Summary
miR-200a alone transforms an immortalized rat epithelial cell line, and miR-200a cooperates with Ras to enhance malignant transformation of an immortalized human epithelial cell line. Thus, miR-200a is pro-oncogenic in transformation of mammalian epithelial cells.
Abstract
Cancer is a multistep disease that begins with malignant cell transformation and frequently culminates in metastasis. MicroRNAs (miRNAs) are small regulatory 21–25 nt RNA molecules and are frequently deregulated in cancer. miR-200a is a member of the miR-200 family, which are known inhibitors of the epithelial-to-mesenchymal transition. As such, the tumor-suppressive role of miR-200a in oncogenesis has been well documented; however, recent studies have found a proliferative role for this miRNA as well as a prometastatic role in the later steps of cancer progression. Little is known about the role of this miRNA in the early stages of cancer, namely, malignant cell transformation. Here, we show that miR-200a alone transforms an immortalized rat epithelial cell line, and miR-200a cooperates with Ras to enhance malignant transformation of an immortalized human epithelial cell line. Furthermore, miR-200a induces cell transformation and tumorigenesis in immunocompromised mice by cooperating with a Ras mutant that activates only the RalGEF effector pathway, but not Ras mutants activating PI3K or Raf effector pathways. This transformative ability is in accordance with miR-200a targeting Fog2 and p53 to activate Akt and directly repress p53 protein levels, respectively. These results demonstrate an oncogenic role for miR-200a and provide a specific cellular context where miR-200a acts as an oncomiR rather than a tumor suppressor by cooperating with an oncogene in malignant cell transformation.
Introduction
Cancer comprises one quarter of all deaths in the United States; however, cancer mortality rates are declining, due largely to improvements in screening and detection. Diagnosis of early stage cancer is strongly associated with better survival (1–4). Thus, it is crucial to understand the molecular events that occur early in this progressive disease. Cell transformation is the initiating step of cancer progression (5). During this process, a cell must bypass senescence and avoid apoptosis, allowing for uncontrolled proliferation, which leads to formation of a primary tumor (6). The hyperproliferative, antiapoptotic phenotypes that arise during transformation are conferred by mutations that upregulate proto-oncogene activity and ablate tumor suppressor gene function (7). The classical model of cell transformation identified the cooperation between the Ras and Myc oncogenes in selecting for a dominant-negative p53 tumor suppressor mutation and transforming primary rodent cells (8). Numerous transforming oncogenes and tumor suppressor mutations have been identified since these landmark studies (9), demonstrating the complexity of cancer initiation.
Recently, non-coding RNAs have garnered interest as mediators of cancer progression (10–12). MicroRNAs (miRNAs) are frequently dysregulated in cancer, and by repressing expression of oncogenes or tumor suppressors, a miRNA may function as a tumor suppressor or oncogene, respectively (10,12,13). Among miRNAs dysregulated in cancer is miR-200a: gene expression profiling reports that miR-200a is frequently downregulated in cancer (14–16). Its most well-studied function is the suppression of Zeb1/2 transcription factors to inhibit the epithelial-to-mesenchymal transition (EMT), implicating it as a tumor suppressor (17–19). However, miR-200a has also been found to promote oncogenesis by promoting the reversal of EMT, the mesenchymal-to-epithelial transition, allowing invasive cells to revert back to a phenotype more conducive to metastatic colonization (20–23). In addition to its involvement in EMT/mesenchymal-to-epithelial transition, studies have shown a hyperproliferative role for miR-200a through suppression of Fog2, a PI3K inhibitor (24,25), and a recent study demonstrates the antiapoptotic function of miR-200a in directly repressing p53 (26). Consistent with its earliest known role as an inhibitor of EMT, miR-200a has been found to be downregulated in breast cancer tissues (27). In addition, miR-200a and its family members are differentially modulated in distinct breast cancer phenotypes (28). The miR-200 family is primarily upregulated in luminal and basal breast cancers, but not malignant myoepithelioma of the breast, which has a more mesenchymal phenotype (29). miR-200a is significantly upregulated in lymph node-positive breast tumors compared to node-negative tumors (30) and in distant metastases compared to primary tumors (31). In this study, we determine the effect of miR-200a overexpression on transformation of both rodent cells and immortalized human MCF10a cells and characterize the underlying mechanism of the ability of miR-200a to cooperate with Ras to transform MCF10a cells.
Materials and methods
Cell culture
RK3E cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum and antibiotics at 37°C with 5% CO2. MCF10A cells were cultured in Dulbecco’s modified Eagle’s medium/F12 (Invitrogen) supplemented with 5% horse serum, 20ng/ml epidermal growth factor, 0.5mg/ml hydrocortisone, 100ng/ml cholera toxin, 10 µg/ml insulin and antibiotics. MCF7 cells were cultured in Eagle’s minimum essential medium supplemented with 10% fetal bovine serum, 10 µg/ml insulin and antibiotics. All three cell lines were obtained directly from ATCC (Manassas, VA) and were passaged in our laboratory for fewer than 6 months after resuscitation.
miRNA screen
RK3E cells were transfected using Lipofectamine LTX/Plus (Invitrogen) reagent according to manufacturer instructions with individual miRNAs from our miRNA library comprised of 366 human miRNA minigenes in the lentiviral PSIF vector (32). After 48 h, wells were visually inspected for three dimensional foci formation.
Viral transduction
Lentiviral transduction was performed as described previously (33), using parental miRNA vector pSIF and packaging and expression vectors pVGV-S and pFIV-34N, respectively. Transduction was carried out three separate times and selection was performed using G418 (200 µg/ml) for five passages. miR-200a expression levels were measured by Taq-Man qRT–PCR expression assay (Invitrogen), ranged from a 5- to 15-fold increase (data not shown). For retrovirus production, retroviral vectors containing constitutively active RasG12V mutant (Addgene 1768), c-Myc (Myc from Addgene plasmid 16011 cloned into plasmid 12269), p53dd (Addgene 9058) or RasG12V effector pathway mutants (Addgene 12274, 12275, 12276) (34) were used.
Acini formation in Matrigel
A total of 5000 positively drug-selected exponentially growing MCF10A cells suspended in 2% Matrigel (Corning, Tewksbury, MA) were layered over a bottom layer of Matrigel in 12-well chamber slides and allowed to grow 5 or 14 days (35). Acini were stained with anti-E-cadherin or cleaved caspase-3 primary antibodies (Cell Signaling, Danvers, MA) and Alexa488-coupled goat anti-rabbit secondary antibodies. Slides were visualized with Olympus FV100 confocal microscope and FluoView software (Olympus, Center Valley, PA).
Colony formation assay
Six well plates were coated with a bottom layer of 0.5% noble agar (Sigma-Aldrich, St Louis, MO) and 2000 MCF10A cells were suspended in triplicate in a top layer of 0.2% noble agar for 2 weeks. Colonies were stained with 0.05% crystal violet, counted and imaged using a dissection microscope coupled to a digital microscope camera (Celestron, Torrance, CA).
Cell cycle and apoptosis analysis
For synchronization, MCF10A cells were progressively deprived of serum and growth factors over 24 h, then stimulated with complete media for 18 h. Cells were stained with propidium iodide, and flow cytometry was performed with a FACS LSRII Flow Cytometer as described previously (33). A minimum of 10 000 cells per sample were collected and the FACS files were analyzed using FlowJo (Tree Star, Ashland, OR) (28). For apoptosis, cells were challenged with 0.5 µM doxorubicin for 24 h to induce apoptotic signaling, stained with Annexin V and propidium iodide using Annexin V FITC Apoptosis Detection Kit (eBioscience, San Diego, CA) and analyzed by flow cytometry with a FACS LSRII and FlowJo software.
Cell migration
Cell migration was assayed as described previously using Transwell chambers (Invitrogen), with a pore size of 8 µm (33). Slides containing migrated cells were imaged as in colony formation assay.
Western blot
Western blot was performed as described previously using phospho-p53, p53, PTEN, phospho-Akt, Akt, phospho-Erk, Erk, Vimentin or E-cadherin antibody (Cell Signaling), Fog2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or β-actin antibody (Sigma-Aldrich) (33,36). Blots were visualized with the SuperSignal West Dura Chemiluminescent Substrate (Thermo Fisher). See Supplemental Material for blotting images.
Mice
Athymic male nude (Foxn1 nu /Foxn1 nu) mice (6 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained in the University of Louisville’s AAALAC-accredited animal facility. All animal studies were conducted in accordance with National Institutes of Health animal use guidelines and a protocol approved by the University of Louisville’s Institutional Animal Care and Use Committee. Exponentially growing cells were injected subcutaneously into nude mice. Mice were injected in each flank with MCF10A cells stably overexpressing a Ras effector mutant alone in one flank and MCF10A cells stably overexpressing the same Ras effector mutant in combination with miR-200a in the other flank. Side of injection was randomized. Tumor size was monitored once per week for 10 weeks before sacrificing. Tumors were harvested and immediately formalin fixed. Tumors were embedded in paraffin, and tissue sections were stained with hematoxylin and eosin.
Statistical analysis
Colony formation data were analyzed by multivariate analysis of variance with T-tests post hoc to look for individual effects with standard errors corrected for multiple comparisons. Cell proliferation data were analyzed by linear regression modeling with T-tests post hoc to determine individual effects using standard errors corrected for multiple comparisons. Cell cycle distribution was analyzed by generalized linear models comparing G1 to combined S/G2 phases with binomial response variables and parameters estimated by maximum likelihood. Log odds ratios were further analyzed by T-tests post hoc to evaluate individual effects using standard errors corrected for multiple comparisons.
Results
miR-200a transforms rat immortalized epithelial cells
The RK3E cell line is an E1A-immortalized rat kidney epithelial cell line, whose defining characteristic is monolayer growth under normal conditions and foci formation under transforming conditions such as Ras activation or Myc overexpression (37–39). In order to determine the role of miRNAs in cell transformation, RK3E cells were transiently transfected with individual miRNAs from our library of 366 miRNA gene constructs (32) and visually screened for foci formation (Figure 1A). A plasmid expressing c-Myc was used as a positive control (40). Of the 366 miRNAs screened, miR-141 and let-7e formed about the same number of foci as c-Myc, and miR-200a formed more foci than c-Myc. We stably infected RK3E cells with lentivirus made from each of these three miRNAs or vector control and assayed them for anchorage-independent growth in soft agar. All three miRNAs induced colony formation in soft agar (Figure 1B). In order to determine tumorigenicity of these miRNAs, RK3E cells stably expressing each of these miRNAs were subcutaneously injected into nude mice. Cells expressing miR-141 or miR-200a formed subcutaneous tumors (Figure 1C), but let-7e-expressing cells did not produce tumors (data not shown). As both miR-200a and miR-141 are miR-200a family members with identical seed sequences, miR-200a was chosen to be analyzed further.
Fig. 1.
miR-200a transforms RK3E cells and stimulates Akt activity but maintains an epithelial phenotype. (A) Schematic of foci formation induced by transient transfection of RK3E cells with individual miRNAs from a library of 366 miRNA minigenes. Cells were visually inspected for foci formation in cell culture. (B) Phase contrast microscopy of three dimensional colonies formed in soft agar by RK3E cells stably overexpressing the indicated miRNAs. A total of 5000 cells per well were plated in 0.2% soft agar in six well plates. (C) Subcutaneous tumor formation of RK3E cells overexpressing the indicated miRNAs. Cells (1×106) were resuspended in Matrigel and injected subcutaneously (n = 5 per group). (D) Left: western blot for the mesenchymal marker Vimentin and the epithelial marker E-cadherin in RK3E cells lentivirally infected with miR-200a or empty vector control. Right: western blot for members of the Akt activation pathway in RK3E cells. (E) Immunofluorescent staining of RK3E tumors; left: E-cadherin expression (green) and right: Vimentin (green) expression.
To determine potential mechanisms underlying cell transformation, we analyzed the expression of markers of epithelial and mesenchymal phenotypes as well as miR-200a target genes (24,41–43). Consistent with the literature, western blot analyses showed that in RK3E cells expressing miR-200a, the epithelial marker, E-cadherin, was upregulated, and the mesenchymal marker, Vimentin, was downregulated compared to vector control (Figure 1D). Furthermore, expression of two mir-200a targets that negatively regulate the PI3K/Akt pathway, Fog2 and Pten, was downregulated, concomitant with an increase in Akt phosphorylation (Figure 1D). We examined E-cadherin and vimentin ratios in RK3E tumors and found that, consistent with western blot results, tumors formed by RK3E cells overexpressing miR-200a expressed high levels of E-cadherin while vimentin was undetectable (Figure 1E).
To determine whether miR-200a acts as a driving force in cell transformation, we employed the classical two-hit model of primary rodent cell transformation by combining miR-200a with Ras, c-Myc or p53 dominant negative (p53dd) in mouse embryonic fibroblasts (MEFs). The Ras construct harbors a G12V mutation, rendering it constitutively active. p53dd is a truncated form of the carboxy terminus of the p53 protein that binds wild-type p53, resulting in a dominant-negative repression of native p53 function (44). miR-200a overexpression in MEFs induced an initial increase in proliferation; however, miR-200a expression alone, or in combination with either Ras or c-Myc, was unable to transform MEFs (data not shown). p53dd is known to immortalize MEFs, however miR-200a was unable to cooperate with this tumor suppressor mutation to transform these cells. As a positive control, Ras and Myc coexpression was able to transform primary MEFs. These results implicate that other genetic alterations in RK3E cells, but not in primary MEFs, may be involved in miR-200a-mediated cell transformation.
miR-200a augments Ras transformation of immortalized human mammary epithelial cells
In order to determine the role of the proliferative effects of miR-200a in human cells, we chose MCF10A cells, an immortalized yet untransformed mammary epithelial cell line isolated from a patient with fibrocystic disease (45). This cell line is readily transformed by overexpression of Ras or Myc, and normal morphology in three dimensional cell culture is disrupted by loss of p53 signaling (46–48). Consistent with our efforts to determine oncogene cooperativity, we stably infected this cell line with miR-200a alone, or in combination with Ras, c-Myc or p53dd. To test for transformation, cells were assayed for anchorage-independent growth in soft agar (Figure 2A and B). MCF10A cells expressing miR-200a alone did not form colonies in soft agar. When miR-200a was expressed in combination with c-Myc or p53dd, colony formation was not significantly increased compared to either oncogene alone. However, colony formation increased >3-fold in cells expressing miR-200a in combination with Ras compared to cells expressing Ras alone. Coexpression of Ras and c-Myc induced colony formation, but their combined effects were not synergistic.
Fig. 2.
miR-200a synergizes with Ras to transform MCF10A cells and inhibit apoptosis. (A) Soft agar colony formation: bright field microscopy of MCF10A cells stably infected with the indicated oncogenes alone (left panels) or in combination with miR-200a (right panels). A total of 4000 cells per well were plated in 0.2% soft agar in six well plates in triplicate. (B) Quantification of soft agar colony formation in A. (C) Cell cycle analysis of MCF10A cells infected with the indicated oncogenes alone or in combination with miR-200a. Cells were stained with propidium iodide and a minimum of 10000 cells per group were analyzed in triplicate by flow cytometry. P values calculated for cell cycle progression out of G1 phase (green bars). (D) Apoptosis assay of MCF10A cells expressing miR-200a and Ras alone or in combination, treated with vehicle control or doxorubicin. (E) Cell proliferation curve of MCF10A cells infected with miR-200a, Ras or miR-200a in combination with Ras. Cells were plated in 96 well plates and analyzed for proliferation by MTT assay at the indicated time points. (F) Transwell cell migration of MCF10A cells infected as in D. Cells (1×105) were seeded in Transwell chambers in 12 well plates in growth factor-deprived low serum media with full media in the lower well to serve as a chemoattractant. (G) Upper left: schematic of three dimensional acinus formed by MCF10A cells in Matrigel. Black line indicates equatorial confocal plane of focus. Upper right: schematic of hollow lumen visualized by confocal microscopy focused at center line depicted in left diagram. Lower: representative image of acinar structures formed by MCF10A stably infected with vector control after 14 days. (H) Three dimensional acinar formation after 5 days in Matrigel of MCF10A cells infected as in E, stained for E-cadherin (green, upper panels) and cleaved caspase-3 (red, lower panels). A total of 5000 cells were plated in 2% Matrigel in 12 well chamber slides. (I) Apoptosis assay of MCF7 cells. Cells were transfected with scrambled control or miR-200a inhibitor (anti-miR-200a) and treated with doxorubicin. Values represent means of three independent experiments ± SE. *P ≤ 0.05 compared to vector or scrambled control, † P ≤ 0.05 compared to miR-200a, ¥ p ≤ 0.05 compared to Ras, § P ≤ 0.05 compared to vehicle.
miR-200a synergizes with Ras to inhibit apoptosis
We next analyzed the role of miR-200a in cell cycle progression. Compared to vector control, all experimental groups caused a decrease in G1 arrest (Figure 2C). Consistent with its effects on colony formation, Ras and Myc coexpression had the smallest effect, causing a small, but statistically significant decrease in G1 arrest. Compared to p53dd alone, miR-200a in combination with p53dd caused the greatest increase in cell cycle progression. Surprisingly, G1 arrest was unchanged between cells expressing Ras alone and cells expressing miR-200a in combination with Ras. Furthermore, compared to miR-200a alone, G1 arrest in cells expressing miR-200a in combination with Ras increased significantly. Compared to c-Myc alone, miR-200a with c-Myc did not change G1 arrest levels. This indicates that induction of cell cycle progression is not responsible for the combinatorial effect of Ras and miR-200a on cell transformation. We next generated a proliferation curve in order to compare growth rates of cells expressing miR-200a alone and cells expressing miR-200a in combination with Ras (Figure 2D). miR-200a alone or in combination with Ras significantly increased rate of proliferation compared to vector control; however, miR-200a combined with Ras did not significantly increase the proliferation rate compared to miR-200a alone. Ras alone showed the greatest increase in proliferation rate. This indicates that stimulation of proliferation is not responsible for the interactive effect between Ras and miR-200a, and that Ras is unable to enhance the proliferative function of miR-200a. It is also possible that miR-200a may be capping the growth rate increase induced by Ras. This is consistent with the smaller colony size observed in cells transformed with miR-200a in combination with Ras, despite an increase in colony number.
We next examined these cells for changes in migratory ability, a cancer hallmark, and a phenotype regulated by miR-200a and its family members. miR-200a decreased transwell cell migration of MCF10A cells compared to vector control (Figure 2E). Ras alone greatly increased cell migration above vector control. Cells expressing Ras in combination with miR-200a showed decreased migration compared to Ras alone; however, these cells formed dense, three dimensional colonies before and after migrating. Inhibition of cell migration is consistent with miR-200a’s established function in regulation of EMT (18,43).
When seeded in Matrigel, MCF10A cells form polarized acinar structures with distinct hollow lumen, which resemble normal breast tissue development (48) (Figure 2G). To gain insight into the potential effects of miR-200a cooperating with Ras on cell growth and morphology in the context of acinar formation, we seeded cells in Matrigel and analyzed three dimensional acinus formation by confocal microscopy (Figure 2H). Acini were examined for changes in three characteristics: morphology, E-cadherin expression and cleaved caspase-3 expression. Acini were stained for E-cadherin to facilitate inspection of morphological and structural changes, as well as changes in E-cadherin expression and subcellular localization (Figure 2H, upper panels). Cells with vector control formed regular spherical structures with hollow lumen. Cells expressing miR-200a alone formed regular spherical structures, with and without regular lumen clearance. Cells expressing Ras alone formed very large, lobular three dimensional structures with loosely packed cells and partial lumen clearance, which resembles a tumor and is consistent with studies using this three dimensional culture model (47). Cells expressing miR-200a in combination with Ras formed large, densely packed, slightly irregular spherical structures with no lumen clearance. This hyperproliferative densely populated structure with only minor disruptions in overall structural shape resembles a non-invasive adenoma or carcinoma in situ (Figure 6). Notably, in acini expressing Ras alone, or Ras with miR-200a, E-cadherin was localized diffusely in the cytoplasm, rather than at the plasma membrane. This is consistent with studies showing disruption of normal adherent junctions in transformed cells, and sequestering of E-cadherin away from the plasma membrane in the cytoplasm (49–51).
In order to determine whether the cause of the loss of lumen clearance in response to miR-200a expression was due to inhibition of apoptosis, we examined levels of cleaved caspase-3 in Matrigel-seeded MCF10A cells allowed to grow for 5 days, at which point normal acini begin to undergo luminal clearance by apoptosis (Figure 2H, lower panels) (35). Acini containing vector or miR-200a expressed high levels of cleaved caspase-3 and showed normal lumen clearance. Acini expressing Ras alone expressed lower levels of cleaved caspase-3. Acini structures expressing Ras in combination with miR-200a showed no lumen clearance and little to no cleaved caspase-3. Taken together, these results indicate that miR-200a alone increases proliferation without decreasing apoptosis; however, miR-200a synergizes with Ras to both increase proliferation, and, more notably, to inhibit apoptosis as evidenced by the dramatic reduction of cleaved caspase-3 compared to Ras alone. While Ras suppresses the apoptotic response and miR-200a represses p53, their individual inabilities to inhibit apoptosis are striking, particularly when juxtaposed to doxorubicin-induced cell apoptosis, which is inhibited by Ras and mir-200a individually (Figure 2D). This indicates that the apoptotic signaling during lumen clearance in acinus formation can overcome repression of miR-200a or Ras alone. The combination of these two signaling pathways—miR-200a suppression of p53 and stimulation of Akt and Ras suppression of apoptotic signaling and stimulation of cell cycle progression pathways—synergizes to produce the phenotypic effect of decreased apoptosis in MCF10A cells. To further examine the effects of miR-200a on apoptosis, we knocked down miR-200a levels in doxorubicin-challenged MCF7 cells (Figure 2I), which express high levels of miR-200a (52). Knockdown of miR-200a significantly inhibits doxorubicin-induced apoptosis in MCF7 cells, corroborating the results seen in the MCF10A cells (Figure 2D).
miR-200a cooperates with the RalGEF Ras effector pathway
Ras activity results in the activation of a wide array of downstream signaling pathways; however, the three main effectors activated by Ras are Raf, PI3K and RalGEF (53). We made use of three Ras mutants to determine the mechanism of miR-200a and Ras cooperation in cell transformation. The T35S mutation activates only the Raf-Erk effector pathway; the Y40C point mutation activates only the PI3K effector pathway, and the E37G point mutation activates only the RalGEF effector pathway (Figure 3A). We expressed miR-200a with each of these individual effector pathway mutations and assayed them for anchorage-independent growth in soft agar (Figure 3B and 3C). Compared to T35S alone, miR-200a in combination with T35S did not significantly increase colony formation. miR-200a in combination with Y40C did not cause an increase in colony formation compared to Y40C alone. miR-200a caused a ~10-fold increase in cells expressing both E37G and miR-200a compared to cells expressing E37G alone. Fully active Ras formed more colonies than any of the effector mutants alone or combined with miR-200a, indicating that while miR-200a synergizes with the E37G mutant to transform MCF10A cells, these two hits do not completely recapitulate the full effect of Ras. We measured cell cycle progression in MCF10A cells expressing Ras effector mutants alone or in combination with miR-200a (Figure 3D). T35S significantly decreased G1 phase arrest, but when miR-200a was added, the effect was ablated. Y40C alone or in combination with miR-200a did not show any change in cell cycle progression, consistent with the lack of effect on colony formation. E37G alone induced a small but significant increase in G1 phase arrest. However, E37G in combination with miR-200a significantly decreased G1 phase arrest, compared to both vector control and E37G alone, indicating that miR-200a induction of cell cycle progression contributes to its cooperation with RalGEF signaling.
Fig. 3.
miR-200a cooperates with the RalGEF Ras effector pathway to transform MCF10A cells. (A) Schematic of the main 3 effector pathways of Ras. Indicated on the leftmost arrows are the activating point mutations that allow for activation of only that pathway. (B) Soft agar colony formation assay of MCF10A cells stably infected with the indicated Ras effector mutants alone (left panels) or in combination with miR-200a (center panels) compared to constitutively activated Ras (right panel). A total of 4000 cells per well were plated in 0.2% soft agar in six well plates. (C) Quantification of soft agar colony formation in B. (D) Cell cycle analysis of MCF10A cells infected with the indicated Ras effector mutants alone or in combination with miR-200a. Cells were stained with propidium iodide and a minimum of 10000 cells per group were analyzed in triplicate by flow cytometry. P values calculated for cell cycle progression out of G1 phase (green bars). (E) Three dimensional acinar formation in Matrigel of MCF10A cells infected with the indicated Ras effector mutants, stained for E-cadherin (green, upper panels) and cleaved caspase-3 (red, lower panels). A total of 5000 cells were plated in 2% Matrigel in chamber slides. Values represent means of three independent experiments ± SE. *P ≤ 0.05 compared to vector control, † P ≤ 0.05 compared to miR-200a, € P ≤ 0.05 compared to E37G.
We next analyzed the three dimensional growth in Matrigel of MCF10A cells expressing E37G Ras effector mutant alone and in combination with miR-200a to determine changes in structure and apoptosis (Figure 3E). Cells expressing E37G Ras mutant alone formed regular, round acini with hollow lumen, but had lower levels of cleaved caspase-3 than the vector control. When miR-200a was added, cells expressing E37G Ras mutant formed large, irregularly shaped structures with tightly packed cells and diffuse E-cadherin staining; punctate cleaved caspase-3 was visible throughout the structures, along with apoptotic cells; however, no lumen clearance was observed. Cleaved caspase-3 levels did not appear dramatically different between cells expressing E37G alone and cells expressing E37G in combination with miR-200a, indicating that miR-200a does not enhance the inhibition of apoptosis by RalGEF signaling; however, structures formed when miR-200a was expressed with E37G were much larger than E37G alone, indicating that miR-200a disrupts proliferation and growth regulation of acini.
miR-200a stimulates Akt and suppresses p53
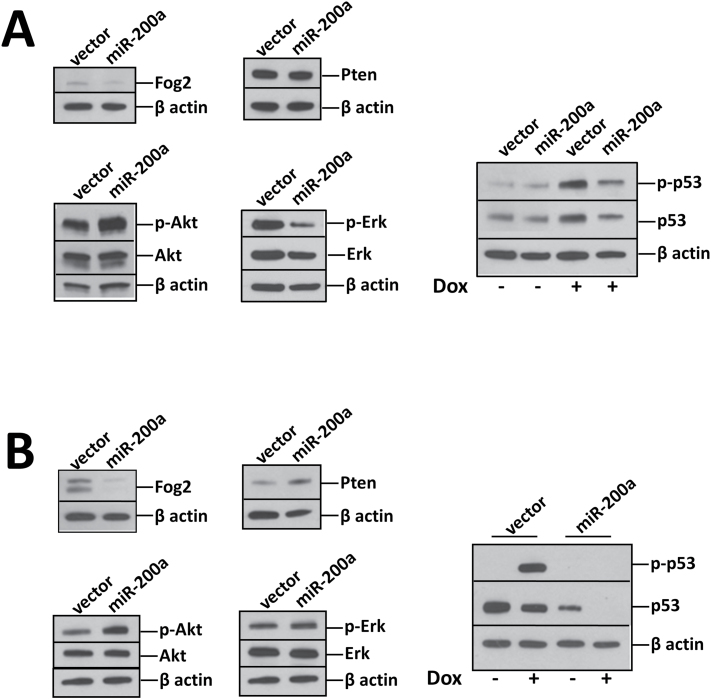
To further elucidate the mechanism of miR-200a in cell transformation, we analyzed MCF10A cells transiently overexpressing miR-200a for known targets as well as potential downstream effectors (Figure 4A). Similar to RK3E cells, Fog2 expression is decreased in MCF10A cells expressing miR-200a, accompanied by an increase in Akt phosphorylation. However, the miR-200a target Pten is not downregulated in MCF10A cells, suggesting species-specific targeting (Figure 1D). Because of the involvement of miR-200a in the Akt pathway and its cooperation with the RalGEF pathway, we also analyzed protein expression and phosphorylation levels of Erk, the third Ras effector pathway. In cells transiently expressing miR-200a, Erk expression and phosphorylation levels decreased compared to vector control. miR-200a directly targets the 3′UTR of p53 (26), so we analyzed p53 protein levels in unchallenged and doxorubicin-challenged MCF10A cells. In untreated cells, basal levels of p53 were slightly repressed in cells expressing miR-200a in comparison with vector control cells. With doxorubicin treatment, p53 protein expression and phosphorylation increased in control cells and the p53 protein expression and phosphorylation also moderately increased in cells expressing miR-200a in response to doxorubicin. In addition, p53 protein expression and phosphorylation were repressed in cells expressing miR-200a (comparing lane 4 and 3; right panel of Figure 4A).
Fig. 4.
miR-200a stimulates Akt and suppresses p53 as determined by western blotting analyses. p53, Pten and Fog2 are directly targeted by miR-200a. Akt is regulated by Fog2 and Pten. Akt and Erk are Ras signaling components. (A) MCF10A cells transiently transfected with miR-200a. (B) MCF10A cells stably infected with virus carrying miR-200a.
In MCF10A cells stably expressing miR-200a through viral transduction (Figure 4B), Fog2 was downregulated and Akt phosphorylation was elevated; however, Pten expression and Erk phosphorylation appeared to be unaffected. Doxorubicin treatment resulted in moderate p53 downregulation (though p53 phosphorylation was elevated) in control cells. We speculate that prolonged drug selection and/or viral infection could evolve cells to adapt a mechanism that may inhibit p53 upregulation by doxorubicin. Nonetheless, miR-200a downregulated p53 protein expression without doxorubicin treatment (comparing lane 3 to 1; right panel, Figure 4B) or with treatment (comparing lane 4 to 2), reaffirming the efficacy of miR-200a-mediated p53 suppression.
miR-200a synergizes with the RalGEF signaling pathway to induce tumorigenesis
We subcutaneously injected nude mice with MCF10A cells expressing miR-200a in combination with the T35S mutant or with the E37G mutant and histologically analyzed resulting tumors (54). MFC10A cells expressing miR-200a in combination with the E37G mutant were the only cells that formed tumors (Figure 5A and B). Transformed MCF10A cells recapitulate human proliferative mammary disease in subcutaneous tumors in nude mice (46); thus, we stained tumor sections with H&E and histologically examined them for abnormal tissue structures (Figure 5C). All tumors were well vascularized, regardless of size (Figure 5C), and duct formation was either absent (Figure 5E) or severely abnormal (Figure 5D). Defined structures of dense connective tissue were present with intermittent sections of loose connective tissue that infrequently contained small structures resembling ducts, but without regular epithelial lining (Figure 5D). The highly irregular tissue structure and loss of normal duct formation resembles the histology seen in human breast cancer.
Fig. 5.
miR-200a synergizes with the RalGEF effector pathway of Ras to induce tumorigenesis in immunocompromised mice. (A) Average tumor size in 6 week old Foxn1 nu /Foxn1 nu mice subcutaneously injected with 5×105 MCF10A cells stably infected with the indicated Ras effector pathways alone or in combination with miR-200a (n = 6 per group). (B) Representative images of tumor formation. Each mouse was injected in each flank with 5×105 MCF10A cells stably overexpressing a Ras effector mutant alone in one flank and 5×105 MCF10A cells stably overexpressing the same Ras effector mutant in combination with miR-200a in the other flank (n = 6 for each group). (C–E) Representative H&E staining of tumors formed by E37G + miR-200a MCF10A cells showing (C) vascularization, (D) abnormal duct formation and (E) irregular tissue architecture in tumors. Tissue structures resembling human mammary histology are labeled as follows: B, blood vessel, L, loose connective tissue, D, dense connective tissue, Dt, duct.
Discussion
miR-200a in cell proliferation and apoptosis
miR-200a is first found to inhibit EMT through suppression of Zeb1/2 and can thus act as a tumor suppressor or tumor promoter (Figure 6). In this work, we identified a new oncogenic role for miR-200a that exists alongside upregulation of epithelial markers. Furthermore, miR-200a suppresses migration of MCF10A cells regardless of Ras expression status. miR-200a synergizes with the RalGEF pathway to transform the epithelial MCF10A cell line into tumor cells that form subcutaneous tumors in nude mice with histology that resembles invasive proliferative breast disease. This is especially notable because miR-200a has been reported to be a crucial player in the maintenance of epithelial cell polarity, particularly in the mammary gland (55). The disruption of tissue structure by the combination of miR-200a overexpression with RalGEF signaling highlights the significance of this interactive effect.
Fig. 6.
Proposed model for the dichotomous roles of miR-200a in cancer progression. miR-200a is labeled in its oncogenic (red) or tumor suppressive (green) roles. (A) miR-200a directly targets p53 and Fog2 to promote malignant cell transformation, the initiating event of oncogenesis. Specifically, miR-200a cooperates with RalGEF to transform MCF10A cells. (B) By targeting Zeb1/2 transcription factors, miR-200a inhibits EMT. Early in metastasis, this is a tumor-suppressive function. (C) After local invasion and intravasation have occurred, promotion of MET by miR-200a enhances metastatic colonization. MET, mesenchymal-to-epithelial transition.
miR-200a directly targets the 3′UTR of Fog2 and positively regulates cell growth (24), and further studies have revealed the important role of the miR-200 family and Fog2 in regulating PI3K activity in the context of insulin signaling, indicating the significance of the miR-200a/Fog2/PI3K axis in human health (56–59). We have demonstrated that miR-200a transforms RK3E cells and augments Ras transformation of immortalized human MCF10A cells while suppressing Fog2. This is consistent with a study demonstrating that activated PI3K complements Ras activation in transformation of human mammary epithelial cells (60).
miR-200a alone significantly increases cell proliferation and cell cycle progression, yet adding Ras does not increase this further, indicating that Ras may activate PI3K/Akt signaling to high levels that cannot be further enhanced by miR-200a suppression of Fog2. Conversely, miR-200a in combination with RalGEF signaling causes a significant decrease in G1 cell cycle arrest compared to RalGEF alone, indicating that miR-200a induction of cell cycle progression, rather than inhibition of apoptosis, is important for cooperation with RalGEF signaling.
miR-200a alone does not decrease cleaved caspase-3 levels in the lumen of acini, despite suppression of p53 and inhibition of apoptosis by miR-200a (26). Although lumen formation is curtailed when Ras is expressed alone, cleaved caspase-3 staining is not ablated, indicating that increased proliferation, rather than loss of apoptosis alone is responsible for the lack of lumen clearance in MCF10A acini with Ras activation. However, when Ras and miR-200a are expressed together, cleaved caspase-3 levels decrease dramatically, indicating that miR-200a synergizes with Ras to inhibit apoptosis. Loss of apoptosis is mechanistically supported by western blot analyses showing downregulation of p53 by miR-200a in MCF10A cells. p53 transactivational targets p21 and PUMA (p53 upregulated modulator of apoptosis) cooperate to modulate β-catenin expression and inhibit EMT (61,62). By repressing p53 expression, miR-200a may promote EMT through this pathway, lending support to its dichotomous role in cancer. In particular, repression of the p53 pathway in the context of increased Ras activity may enhance oncogenic signaling and cell transformation. miR-25, another p53-suppressive miRNA, is upregulated in multiple types of cancer (63–65). Similar to miR-200a (30), miR-25 is overexpressed in breast cancer and is strongly associated with the high proliferation that marks lymph node-positive breast cancer (66). The pathological overlap of miR-200a and miR-25 indicates the potential for functional integration of these p53-targeting miRNAs during tumorigenesis.
miR-200a cooperates with RalGEF
The Raf-Erk effector pathway is largely responsible for Ras-mediated transformation of murine cells (67,68), but not human cells; instead, the RalGEF effector pathway alone is able to recapitulate ~60% of Ras-induced transformation of human embryonic kidney cells, but is not able to induce tumorigenesis in vivo (69). Consistent with this, our study shows that activation of the RalGEF pathway alone in MCF10A cells is unable to induce tumorigenesis. However, we showed that the combination of miR-200a overexpression and RalGEF pathway activation is enough to induce transformation and subcutaneous tumor formation in vivo. This effect is not seen when miR-200a is combined with mutants activating either the PI3K or the Raf/Erk pathways. miR-200a overexpression alone is insufficient to induce colony formation or tumorigenesis, indicating that the combination of Akt activation and loss of p53 is not enough to effect transformation. Overall, this shows that miR-200a overexpression is sufficient to transform immortalized rat RK3E cells, but not immortalized human cells; furthermore, these data indicate that miR-200a activation of Akt and suppression of p53 cooperate with RalGEF signaling to transform immortalized human MCF10A cells and induce tumorigenesis (Figure 6).
The dichotomous roles of miR-200a
miR-200a has a well-studied dichotomous role in cancer, and both its overexpression and downregulation have been associated with increased tumorigenesis (16,70), emphasizing the potential for context-specific signaling and pathway interactions. In particular, miR-200a has been shown to be dysregulated in pancreatic cancer (70,71). In one study, miR-200a was found to be expressed at high levels in early pancreatic neoplasms compared to normal controls (71). Another study found that the miR-200a cluster is hypomethylated pancreatic tumors, leading to increased expression levels in the tumor as well as in circulation. Pancreatic cancer relies on overactive Ras signaling, and >90% of pancreatic cancers harbor activating K-Ras mutations (72). miR-200a is also dysregulated in ovarian cancers, which frequently harbor K-Ras mutations that are associated with less aggressive versions of the disease (73–75). Higher levels of miR-200a is reported in ovarian tumors compared to benign ovarian cysts, and when further stratified, higher levels of miR-200a were seen in non-metastatic tumors versus metastatic tumors (74). This is consistent with the hyperproliferative, non-migratory phenotype of miR-200a-overexpressing cells we observed in our study. These gene expression profiling studies reveal potential cross-talk of miR-200a dysregulation and K-Ras mutations in human cancer. We illuminate a model that represents the oncogenic and tumor-suppressive roles of miR-200a when it is expressed throughout cancer progression (Figure 6). On one hand, miR-200a inhibits EMT to prevent metastatic dissemination (16). On the other hand, miR-200a is upregulated in invasive forms of breast cancer and may promote metastatic colonization (22,30). Our data support the oncogenic role of miR-200a during cancer initiation, illustrating the dichotomous nature of miR-200a in cancer.
In summary, our results show that miR-200a enhances Ras-mediated transformation of human cells. This is the first study that delineates miR-200a’s function in Ras-mediated cell transformation. We also show that miR-200a synergizes with the RalGEF-activating E37G Ras effector mutant to transform MCF10A cells and induce tumorigenesis in vivo, providing mechanistic insight into this dichotomous miRNA. By determining that cooperation with RalGEF is necessary for miR-200a-mediated transformation, we have illuminated a new, specific role for miR-200a in cell transformation by activating Akt and suppressing p53. Future studies examining the concomitant genetic changes occurring during cell transformation will reveal the role of miR-200a in cancer initiation and bespeak its translational potential.
Supplementary Data
Supplementary Material (full film scans of western blots of Figure 1D and Figure 4) can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (NIH) (R01 CA138688) and Kentucky Lung Cancer Research Program (to Y.L.). National Institute of Environmental Health Sciences (NIEHS) training grant (T32-ES011564 to L.E.B.).
Supplementary Material
Acknowledgements
We would like to thank Dr Damien Wilburn for his expertise in statistical analyses and Randy Becker for figure formatting. L.E.B. designed and performed experiments and prepared the manuscript. A.A.L.T. designed and performed experiments and approved the manuscript. Y.L. designed experiments and prepared the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- EMT
epithelial-to-mesenchymal transition
- MEF
mouse embryonic fibroblast
- miRNA
microRNA.
References
- 1. Paci E., et al. (2005). Early diagnosis, not differential treatment, explains better survival in service screening. Eur. J. Cancer, 41, 2728–2734. [DOI] [PubMed] [Google Scholar]
- 2. Sant M., et al. ; EUROCARE Working Group. (2003). EUROCARE-3: survival of cancer patients diagnosed 1990-94–results and commentary. Ann. Oncol., 14 (suppl. 5), v61–118. [DOI] [PubMed] [Google Scholar]
- 3. Siegel R., et al. (2013). Cancer statistics, 2013. CA. Cancer J. Clin., 63, 11–30. [DOI] [PubMed] [Google Scholar]
- 4. Chambers A.F., et al. (2002). Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer, 2, 563–572. [DOI] [PubMed] [Google Scholar]
- 5. Mcallister R.M. (1965). Viruses and cancer. Calif. Med., 102, 344–352. [PMC free article] [PubMed] [Google Scholar]
- 6. Hanahan D., et al. (2000). The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 7. Knudson A.G. (2001). Two genetic hits (more or less) to cancer. Nat. Rev. Cancer, 1, 157–162. [DOI] [PubMed] [Google Scholar]
- 8. Boehm J.S., et al. (2005). Transformation of human and murine fibroblasts without viral oncoproteins. Mol. Cell. Biol., 25, 6464–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanahan D., et al. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 10. Chen P.S., et al. (2012). Dysregulation of microRNAs in cancer. J. Biomed. Sci., 19, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fabbri M., et al. (2013). Epigenetic regulation of miRNAs in cancer. Adv. Exp. Med. Biol., 754, 137–148. [DOI] [PubMed] [Google Scholar]
- 12. Lee Y.S., et al. (2009). MicroRNAs in cancer. Annu. Rev. Pathol., 4, 199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calin G.A., et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA, 101, 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hill L., et al. (2013). ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer, 132, 745–754. [DOI] [PubMed] [Google Scholar]
- 15. van Kempen L.C., et al. (2012). Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch., 461, 441–448. [DOI] [PubMed] [Google Scholar]
- 16. Xia H., et al. (2010). miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem. Biophys. Res. Commun., 391, 535–541. [DOI] [PubMed] [Google Scholar]
- 17. Bracken C.P., et al. (2008). A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res., 68, 7846–7854. [DOI] [PubMed] [Google Scholar]
- 18. Burk U., et al. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep., 9, 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park S.M., et al. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev., 22, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wells A., et al. (2008). E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin. Exp. Metastasis, 25, 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bendoraite A., et al. (2010). Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol. Oncol., 116, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dykxhoorn D.M., et al. (2009). miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE, 4, e7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korpal M., et al. (2011). Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med., 17, 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hyun S., et al. (2009). Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell, 139, 1096–1108. [DOI] [PubMed] [Google Scholar]
- 25. Petrelli A., et al. (2014). MicroRNA/gene profiling unveils early molecular changes and nuclear factor erythroid related factor 2 (NRF2) activation in a rat model recapitulating human hepatocellular carcinoma (HCC). Hepatology, 59, 228–241. [DOI] [PubMed] [Google Scholar]
- 26. Becker L.E., et al. (2012). A systematic screen reveals MicroRNA clusters that significantly regulate four major signaling pathways. PLoS ONE, 7, e48474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao J., et al. (2014). microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor A in breast cancer. DNA Cell Biol., 33, 291–300. [DOI] [PubMed] [Google Scholar]
- 28. Castilla M.A., et al. (2012). MicroRNA-200 family modulation in distinct breast cancer phenotypes. PLoS One, 7, e47709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bockmeyer C.L., et al. (2011). MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res. Treat., 130, 735–745. [DOI] [PubMed] [Google Scholar]
- 30. Wang B., et al. (2014). miRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life, 66, 371–377. [DOI] [PubMed] [Google Scholar]
- 31. Gravgaard K.H., et al. (2012). The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res. Treat., 134, 207–217. [DOI] [PubMed] [Google Scholar]
- 32. Lu Z., et al. (2011). miR-301a as an NF-?B activator in pancreatic cancer cells. EMBO J., 30, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takwi A.A., et al. (2012). A statin-regulated microRNA represses human c-Myc expression and function. EMBO Mol. Med., 4, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rangarajan A., et al. (2004). Species- and cell type-specific requirements for cellular transformation. Cancer Cell, 6, 171–183. [DOI] [PubMed] [Google Scholar]
- 35. Debnath J., et al. (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods, 30, 256–268. [DOI] [PubMed] [Google Scholar]
- 36. Ma X., et al. (2011). Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl Acad. Sci. USA, 108, 10144–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kolligs F.T., et al. (1999). Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell. Biol., 19, 5696–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Foster K.W., et al. (1999). Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ., 10, 423–434. [PubMed] [Google Scholar]
- 39. Louro I.D., et al. (2002). Comparative gene expression profile analysis of GLI and c-MYC in an epithelial model of malignant transformation. Cancer Res., 62, 5867–5873. [PubMed] [Google Scholar]
- 40. Meyer N., et al. (2008). Reflecting on 25 years with MYC. Nat. Rev. Cancer, 8, 976–990. [DOI] [PubMed] [Google Scholar]
- 41. Li R., et al. (2014). MiR-200a is involved in proliferation and apoptosis in the human endometrial adenocarcinoma cell line HEC-1B by targeting the tumor suppressor PTEN. Mol. Biol. Rep., 41, 1977–1984. [DOI] [PubMed] [Google Scholar]
- 42. Shen L.J., et al. (2013). Mmu-microRNA-200a overexpression leads to implantation defect by targeting phosphatase and tensin homolog in mouse uterus. Reprod. Sci., 20, 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korpal M., et al. (2008). The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem., 283, 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bowman T., et al. (1996). Tissue-specific inactivation of p53 tumor suppression in the mouse. Genes Dev., 10, 826–835. [DOI] [PubMed] [Google Scholar]
- 45. Soule H.D., et al. (1990). Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res., 50, 6075–6086. [PubMed] [Google Scholar]
- 46. Dawson P.J., et al. (1996). MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol., 148, 313–319. [PMC free article] [PubMed] [Google Scholar]
- 47. Imbalzano K.M., et al. (2009). Increasingly transformed MCF-10A cells have a progressively tumor-like phenotype in three-dimensional basement membrane culture. Cancer Cell Int., 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., et al. (2011). Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J. Biol. Chem., 286, 16218–16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bryant D.M., et al. (2004). The ins and outs of E-cadherin trafficking. Trends Cell Biol., 14, 427–434. [DOI] [PubMed] [Google Scholar]
- 50. Cavallaro U., et al. (2004). Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer, 4, 118–132. [DOI] [PubMed] [Google Scholar]
- 51. Li Q., et al. (2008). Restoration of E-cadherin cell-cell junctions requires both expression of E-cadherin and suppression of ERK MAP kinase activation in Ras-transformed breast epithelial cells. Neoplasia, 10, 1444–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu S.J., et al. (2013). MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin. Cancer Res., 19, 1389–1399. [DOI] [PubMed] [Google Scholar]
- 53. Castellano E., et al. (2011). RAS interaction with PI3K: more than just another effector pathway. Genes Cancer, 2, 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miller F.R., et al. (1993). Xenograft model of progressive human proliferative breast disease. J. Natl. Cancer Inst., 85, 1725–1732. [DOI] [PubMed] [Google Scholar]
- 55. Nagaoka K., et al. (2013). Epithelial cell differentiation regulated by MicroRNA-200a in mammary glands. PLoS ONE, 8, e65127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crépin D., et al. (2014). The over-expression of miR-200a in the hypothalamus of ob/ob mice is linked to leptin and insulin signaling impairment. Mol. Cell. Endocrinol., 384, 1–11. [DOI] [PubMed] [Google Scholar]
- 57. Wullschleger S., et al. (2006). TOR signaling in growth and metabolism. Cell, 124, 471–484. [DOI] [PubMed] [Google Scholar]
- 58. Park J.T., et al. (2013). FOG2 protein down-regulation by transforming growth factor-ß1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J. Biol. Chem., 288, 22469–22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dou L., et al. (2013). miR-200s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J. Biol. Chem., 288, 22596–22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao J.J., et al. (2003). Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell, 3, 483–495. [DOI] [PubMed] [Google Scholar]
- 61. Rieber M., et al. (2014). p53 inactivation decreases dependence on estrogen/ERK signalling for proliferation but promotes EMT and susceptibility to 3-bromopyruvate in ERα+ breast cancer MCF-7 cells. Biochem. Pharmacol., 88, 169–177. [DOI] [PubMed] [Google Scholar]
- 62. Zhang Y., et al. (2013). PUMA cooperates with p21 to regulate mammary epithelial morphogenesis and epithelial-to-mesenchymal transition. PLoS ONE, 8, e66464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Corthals S.L., et al. (2011). MicroRNA signatures characterize multiple myeloma patients. Leukemia, 25, 1784–1789. [DOI] [PubMed] [Google Scholar]
- 64. Chi J., et al. (2011). MicroRNA expression in multiple myeloma is associated with genetic subtype, isotype and survival. Biol. Direct, 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deng Q., et al. (2014). The dichotomy of p53 regulation by noncoding RNAs. J. Mol. Cell Biol., 6, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jonsdottir K., et al. (2012). Validation of expression patterns for nine miRNAs in 204 lymph-node negative breast cancers. PLoS ONE, 7, e48692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bonner T.I., et al. (1985). Structure and biological activity of human homologs of the raf/mil oncogene. Mol. Cell. Biol., 5, 1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leevers S.J., et al. (1994). Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature, 369, 411–414. [DOI] [PubMed] [Google Scholar]
- 69. Hamad N.M., et al. (2002). Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev., 16, 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li A., et al. (2010). Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res., 70, 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. du Rieu M.C., et al. (2010). MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin. Chem., 56, 603–612. [DOI] [PubMed] [Google Scholar]
- 72. Vincent A., et al. (2011). Pancreatic cancer. Lancet, 378, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen Y., et al. (2013). Candidate microRNA biomarkers in human epithelial ovarian cancer: systematic review profiling studies and experimental validation. Cancer Cell Int., 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu S., et al. (2014). The biphasic expression pattern of miR-200a and E-cadherin in epithelial ovarian cancer and its correlation with clinicopathological features. Curr. Pharm. Des., 20, 1888–1895. [DOI] [PubMed] [Google Scholar]
- 75. Kohn E.C., et al. (2013). Ovarian cancer: making its own rules-again. Cancer, 119, 474–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.