Summary
The present study showed, for the first time, that aberrant cfmiRNAs may contribute to survival outcome of esophageal adenocarcinome (EA) and H. pylori infection status may modify the association between cfmiRNAs and EA survival.
Abstract
Cell free circulating microRNAs (cfmiRNAs) have been recognized as robust and stable biomarkers of cancers. However, little is known about the prognostic significance of cfmiRNAs in esophageal adenocarcinoma (EA). In this study, we explored whether specific cfmiRNA profiles could predict EA prognosis and whether Helicobacter pylori (HP) infection status could influence the association between cfmiRNAs and EA survival outcome. We profiled 1075 miRNAs in pooled serum samples from 30 EA patients and 30 healthy controls. The most relevant cfmiRNAs were then assessed for their associations with EA survival in an independent cohort of 82 patients, using Log-rank test and multivariate Cox regression models. Quantitative real-time PCR (qRT-PCR) was used for cfmiRNA profiling. HP infection status was determined by immunoblotting assay. We identified a panel of 18 cfmiRNAs that could distinguish EA patients from healthy subjects (P = 3.0E–12). In overall analysis and in HP-positive subtype patients, no cfmiRNA was significantly associated with EA prognosis. In HP-negative patients, however, 15 cfmiRNAs were significantly associated with overall survival (OS) (all P < 0.05). A combined 2-cfmiRNA (low miR-3935 and high miR-4286) risk score was constructed; that showed greater risk for worse OS (HR = 2.22, P = 0.0019) than individual cfmiRNA alone. Patients with high-risk score had >10-fold increased risk of death than patients with low risk score (P = 0.0302; HR = 10.91; P = 0.0094). Our findings suggest that dysregulated cfmiRNAs may contribute to EA survival outcome and HP infection status may modify the association between cfmiRNAs and EA survival.
Introduction
Esophageal adenocarcinoma (EA) is one of the most aggressive gastrointestinal cancers. Despite advances in diagnostic and therapeutic strategies, the prognosis of EA remains relatively poor, with 5-year overall survival rate approximately 10% in Western countries (1). Moreover, the incidence of EA is steadily increasing, with 4- to 5-fold increase in the North America in the last four decades (2). At present, the most important prognostic factor for EA is histological stage (The TNM staging system [3]). However, large variations in the clinical outcomes of patients with the same pathological stage have been observed, suggesting that the histological staging system is inadequate for accurately defining prognosis. Moreover, the TNM cancer staging systems predict survival on the basis of anatomic extent of the tumor rather than on molecular changes, providing little information for developing novel therapeutic strategies. Recent studies suggest that several broad categories of molecules, including gene expression, protein biomarkers and genetic polymorphisms may contribute to EA prognosis (4); but most of these biomarkers had moderate predictive powers and were not cell- or tissue-type specific. Thus, there is an urgent need to identify novel biomarkers that are more crucial to EA prognosis.
MicroRNAs (miRNAs) are endogenous non-coding RNAs that post-trancriptionally control gene expression and regulate various biologic functions, such as cellular proliferation, differentiation and apoptosis (5). Aberrant miRNA expression in tumor tissues has been associated with the development and progression of various types of cancers (6), including EA (7). However, the invasive procedure of obtaining tumor tissue samples limits the application of tumor tissue for miRNA biomarker studies. Recently, increasing evidences have shown that tumor cells can release miRNAs into the circulation (8) and profiles of cell free circulating miRNAs (cfmiRNA) in plasma and serum have been found to be altered in cancers and other benign diseases (9), suggesting broad opportunities for development of cfmiRNAs as less-invasive biomarkers. Importantly, cfmiRNAs stable in serum or plasma, and freeze/thaw as well as prolonged storage at room temperature do not affect cfmiRNA levels (10). Because of the stability in circulation and evidence for their association with pathological changes, growing attentions have been paid to the study of cfmiRNAs as biomarkers of cancer diagnosis and prognosis. Indeed, many studies have shown that individual cfmiRNAs are potential biomarkers for prognosis of cancers (11), including esophageal squamous cell carcinoma (12,13). However, few studies have comprehensively investigated the roles of cfmiRNAs in cancer prognosis on an epigenome-wide scale. Little is known about the clinical utility of cfmiRNAs in EA (6,9).
Currently, more than 2000 human miRNAs have been identified and each of these may target around 1000 genes, leading to complex layer of control of signally pathways important to the development or progression of cancers (14,15). Thus, high-throughput analysis is clearly important to our understanding of miRNA functions in disease. However, studies on high-throughput platforms usually require large sample size to provide adequate statistical power for association analyses, limiting its applications in uncommon diseases where large samples are extremely difficult to obtain. One of the most useful solutions that have been applied to high-dimensional biological data is sample pooling (16). It is a technique where subsets of samples are randomly selected and pooled within each group, for estimating average levels of biomarkers within a group. Pooling helps to reduce cost, time, and amount of starting material required (17).
Helicobacter pylori (HP) is a gram-negative bacterium causally implicating in many gastric diseases, including duodenitis, gastritis, peptic ulcers and gastric cancer (18). In contrast to its association with gastric disorders, numerous studies have demonstrated that HP infection is inversely associated with the development of EA. Results of several meta-analyses consistently showed that HP infection is associated with a nearly 50% reduction in the risk of EA (19,20). While the inverse association of HP infection with EA risk is well-recognized, little is known about the impact of HP infection on survival outcomes in patients with EA. Recent studies suggest that HP infection can modify the biological functions of miRNAs (21,22). However, the contribution of HP-related miRNA dysregulation to EA prognosis remains unclear.
In this study, we applied miRNome arrays to screen for candidate cfmiRNAs that can differentiate EA patients from healthy individuals, using pooled serum samples from EA patients and controls. We then used custom miRNA arrays to investigate whether EA-associated cfmiRNA profiles could serve as prognostic biomarkers for EA. Specifically, we tested the hypothesis that HP infection status may influence the associations of aberrant cfmiRNA profiles with EA prognosis.
Materials and methods
Study design
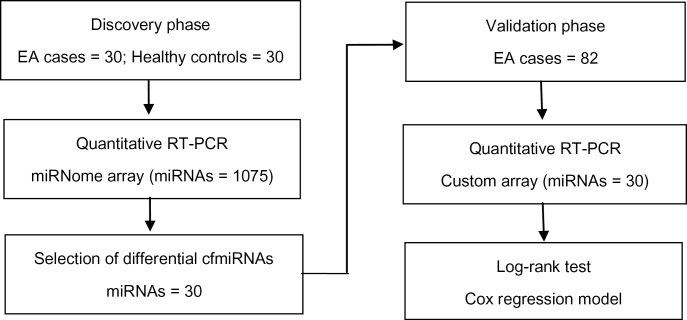
A two-phase test was designed to identify prognostic cfmiRNAs for EA. In the discovery phase, equivalent amounts of serum RNAs from 30 EA cases and 30 healthy controls were randomly divided into 3 EA pools and 3 control pools. Each pool contained serum RNAs from 10 samples. Serum levels of miRNAs in pooled samples were quantified by quantitative real-time PCR (qRT-PCR), using the miRNome array that contains probes for 1075 human miRNAs. Subsequently, the associations of the most relevant cfmiRNAs with overall survival (OS) of EA were further studied in a cohort of 82 EA cases (Figure 1).
Fig. 1.
Study flow chart.
Study population
This study was approved by the Human Subjects Committees of Massachusetts General Hospital (MGH) and Harvard School of Public Health (HSPH). Study subjects were selected from an existing case-control study of EA at MGH. Details of this study population have been previously reported (23). All subjects provided informed, written consents to the participation of the study. Blood samples from EA patients were taken at diagnosis before treatment. Blood samples from controls were obtained at recruitment. EA cases were incident patients with newly diagnosed and histologically confirmed EA that were recruited prospectively at MGH between 1992 and 1996. For this study, EA was defined as a tumor center located at or above the gastroesophageal junction and had at least two-thirds of the bulk tumor located in the esophagus. Subjects mixed with Barrett’s esophagus (BE) or BE patients were excluded from the present study. Controls were accrued from a common control pool originally recruited for multiple case-control studies of lung and esophageal cancers (23). Controls were healthy friends and non-blood-related family members who were visiting hospital patients. All controls had never had received a diagnosis of cancer.
Circulating RNA extraction
Peripheral venous blood sample was drawn from each subject and serum sample was separated within 2h. Serum was isolated by centrifugation at 2000 r.p.m. for 10min at 4°C and stored at −80°C until analysis. Total RNA was extracted from 200 µl aliquots of serum using the miRNeasy Serum/Plasma Kit (Qiagen, Valenca, CA), according to the manufacturer’s instruction. For normalization of sample-to-sample variation during the RNA isolation procedures, synthetic cel-miR-39 mimic was added to each serum sample before RNA extraction. Extracted RNA was eluted in 14 μl of RNase-free water and then stored at −80°C. Concentration and purity of RNA was measured by a GenQuant spectrophotometry (Pharmacia Biotech, Piscataway, NJ). The ratios of A260/280 were used to indicate the purity of total RNA. RNA yield was expressed as ng/µl serum.
Serum miRNA profiling and data normalization
For qRT-PCR assay, 4 µl enriched RNAs from the 20 µl elute of RNA isolation was reverse-transcribed to cDNA using the miScript II RT Kit (Qiagen Sciences, Germantown, MD) in a total reaction volume of 10 µl. A 1:20 dilution of RT products was used as template for the PCR stage. cfmiRNA profiles in pooled samples were measured using the CS2041 Human whole-miRNome Array by qRT-PCR and the miScript SYBR green PCR kit (Qiagen Sciences) following the manufacturers’ instructions.
Custom miRNA arrays for analysing the selected cfmiRNAs were ordered from Qiagen (Qiagen Sciences), using the custom Qiagen plates with specific primer probes. Candidate cfmiRNA levels were analysed by qRT-PCR using the same miScript SYBR green PCR kit (Qiagen) and protocols as that for pooled samples.
All qRT-PCR reactions were carried out in 384-well plates including synthetic miRNAs miRTC and positive PCR control as internal references. All qRT-PCR reaction was conducted on the ABI 7900HT Real-Time PCR System (Applied Biosystems) under the following conditions: 15min at 95°C and 40 cycles of 15 s at 94°C, 30 s at 60°C and 30 s at 72°C.
Raw data were normalized against the reference miRNAs that were included in all plates. Only values below a minimum threshold (CT < 32) were normalized in order to avoid artefactual regulation due to sample normalization. To exclude the possibility that results were influenced by the normalization method, we repeated normalization analysis with a different method: the median normalization method in which each sample was against the median value of all miRNA samples. The two different normalization methods yielded similar results (r 2 = 98; P < 2.2e−16).
Detection of Helicobacter pylori infection
Serum HP infection status was determined using the Helicoblot 2.1 kit (Genelabs Diagnostics®, Singapore) according to manufacturer’s instructions (24). This immunoblotting assay is known to have high sensitivity and specificity in detecting the IgG of HP in serum samples stored for up to 15 years (25). Positive and negative controls and blinded duplicate samples were run for each reaction.
Statistical analysis
Quantitative data was expressed as mean ± standard deviation. Fisher’s exact chi-square tests were applied to categorical variables, and the student’s t test was used to compare the differences between continuous data. The relative levels of miRNA were quantified using the 2-ΔΔCT method, where ΔCT = CTtarget − CTreference.
The primary outcome of this study was OS, measured from the date of pathological diagnosis to the date of death (event) or last known to be alive (censored). Associations between cfmiRNAs and outcomes (OS) were estimated using the method of Kaplan–Meier to generate survival curves and assessed using the log-rank tests. Cox proportional hazards models were used as our primary analyses, adjusting for age, gender, stage, performance status, smoking status, BMI and HP infection status. Censoring refers to the patients who may drop out or still are alive at the end of the study. All statistical tests were two-sided, and a P-value less than 0.05 was considered significant.
Results
Characteristics of study population
Supplementary Table I, available at Carcinogenesis online, and Table I list the clinical characteristics of subjects in the discovery and validation sets, respectively. Age, gender, smoking status and HP infection status did not differ significantly between EA patients and controls in the discovery set. Moreover, no significant differences between HP-positive and HP-negative patients were observed in terms of age, pathological stage and smoking.
Table I.
Characteristics of EA patients
| Characteristics | All cases | HP positive (n = 24) | HP negative (n = 58) | P-valuea |
|---|---|---|---|---|
| Age | 63.31±10.57 | 65.03±10.70 | 62.61±10.53 | 0.8887 |
| Sex | ||||
| Male | 69 | 17 | 52 | |
| Female | 13 | 7 | 6 | 0.0474 |
| Stage | ||||
| I | 7 | 2 | 5 | 0.7708 |
| II | 36 | 9 | 27 | |
| III | 21 | 6 | 15 | |
| IV | 18 | 7 | 11 | |
| Smoking | ||||
| Never | 17 | 3 | 14 | 0.2892 |
| Former | 44 | 16 | 28 | |
| Current | 21 | 5 | 16 | |
| Packyears | 30.0 (0–133) | 35 | 25 | 0.1872 |
| GERD | ||||
| Yes | 40 | 11 | 29 | 0.8105 |
| No | 42 | 13 | 29 | |
| HP infection status | ||||
| Positive | 24 | |||
| Negative | 58 |
Data are means ± SD for age and frequency for other variables.
aComparing HP positive patients versus HP negative patients.
Identification of differential cfmiRNA profiles between pooled EA and control samples
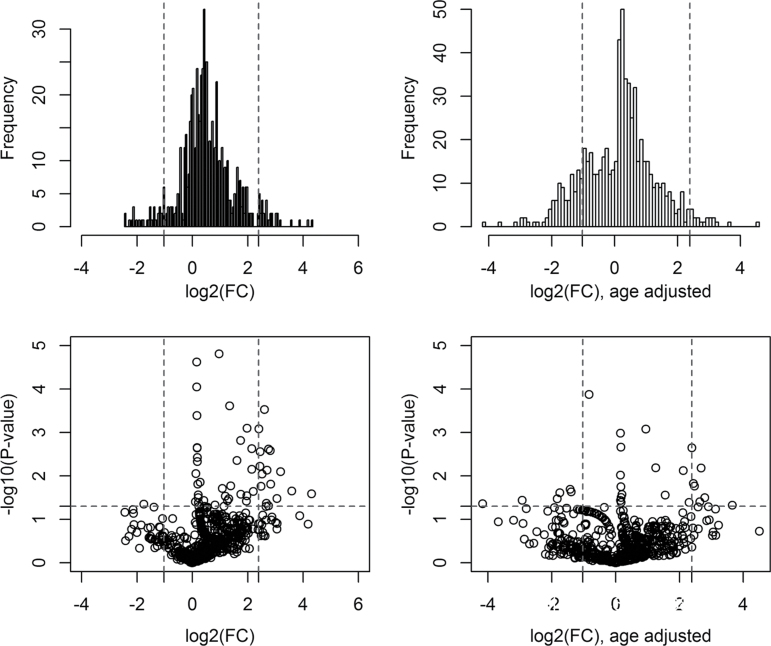
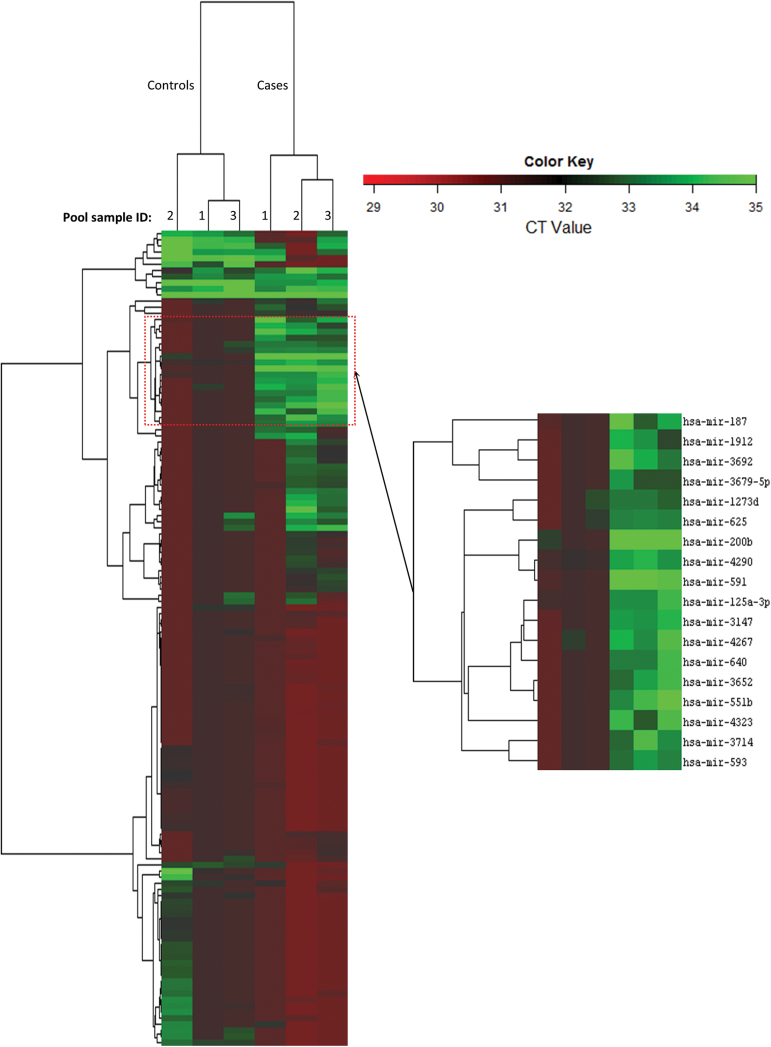
To derive a relative concentration for a given miRNA, we normalized the CT values against the mean CT of miRNAs that were expressed in both case and control pools and had a CT < 32, where CT is the fractional cycle number at which the fluorescence signal exceeds background. The fold change (FC) value was estimated by: FC = 2(ΔCt,control–ΔCt,case). After data filtering, a total of 657 cfmiRNAs were stably detectable in both the case pools and control pools. Among them, 301 cfmiRNAs (fold changes between EA pools and control pools were >2, >4, and >6 for 184, 56 and 23 cfmiRNAs, respectively) with FC >2.0 were selected for volcano plot analysis. We used the following criteria to identify the most differentiated cfmiRNAs (i) the fold change between EA pools and control pools was over the upper 95th percentile or below the lower 5th percentile; (2) the P values of t test were less than 0.05. cfmiRNAs that met both two criteria were selected as candidate cfmiRNAs. Based on these criteria, we identified 18 cfmiRNAs that were significantly differentiated between EA and control (median FC = 5.76). (Figure 2, Supplementary Table II, available at Carcinogenesis online) The effectiveness of these aberrant cfmiRNAs in differentiating EA from control was further analysed by hierarchical clustering model. Multivariate analysis (Hotelling T test) showed that this signature of 18 cfmiRNAs clearly separated the EA samples from control samples (P = 3.0E–12) (Figure 3).
Fig. 2.
Volcano plot illustrates the frequency distribution of FC values and the distribution of FC with corresponding P-values (all values were log transformed). The vertical dash lines represent the cut-off levels of upper 95th percentile and lower 5th percentile of FC values, respectively. The horizontal dash line indicates the significant level (0.05) in log10 scale. cfmiRNAs with FCs either above the 95th percentile or below the 5th percentile and with P-values <0.05 were selected as the most differentiated cfmiRNAs. FC, fold change.
Fig. 3.
Hierarchical cluster analysis demonstrates that a panel of 18 cfmiRNAs could clearly distinguish EA cases from controls (P = 3.0E–12, Hotelling T test).
Association of aberrant cfmiRNAs with survival outcomes EA
We constructed custom arrays that included probes for: (1) 18 miRNAs identified by volcano plot and hierarchical clustering model (Figure 2I); (2) 12 miRNAs whose average FC levels between EA pools and control pools were greater than 6. We used these 30 miRNA custom arrays to quantify cfmiRNA levels in an independent cohort of 82 EA patients. In multivariate Cox regression analysis in the entire cohort, no candidate cfmiRNA was significantly associated with OS (Table II). Moreover, HP infection was not found to be associated with OS of EA (HR = 0.645; 95%CI: 0.16–2.54; P = 0.5302). To investigate whether HP infection status may influence the associations of cfmiRNAs with EA survival outcome, we divided subjects into two groups: HP-positive patients (n = 24) and HP-negative patients (n = 58). In HP-positive subgroup patients, no cfmiRNAs were found to be associated with EA survival outcome. In patients without HP infection, however, 15 cfmiRNAs were significantly associated with EA survival outcomes (all P values <0.05, Table II). The associations remain significant even after adjusting for age, gender, stage, performance status, smoking and gastroesophageal reflux disease symptoms.
Table II.
Associations of cfmiRNAs with OS of EA
| cfmiRNAs | All EA patients (n = 82) | HP positive EA (n = 24) | HP negative EA (n = 58) | All EA patients (n = 82) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P a | HR | 95% CI | P a | HR | 95% CI | P a | P b Interaction | |
| hsa.mir.1253 | 0.84 | 0.59–1.13 | 0.2190 | 1.16 | 0.45–2.98 | 0.7559 | 0.42 | 0.23–0.76 | 0.0042 | 0.0232 |
| hsa.mir.125a.3p | 0.97 | 0.70–1.34 | 0.2973 | 0.95 | 0.59–1.53 | 0.8358 | 1.83 | 0.81–4.11 | 0.1450 | 0.7559 |
| hsa.mir.1273d | 1.05 | 0.80–1.37 | 0.7413 | 0.49 | 0.19–1.28 | 0.1416 | 1.75 | 1.09–2.80 | 0.0196 | 0.0025 |
| hsa.mir.187.5p | 0.88 | 0.65–1.18 | 0.3916 | 1.37 | 0.72–2.58 | 0.3370 | 0.52 | 0.30–0.90 | 0.0187 | 0.0864 |
| hsa.mir.1912 | 1.07 | 0.84–1.37 | 0.5788 | 0.46 | 0.17–1.23 | 0.1213 | 1.51 | 1.14–2.01 | 0.0046 | 0.0256 |
| hsa.mir.200b.5p | 1.22 | 0.94–1.59 | 0.1352 | 0.78 | 0.32–1.90 | 0.5820 | 1.44 | 1.10–1.90 | 0.0087 | 0.1966 |
| hsa.mir.2276 | 1.08 | 0.83–1.42 | 0.3519 | 1.12 | 0.79–1.71 | 0.5920 | 0.97 | 0.61–1.53 | 0.8818 | 0.7646 |
| hsa.mir.3147 | 1.14 | 0.91–1.45 | 0.2592 | 0.70 | 0.47–1.05 | 0.0824 | 1.40 | 1.10–1.78 | 0.0062 | 0.0010 |
| hsa.mir.3149 | 0.86 | 0.67–1.12 | 0.2640 | 0.57 | 0.29–1.14 | 0.1130 | 1.36 | 0.83–2.20 | 0.2192 | 0.0812 |
| hsa.mir.3200.5p | 0.98 | 0.73–1.34 | 0.9321 | 1.04 | 0.63–1.71 | 0.8834 | 1.09 | 0.59–2.00 | 0.7839 | 0.7633 |
| hsa.mir.324.3p | 1.09 | 0.76–1.57 | 0.6335 | 0.96 | 0.45–2.04 | 0.9166 | 2.14 | 1.14–4.04 | 0.0185 | 0.6142 |
| hsa.mir.326 | 1.03 | 0.78–1.37 | 0.8164 | 0.86 | 0.52–1.42 | 0.5518 | 1.83 | 1.12–3.00 | 0.0161 | 0.1263 |
| hsa.mir.3652 | 1.13 | 0.88–1.47 | 0.3419 | 0.53 | 0.22–1.28 | 0.1589 | 1.52 | 1.12–2.08 | 0.0082 | 0.0185 |
| hsa.mir.3679.5p | 0.92 | 0.72–1.17 | 0.4879 | 0.44 | 0.17–1.16 | 0.0958 | 1.90 | 0.80–1.76 | 0.3980 | 0.0097 |
| hsa.mir.3692.5p | 1.16 | 0.90–1.49 | 0.2658 | 1.28 | 0.71–2.29 | 0.4109 | 1.43 | 0.91–2.26 | 0.1223 | 0.6193 |
| hsa.mir.3714 | 0.85 | 0.65–1.10 | 0.2116 | 0.27 | 0.07–1.09 | 0.0655 | 1.18 | 0.79–1.78 | 0.4206 | 0.0021 |
| hsa.mir.3935 | 0.80 | 0.50–1.28 | 0.3496 | 1.99 | 0.64–6.19 | 0.2357 | 0.43 | 0.19–0.96 | 0.0393 | 0.2159 |
| hsa.mir.4252 | 1.07 | 0.81–1.43 | 0.6212 | 1.15 | 0.74–1.77 | 0.5414 | 1.15 | 0.89–2.35 | 0.1332 | 0.7551 |
| hsa.mir.4267 | 1.23 | 0.81–1.79 | 0.2888 | 1.09 | 0.57–2.11 | 0.7891 | 1.99 | 1.14–3.47 | 0.0162 | 0.6863 |
| hsa.mir.4274 | 1.06 | 0.78–1.43 | 0.1345 | 0.75 | 0.28–2.03 | 0.5732 | 1.62 | 1.04–2.53 | 0.0340 | 0.2452 |
| hsa.mir.4286 | 1.20 | 0.90–1.58 | 0.2170 | 0.81 | 0.44–1.50 | 0.5054 | 2.31 | 1.29–4.12 | 0.0043 | 0.0563 |
| hsa.mir.4290 | 0.89 | 0.65–1.21 | 0.4441 | 0.46 | 0.16–1.34 | 0.1550 | 1.23 | 0.73–2.07 | 0.4299 | 0.0284 |
| hsa.mir.4323 | 1.09 | 0.84–1.41 | 0.5268 | 0.55 | 0.25–1.24 | 0.1497 | 1.60 | 1.11–2.34 | 0.0128 | 0.0203 |
| hsa.mir.4325 | 1.01 | 0.85–1.21 | 0.8831 | 0.99 | 0.43–2.25 | 0.9710 | 1.26 | 0.99–1.61 | 0.0566 | 0.9303 |
| hsa.mir.551b.5p | 0.95 | 0.72–1.23 | 0.6730 | 0.91 | 0.43–1.93 | 0.8078 | 1.28 | 0.83–1.96 | 0.2695 | 0.5441 |
| hsa.mir.591 | 0.89 | 0.71–1.10 | 0.2740 | 0.43 | 0.17–1.70 | 0.4360 | 1.60 | 0.97–2.65 | 0.0650 | 0.0036 |
| hsa.mir.593.5p | 0.82 | 0.59–1.14 | 0.2378 | 0.71 | 0.29–1.70 | 0.4360 | 1.14 | 0.63–2.07 | 0.6631 | 0.2131 |
| hsa.mir.625.5p | 1.06 | 0.88–1.28 | 0.5157 | 1.00 | 0.75–1.34 | 0.9901 | 1.29 | 0.97–1.96 | 0.0840 | 0.3656 |
| hsa.mir.640 | 1.20 | 0.98–1.47 | 0.0866 | 1.00 | 0.61–1.58 | 0.9449 | 1.48 | 1.11–1.96 | 0.0069 | 0.2523 |
aAdjusted for age, sex, smoking sort, stages, body mass index at diagnosis, performance status and GERD symptoms.
b P-value for interaction between cfmiRNA and HP infection status.
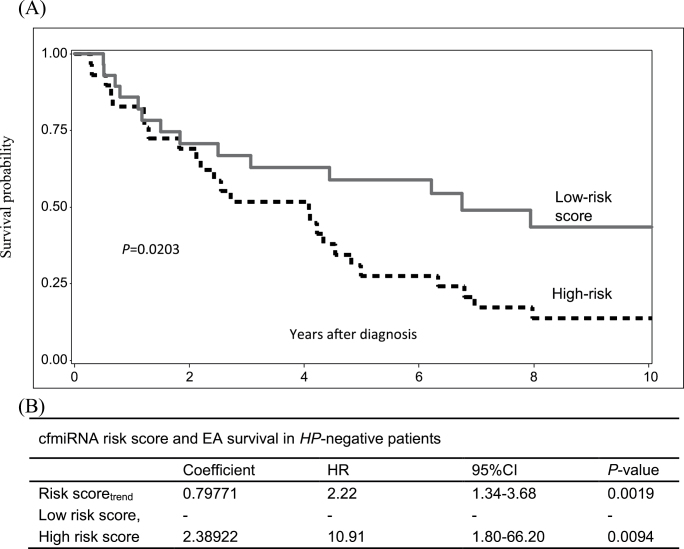
To explore the combined effect of prognostic cfmiRNAs on OS of EA, we developed a cfmiRNA score to predict EA survival outcome. We constructed correlation structure using the 15 candidate cfmiRNAs. We selected cfmiRNAs that had low correlations with other cfmiRNAs (r2 < 0.7) (Supplementary Table III, available at Carcinogenesis online) to calculate a risk score for each patient. Two cfmiRNAs (miR-3935, miR-4286) were selected to build the risk score. A patient’s risk score was calculated as the sum of expression level of each cfmiRNA, multiplied by the corresponding multivariate Cox regression coefficients (Risk score = −0.80287*mir3935 + 0.83495*mir4286). Patients were classified as having a high-risk cfmiRNA score or a low-risk cfmiRNA score, with the median value of the risk score (−2.29) as the threshold value. The median risk score was chosen as the threshold value in order to eliminate the effect of extreme values in the cohort. The cfmiRNA score showed a better prediction of survival than did individual cfmiRNA alone (HRtrend = 2.22; 95%CI: 1.34–3.68; P = 0.0019). Compared with low-risk group, patients in high score group had a significantly worse OS (HR = 10.91; 95%CI: 1.80–66.12; P = 0.0094), (Figure 4).
Fig. 4.
Survival analysis of the cfmiRNA score. (A) Kaplan–Meier estimates of the cfmiRNA score identified by Cox regression model. Patients were classified into low risk group (reference) and high risk group by median stratification. (B) Cox regression analysis of the cfmiRNA risk score in HP-negative patients, adjusting for age, sex, stages, performance status, smoking sort, body mass index at diagnosis and GERD symptoms.
Given the distinct difference in the association of cfmiRNAs with OS of EA between HP-positive and HP-negative patients, we further explored whether interactions existed between cfmiRNAs and HP infection status. To estimate P-values for interaction, a multiplicative interaction term was included in the Cox regression models, accounting for multiple covariates. A total of 10 cfmiRNAs were found to be significantly associated with OS of EA through interactions with HP infection status (Table II).
Discussion
Our study suggests that epigenome-wide miRNA profiling is able to identify specific cfmiRNA pattern that could distinguish EA patients from healthy controls. We also demonstrate that aberrant cfmiRNA profiles are potential biomarkers of EA prognosis. More interestingly, we have found that HP infection status may modify the associations of cfmiRNA with EA prognosis, suggesting that miRNA-HP interactions may play an important role in the prognosis of EA.
In recent years, several high-throughput methodologies have developed to determine miRNA profiles including miRNome array and next-generation sequencing methodologies. Such highly quantitative miRNA analyses are clearly vital to improve our understanding about the role of miRNA dysregulation in complex diseases. Nevertheless, lessons learnt from large-scale genetic association studies suggest that extremely large sample size may be crucial in detecting the small effects expected in the highly complex disorders that contribute most to the global burden of disease (16). While systematic assessment of miRNAs has the potential to revolutionize our knowledge about the pathogenesis of complex disorders, the difficulty of conducting such large-scale research remains prohibitive to many researchers. Validated pooling techniques have been widely employed in studies of DNA sequence variation (26), gene expression (27) and DNA methylation (16), and have allowed researchers to identify candidate biomarkers using relatively small sample sizes. To date, however, very few studies have systematically explored the application of RNA pooling for the analysis of miRNAs. Rather than generating individual results and averaging them within a group, such approaches combine miRNA of different individuals to generate estimates of their average results. These estimates can then be used to compare case and control groups. In studies of DNA sequence variation, screens of DNA pools across thousands of loci have been used to identify regions of the genome for further study via individual genotyping (16). Conversely, some screens have reported no group differences, indicating that the time and expense of further investigation at the individual-sample level may be unjustified (28). In the present study, we applied the miRNome array to the analysis of pooled RNAs, for estimating average cfmiRNA levels within a group. This method can be used to detect group differences and estimate group miRNA averages. As it reduces the amount of RNA starting material required, such an approach may be especially useful to researcher with limited RNA stocks. In larger scale studies involving multiple candidate regions, this method will also prove valuable in highlighting those markers which warrant further study at the individual-sample level.
Our data suggest that specific cfmiRNAs are associated with OS of EA only in HP -negative patients. The striking difference in the association of cfmiRNA with EA outcomes between HP+ and HP− EA patients was unlikely to be due to the differences in age, gender, smoking status, pathological stages or GERD between HP infection statuses (Table II). Moreover, the associations of particular cfmiRNAs with EA prognosis seemed to be independent of the influence of these clinicopathological factors, because after adjusting for these covariates, these cfmiRNAs remained significantly associated with EA prognosis in HP− EA patients. This observation is in line with previous reports describing the influence of HP infection on miRNA dysregulation. For instance, HP induces aberrant silencing of let-7 expression, leading to upregulation of Ras oncoprotein expression (19). HP-induced inflammatory cytokines (IL-8, TNF-α and IL-1β) were able to increase the expression of miR-146a, which in turn reduced the expression of IL-8, TNF-α and IL-1β (29). HP suppressed the expression of miR-320 and upregulated expression of the antiapoptotic protein Mcl-1, leading to decreased apoptosis in a cagA-dependent manner (30). Persistent HP infection may affect expression of miRNAs via chronic inflammatory cytokines, including IL-1β, IL-6 and TNF-α(29,31). While these observations strongly suggest the involvement of HP in miRNA regulation, information in this regard is mostly from studies in gastric disorders. Very little is known about cfmiRNA profiles in EA with special reference to HP infection status. The association of cfmiRNA profile with HP infection status identified in the present study agrees with those reports showing the impact of HP on miRNA expression in other tissues. Our finding that cfmiRNAs are associated with EA OS only in HP-negative subgroup suggests that the contribution of aberrant cfmiRNAs to EA prognosis may be modified by HP infection status. Further studies in larger sample sizes are required to elucidate the molecular mechanisms underlying the interaction of miRNAs and HP infection in the prognosis of EA.
In the present study, we identify 15 candidate cfmiRNAs that are significantly associated with EA prognosis in HP-negative subjects. An important question in this regards is whether these cfmiRNAs may play any role in the pathogenesis of EA. Although the current study did not provide direct evidence demonstrating that these miRNAs are involved in EA prognosis, evidence from previous studies in multiple cancer-related pathological processes may indirectly support our findings. MiR-3652 and miR-640 are able to bind to oncogenes and significantly regulate oncogene expressions (32). Downregulation of miR-1273d has been observed in gastric cancer cell lines (33). Further, it has been shown that miR-1253 is involved in the regulation of multiple drug resistance (MDR) in cancers (34). There is evidence that miR-200 family expression is downregulated in both EA and Barrett’s epithelium (35). It has been reported that circulating miR-200b-5p levels can discriminate lung cancer from controls (36); whereas serum miR-3147 expression levels can predict metastasis in patients with early-stage cervical squamous cell carcinoma (37). Similarly, blood miR-4286 concentrations are associated with metastasized seminoma (38). Serum miR-324-3p was differentially expressed between breast cancer and controls (39). miR-326 was involved in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1 (40).
Compared with those studies on circulating microRNAs for cancer prognosis, our study is unique for the following reasons: first, we screened a large number of serum microRNAs via a miRNome array covering a complete list of human miRNA, which enabled us to have better chance to identify potential aberrant markers. Furthermore, we took the impact of HP infection into account in analyzing the association of cfmiRNAs with EA survival. This integrated analysis approach provide new insight into the mechanisms underlying EA progression. Furthermore, all serum samples were collected at diagnosis before treatment and all subjects had complete demographics, clinical and follow-up information, allowing us to control for the influence of multiple confounding factors and minimize bias of results due to incomplete information.
In summary, we found that specific cfmiRNAs were associated with EA survival outcomes. More importantly, we revealed that the associations of cfmiRNA with EA prognosis may be influenced by HP infection status. This adds new knowledge for better understanding the molecular mechanisms underlying the association between miRNAs and EA prognosis. Although our results need to be confirmed in larger, prospective studies, we conclude that cfmiRNAs hold a great promise to become non-invasive biomarkers for EA prognosis.
Supplementary material
Supplementary Tables I–III can be found at http://carcin.oxfordjournals.org/
Funding
Harvard-NIEHS Center (ES000002 to R.Z); National Institute of Health (CA92824, CA74386, CA90578, and CA119650 to D.C.C).
Supplementary Material
Acknowledgements
We thank the staff of the Massachusetts General Hospital Thoracic Oncology Center for participant recruitment and Andrea Shafer and Salvatore Mucci for data collection and management.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- cfmiRNA
cell free circulating microRNA
- EA
esophageal adenocarcinoma
- FC
fold change
- miRNA
microRNA
- HP
Helicobacter pylori
- OS
overall survival
- qRT-PCR
quantitative real-time PCR.
References
- 1. Lagarde S.M., et al. (2006). Prognostic factors in adenocarcinoma of the esophagus or gastroesophageal junction. J. Clin. Oncol., 24, 4347–4355. [DOI] [PubMed] [Google Scholar]
- 2. Brown L.M., et al. (2008). Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J. Natl. Cancer Inst., 100, 1184–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Enzinger P.C., et al. (2003). Esophageal cancer. N. Engl. J. Med., 349, 2241–2252. [DOI] [PubMed] [Google Scholar]
- 4. Lagarde S.M., et al. (2007). Molecular prognostic factors in adenocarcinoma of the esophagus and gastroesophageal junction. Ann. Surg. Oncol., 14, 977–991. [DOI] [PubMed] [Google Scholar]
- 5. Iorio M.V., et al. (2012). microRNA involvement in human cancer. Carcinogenesis, 33, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakai N.S., et al. (2013). A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin. Cancer Biol., 23(6 Pt B), 512–521. [DOI] [PubMed] [Google Scholar]
- 7. Mathé E.A., et al. (2009). MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin. Cancer Res., 15, 6192–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wittmann J., et al. (2010). Serum microRNAs as powerful cancer biomarkers. Biochim. Biophys. Acta, 1806, 200–207. [DOI] [PubMed] [Google Scholar]
- 9. Ichikawa D., et al. (2012). Circulating microRNA in digestive tract cancers. Gastroenterology, 142, 1074–1078. [DOI] [PubMed] [Google Scholar]
- 10. Mitchell P.S., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A., 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allegra A., et al. (2012). Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int. J. Oncol., 41, 1897–1912. [DOI] [PubMed] [Google Scholar]
- 12. Komatsu S., et al. (2011). Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer, 105, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Komatsu S., et al. (2012). Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin. Biol. Ther., 12 Suppl 1, S53–S59. [DOI] [PubMed] [Google Scholar]
- 14. Bartel D.P. (2009). MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song J.H., et al. (2012). MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology, 143, 35–47.e2. [DOI] [PubMed] [Google Scholar]
- 16. Docherty S.J., et al. (2010). DNA methylation profiling using bisulfite-based epityping of pooled genomic DNA. Methods, 52, 255–258. [DOI] [PubMed] [Google Scholar]
- 17. Docherty S.J., et al. (2009). Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenetics Chromatin, 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cover T.L., et al. (2009). Helicobacter pylori in health and disease. Gastroenterology, 136, 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Islami F., et al. (2008). Helicobacter pylori and esophageal cancer risk: a meta-analysis. Cancer Prev. Res. (Phila)., 1, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhuo X., et al. (2008). Helicobacter pylori infection and oesophageal cancer risk: association studies via evidence-based meta-analyses. Clin. Oncol. (R. Coll. Radiol)., 20, 757–762. [DOI] [PubMed] [Google Scholar]
- 21. Hayashi Y., et al. (2013). CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut, 62, 1536–1546. [DOI] [PubMed] [Google Scholar]
- 22. Noto J.M., et al. (2013). Helicobacter pylori promotes the expression of Krüppel-like factor 5, a mediator of carcinogenesis, in vitro and in vivo . PLoS One, 8, e54344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhai R., et al. (2010). Interactions among genetic variants in apoptosis pathway genes, reflux symptoms, body mass index, and smoking indicate two distinct etiologic patterns of esophageal adenocarcinoma. J. Clin. Oncol., 28, 2445–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye W., et al. (2004). Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. J. Natl. Cancer Inst., 96, 388–396. [DOI] [PubMed] [Google Scholar]
- 25. Aucher P., et al. (1998). Use of immunoblot assay to define serum antibody patterns associated with Helicobacter pylori infection and with H. pylori-related ulcers. J. Clin. Microbiol., 36, 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirov G., et al. (2006). Pooled DNA genotyping on Affymetrix SNP genotyping arrays. BMC Genomics, 7, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kendziorski C., et al. (2005). On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. U. S. A., 102, 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Butcher L.M., et al. (2008). The nature of nurture: a genomewide association scan for family chaos. Behav. Genet., 38, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li N., et al. (2012). H. pylori related proinflammatory cytokines contribute to the induction of miR-146a in human gastric epithelial cells. Mol. Biol. Rep., 39, 4655–4661. [DOI] [PubMed] [Google Scholar]
- 30. Noto J.M., et al. (2011). The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis. Front. Cell. Infect. Microbiol., 1, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Isomoto H., et al. (2012). Interweaving microRNAs and proinflammatory cytokines in gastric mucosa with reference to H. pylori infection. J. Clin. Immunol., 32, 290–299. [DOI] [PubMed] [Google Scholar]
- 32. Ivashchenko A.T., et al. (2013). miR-1279, miR-548j, miR-548m, and miR-548d-5p binding sites in CDSs of paralogous and orthologous PTPN12, MSH6, and ZEB1 Genes. Biomed Res. Int., 2013, 902467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y., et al. (2013). microRNA expression profiling in multidrug resistance of the 5Fu-induced SGC-7901 human gastric cancer cell line. Mol. Med. Rep., 7, 1506–1510. [DOI] [PubMed] [Google Scholar]
- 34. Li H., et al. (2013). The role of microRNA in chemotherapy resistance. Acta. Pharmacol. Sin, 34, 87087–87089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith C.M., et al. (2010). MicroRNAs, development of Barrett’s esophagus, and progression to esophageal adenocarcinoma. World J. Gastroenterol., 16, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cazzoli R., et al. (2013). microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol., 8, 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen J., et al. (2013). Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int. J. Mol. Med., 32, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruf C.G., et al. (2014). Small RNAs in the peripheral blood discriminate metastasized from non-metastasized seminoma. Mol. Cancer, 13, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu Z., et al. (2012). Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis, 33, 828–834. [DOI] [PubMed] [Google Scholar]
- 40. Liang Z., et al. (2010). Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem. Pharmacol., 79, 817–824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.