McIver CJ, Taylor P. 2008. Multidrug-resistant Salmonella strains expressing emerging antibiotic resistance determinants. Clin Infect Dis 46:324–325. doi: 10.1086/524898. [DOI] [PubMed] [Google Scholar]
- 8.Fischer J, Rodriguez I, Schmoger S, Friese A, Roesler U, Helmuth R, Guerra B. 2012. Salmonella enterica subsp. Enterica producing VIM-1 carbapenemase isolated from livestock farms. J Antimicrob. Chemother 68:478–480. doi: 10.1093/jac/dks393. [DOI] [PubMed] [Google Scholar]
- 9.Savard P, Gopinath R, Zhu W, Kitchel B, Rasheed JK, Tekle T, Roberts A, Ross T, Razeq J, Landrum BM, Wilson LE, Limbago B, Perl TM, Carroll KC. 2011. First NDM-positive Salmonella sp. strain identified in the United States. Antimicrob Agents Chemother 55:5957–5958. doi: 10.1128/AAC.05719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Wang M, Ding H, Ye M, Hu F, Guo Q, Xu X, Wang M. 2013. New Delhi metallo-β-lactamase-1 in carbapenem resistant Salmonella strain, China. Emerg Infect Dis 19:2049–2051. doi: 10.3201/eid1912.130051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimont PAD, Weill FX. 2007. Antigenic formulas of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 13.British Society of Antimicrobial and Chemotherapy. 2012. BSAC methods for antimicrobial susceptibility testing, version 11.1. http://bsac.org.uk/wp-content/uploads/2012/02/Version-11.1-2012-Final-.pdf.
- 14.Stuart CJ, Leverstein-Van Hall MA. 2010. Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents 36:205–210. doi: 10.1016/j.ijantimicag.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob. Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 16.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 17.EUCAST. 2003. The Salm-gene project—a European collaboration for DNA fingerprinting for food-related salmonellosis. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=401. [DOI] [PubMed]