Summary
Accurate assessment of the biological impact of xenoestrogens could assist breast cancer prevention. Effects on DNA integrity of breast epithelium, which might be missed in common chemical test screens, underscore the importance of endpoints beyond estrogen receptor interaction and cell proliferation.
Abstract
Identification of early perturbations induced in cells from non-cancerous breast tissue is critical for understanding possible breast cancer risk from chemical exposure. We have demonstrated previously that exposure to the ubiquitous xenoestrogens, bisphenol A (BPA) and methyl paraben, promotes the hallmarks of cancer in non-malignant human high-risk donor breast epithelial cells (HRBECs) isolated from several donors. Here we show that terephthalic acid (TPA), a major chemical precursor of polyethylene terephthalate (PET) containers used for the storage of food and beverages, increased the ERα: ERβ ratio in multiple HRBEC samples, suggesting an estrogenic effect. Although, like BPA and methyl paraben, TPA also promoted resistance to tamoxifen-induced apoptosis, unlike these chemicals instead of inducing an increased S-phase fraction, TPA treatment arrested cell proliferation. DNA-PK, ATM and members of the MRN complex, known to be involved in DNA damage sensor and effector proteins, were elevated indicating induction of DNA strand breaks. Early DNA damage checkpoint response, mediated through p53/p21, led to G1 arrest in TPA-exposed cells. Removal of TPA from the growth medium resulted in the rapid induction of BCL2, increasing the ratio of anti-: pro-apoptotic proteins, together with overexpression of Cyclin A/CDK2 proteins. Consequently, despite elevated p53pSer15 and H2AXpSer139, indicating sustained DNA damage, TPA exposed cells resumed robust growth rates seen prior to TPA exposure. The propensity for the perpetuation of DNA aberrations that activate DNA damage pathways in non-malignant breast cells justifies careful consideration of human exposure to TPA, particularly at vulnerable life stages.
Introduction
Due to its high-impact resistance, polyethylene terephthalate (PET) is the commercial plastic of choice for packaging a wide variety of foods and beverages for human consumption, as well as the manufacture of medical supplies and implants. Also known as terephthalic acid-ethylene glycol polyester, the major chemical constituents in the synthesis of PET are terephthalic acid (TPA) and ethylene glycol (1). The importance of TPA as the main raw material for the production of polyester fiber and its use as such has led to an unprecedented increase in its demand as an industrial chemical. Expectedly, with rising global production, TPA discharge into the environment is inevitable, and growing public health issues are foreseeable. Pertinent to this are reports of the release of unpolymerized monomers from polymer resins (2,3), including TPA from PET (4). Dietary TPA promotes chemically induced urinary bladder tumors in rodents (5,6). However, the carcinogenic potential of TPA in humans is currently unknown.
Food and beverage packaging materials are known to leach chemicals that display endocrine disruptive properties (7,8). Previous studies reviewed by Bach et al. (9) have examined the biological effects of migrated PET components using a variety of prokaryotic and eukaryotic models. Although presently uncertain (8,10), a possible role for TPA as an estrogen mimic has critical implications for breast carcinogenesis. Here, we have directly investigated the effects of TPA on well-characterized non-malignant human breast epithelial cells (11–13) using well-accepted in vitro endpoints of cancer progression (14). Since plasma concentrations of TPA are known to reach between 8 and 10 µg/ml following oral administration in experimental animals (15), we tested the cellular effects of TPA at a wide range of concentrations incorporating this exposure level (100 pM to 100 µM: equivalent to 0.032 µg to 32mg). Multiple assays were implemented to provide quantitative measures of a wide spectrum of TPA-induced effects. Our consistent observations of perturbations arising within TPA-treated cultures of non-malignant cells isolated from the breast tissue of multiple donors suggests that ascertaining safe levels of TPA exposure must be considered a high priority.
Materials and methods
Cell culture and TPA treatment
Spontaneously immortalized high-risk donor-derived breast epithelial cell (HRBEC) lines, designated as PA024, PA025 and PA115 (13) were isolated previously by us from primary cultures of donor-derived random periareolar fine needle aspirates (RPFNA) of the unaffected contralateral breast of patients undergoing surgical procedures for benign or malignant disease. HRBEC lines were used at passage 22–27, and propagated as described previously (11–13). Immediately prior to initiating the TPA exposure studies described here, total DNA from each line was amplified with nine sets of widely used DNA fingerprinting primers, and the PCR products resolved in 10% TBE–PAGE gels. Each HRBEC cell line was confirmed to display a unique short tandem repeat (STR) profile, distinct from the cancer cell lines used here. Additional primary HRBECs were isolated from RPFNA samples acquired from human donors with written informed consent approved by the California Pacific Medical Center Institutional Review Board. Established breast cancer cell lines, T47D, MDA231 and SKBR3, were expanded in RPMI+10% fetal bovine serum (FBS) and MCF7 in DMEM+10% FBS.
A TPA stock solution was prepared at 0.1M (1000×) in 0.5M NaOH, which was completely miscible in growth medium without altering the optimal pH. For TPA exposure, cells were seeded either in six-well or 100mm plates and treated for 7 days at indicated concentrations. Under test conditions, cells were maintained in phenol red-free formulations of their optimal growth medium, supplemented with charcoal-stripped FBS at reduced levels that did not affect baseline cell survival (0.2% for HRBECs; 1% for T47D, MDA231 and SKBR3; and 5% for MCF7). TPA, 17β-estradiol (E2), bisphenol A (BPA), 4-hydroxytamoxifen (OHT), bromodeoxyuridine (BrdU) and hydrogen peroxide (H2O2) were purchased from Sigma–Aldrich (St Louis, MO).
Protein isolation and western blot analysis
Cells were lysed in a non-denaturing buffer to generate whole cell protein lysates (150mM NaCl, 50mM Tris–HCl pH 7.4, 0.1% NP40, 1mM dithiothreitol, 5% glycerol containing protease and phosphatase inhibitors). Insoluble pellets isolated from the centrifuged lysates were resuspended in NP40 buffer + 6M Urea and sonicated to recover DNA-bound proteins. Lysates resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis were transferred to polyvinylidene difluoride membranes, incubated with appropriate primary and secondary antibodies, detected by enhanced chemiluminescence and recorded on X-ray film.
Primary antibodies directed at 20 different proteins in TPA-treated or vehicle only cell lysates and their commercial source include: BAX (GTX109683) and ERβ (GTX112927) from Genetex (Irvine, CA); p53pSer15 (9286), ATMpSer1981 (4602), CDK2 (2546), CDK4 (12790), Cyclin A (4656), Cyclin D1 (2978), DNA-PK (4602), H2AX pSer139 (9718), MRE11 (4602), RAD50 (4602) and pRBpSer807/811 (8516) from Cell Signaling Technology (Danvers, MA); p21Waf1 (sc-397), p53 (sc-126), BCL2 (sc492), ERα (sc-543), MLH1 (sc-582), PCNA (sc25280) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (sc-365062) from Santa Cruz Biotechnology (Santa Cruz, CA). Densitometric analysis of western blots was done using AlphaEaseFC software (Alpha Innotech, Santa Clara, CA). To evaluate quantitative changes induced by TPA exposure on cellular proteins, treated and untreated samples of each test cell line were processed in parallel and resolved in adjacent lanes within the same gel. GAPDH was used as a loading control for protein quantitation in every blot. Expanded gels/blots, showing antibody-specificity for each of the 20 above-mentioned proteins in all test cell lines, are included (Supplementary Western Blot Data, available at Carcinogenesis Online).
Quantitation of apoptotic cell death
Apoptosis was induced by 24h treatment with 10 μM OHT. Unfixed cells were stained with fluorescein isothiocyanate (FITC)-conjugated Annexin V (BD Biosciences, San Jose, CA) following manufacturer’s instructions and analyzed by fluorescence-activated cell sorting (FACS) with a FACScan using CellQuest software (BD Biosciences). Vehicle only controls without OHT treatment provided baseline apoptosis in each cell line.
Cell cycle analysis and determination of growth rate
DNA synthesis was quantitated by incorporation of 10 µM BrdU (Sigma–Aldrich) for 1h into cell cultures, subsequently fixed with 70% ethanol. Fixed cells were stained with anti-BrdU (Santa Cruz Biotechnology), FITC-conjugated secondary antibody (Life Technologies, Grand Island, NY), counterstained with propidium iodide (PI) and analyzed by FACS.
Quantitation of DNA damage
Two to four replicates of methanol-fixed cells from indicated treatments were permeabilized with 0.1% Triton X-100 and incubated with anti-p53pSer15 or anti-H2AXpSer139 primary antibody (Cell Signaling Technology) followed by Alexa Fluor® 488 anti-rabbit (Invitrogen), counterstained with PI and analyzed by FACS. Mean fluorescence intensity (MFI) values were acquired for all samples.
For microscopic analysis, cells were seeded into eight-well Millicell slides (Millipore) and treated as indicated in duplicate wells. After fixation with paraformaldehyde, cells were permeabilized with 0.1% Triton X-100, processed with primary and secondary immune reagents and counterstain as detailed above for the FACS assay and analyzed by fluorescence microscopy. Exposure times for each color channel were maintained at a constant setting throughout the acquisition of fluorescent images from all cell lines.
Statistical analysis
All FACS-derived data were summarized as mean ± SD. Response of cells with or without TPA pretreatment was compared using one-way analysis of variance (ANOVA), or two-tailed Student’s t-test. The statistics were analyzed using STATA version 12.1, with P < 0.05 considered to be statistically significant.
Results
Endpoints of TPA-induced cellular perturbation described below were repeatedly evaluated in three described previously non-malignant breast epithelial cell lines—PA024, PA025 and PA115, isolated from independent RPFNAs from cancer-free breast tissue of individuals with an elevated risk of breast cancer (13). These spontaneously immortalized HRBEC lines represent rare samples (3/200), confirmed by cytopathological criteria to be non-malignant at the time of RPFNA collection from the donor. In addition to these cell lines, early passage primary cultures of finite-lifespan HRBECs from other donors were tested whenever sufficient cell numbers were available for the assays employed in this study.
TPA treatment alters estrogen receptor levels in non-malignant cells
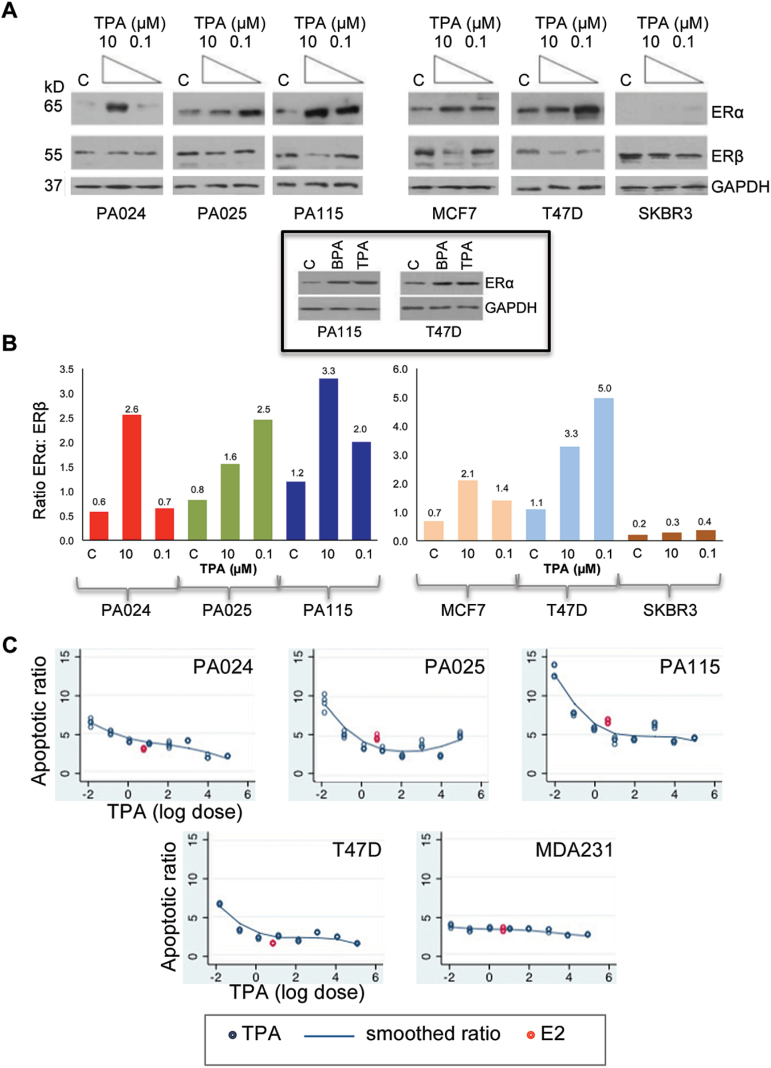
Unlike other reported immortalized non-malignant breast epithelial cell lines, the HRBEC lines—PA024, PA025 and PA115 uniquely maintain estrogen receptor (ER)-positive status in vitro. Cultures exposed to 7 days TPA-treatment at increasing doses were evaluated for ERα and ERβ expression by western blotting (Figure 1A). ERα-positive (T47D, MCF7) and ERα-negative (SKBR3) breast cancer cell lines were included for comparison or for reference. In the presence of TPA, ERα expression was elevated compared with untreated controls in 5/5 ERα-positive cell lines. Induction of ERα protein varied among the test cells lines with PA025 and T47D being the most sensitive at 100nM TPA. Other test cell lines showed a similar level of induction at 10 µM TPA, which appeared to be toxic to PA025 cells (Supplementary Figure 1 is available at Carcinogenesis Online). In contrast, ERβ was detectably decreased by TPA exposure in all cell lines with the exception of PA024. Altogether, the ERα: ERβ ratio was perturbed by TPA exposure in all ERα-positive test cells (Figure 1B). ERα protein induction by TPA or by another known XE–BPA, at the same concentration (100nM), occurred to a similar level (Figure 1A insert).
Fig. 1.
TPA induces ERα, and confers resistance to programmed cell death. (A) Western blot analysis of non-malignant HRBEC lines (PA024, PA025 and PA115). Each subpanel represents adjacent lanes within the same gel. Seven days exposure to TPA results in higher ERα and unchanged or lower ERβ protein levels. Boxed insert shows comparable ERα protein induced by TPA or BPA under comparable exposure conditions in representative cell lines. (B). The ratio of ERα:ERβ expression in ER-positive malignant and non-malignant cells, derived from quantitation of band density in panel A, is consistently increased in the presence of TPA compared with control. (C) Apoptotic ratios of TPA-treated HRBEC and breast cancer cell lines derived by FACS analysis of Annexin V-FITC stained samples. Cells were pretreated with increasing TPA or with E2 for 7 days prior to the apoptotic stimulus of OHT treatment for 24h. X-axis is log(base 10) of TPA (µM) with logdose = −2 representing no TPA. The full range of TPA doses includes 0.0001 to 100 μM and E2 at 5nM (red). Y-axis values represent the ratio of apoptotic cells in OHT: non-OHT treated cultures. Each data point represents three or more replicates. Smoothed lines are lowess (locally weighted smoothing). Significant variability is evident across cell lines in both sensitivity to the apoptotic stimulus, as well as induction of apoptosis evasion by TPA and E2.
TPA-treated cells are resistant to programmed cell death
Next, we determined whether elevated ERα levels were accompanied by suppression of apoptotic cell death in HRBECs. Cells pretreated with TPA were exposed to 10 µM OHT for apoptosis induction and detected by Annexin V binding. We used the ratio of OHT-treated: untreated values from three HRBEC lines and two breast cancer cell lines over a dose range of 0.1nM to 100 µM TPA for pairwise comparisons. Ratios of each cell line were plotted against TPA dose using a model that included dose and cell line (as discrete factors), as well as the interaction between these two factors to determine whether the fraction of apoptotic cells was altered by treatment. In ER-negative MDA231 cancer cells, OHT treatment had a minimal effect on apoptosis induction, and consequently apoptosis evasion was minimal as well. However, in 4/4 ER-positive cell lines, even the lowest dose of TPA (0.1nM) was found to reduce OHT-induced apoptosis by 27–53% compared with non-TPA treatment controls. A logarithmic increase in TPA dose did not necessarily result in a proportional increase in apoptosis evasion. Statistical t-tests for all non-interaction coefficients were significant at P < 0.001 (Figure 1C).
Similarly, the apoptotic fraction was significantly reduced by treatment of ER-positive cells with E2. In PA024, apoptosis evasion mediated by E2 (5nM) was comparable to a 10–100 μM TPA dose whereas in PA025 and PA115, apoptosis evasion was induced by TPA at concentrations lower (0.1–1nM) than E2, and in T47D cells, E2 and TPA equivalence was at an intermediate level (100nM). E2-treated or untreated MDA231 cells were equally resistant to OHT (Figure 1C). These data demonstrate a close similarity in the effects of TPA to natural and synthetic estrogens in triggering apoptosis evasion in breast epithelial cells.
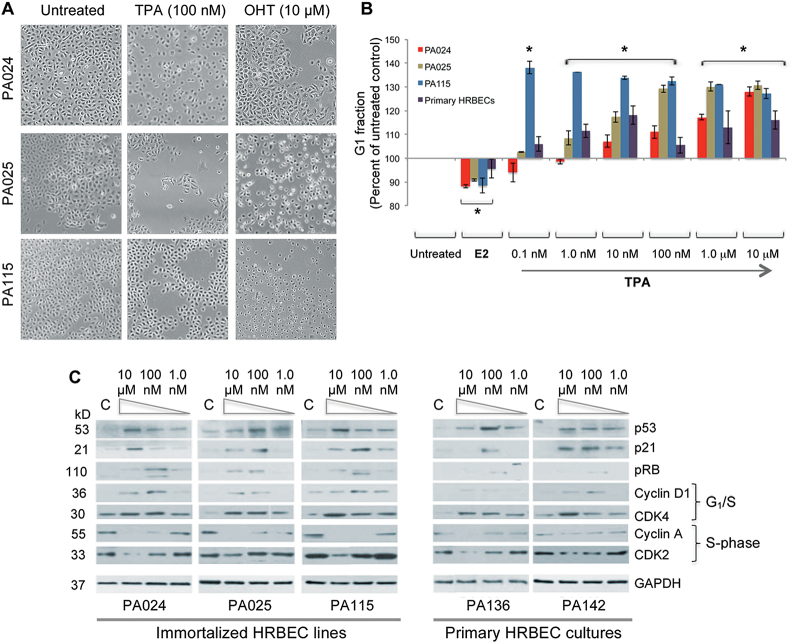
TPA activates the G1/S checkpoint leading to growth arrest
As shown previously by us, the XEs, BPA and methyl paraben stimulate S-phase in HRBECs (12). In contrast, TPA treatment was found to induce growth cessation similar to that displayed in the presence of OHT (Figure 2A). Albeit to different degrees, all TPA-treated HRBECs, both cell lines and finite-life HRBECs (n = 3), demonstrated a cell cycle arrest in G1 accompanied by a decline in the BrdU-labeled S-phase fraction across the range of TPA doses tested. Growth arrest was less striking in finite-life HRBECs since their baseline proliferation rate is relatively slow. As is typical with increased cell proliferation, all cultures treated with E2 displayed a reduction in the G1 fraction concurrently with an increase in the S-phase population (Figure 2B).
Fig. 2.
TPA arrests cell proliferation in immortalized and finite-life HRBECs. (A) Reduction in cell growth compared with untreated controls is evident in cultures exposed to 100nM TPA for 7 days. For comparison, growth arrest induced by 10 μM OHT treatment for 24h is shown (Brightfield images—×4 objective). (B) G1 arrest of the cell cycle measured by FACS analysis of BrdU-labeled HRBEC lines and finite-life primary HRBEC cultures (combined average values shown for samples PA124, PA132 and PA142) pretreated with TPA concentrations ranging from 0.1nM to 10 μM. Untreated cells served as a negative control and cells treated with 5nM E2 as the positive control. Data are represented as mean ± SD of two independent experiments performed in triplicate. Statistically significant induction (or reduction) of the G1 fraction is indicated by asterisks (two-tailed P value < 0.05). (C) Western blot analysis of cell cycle checkpoint proteins in TPA-treated lysates of HRBEC lines and in finite-lifespan primary HRBEC cultures (samples PA136 and PA142). Each set of four lanes represents untreated control (C), 10 μM, 100nM and 1nM TPA treatment. Induction of p53, p21WAF1, CDK4 and Cyclin D1 proteins and phosphorylation of pRB is evident at one or more TPA concentration in all samples. Conversely, Cyclin A and CDK2 proteins required for passage through S-phase, are reduced by one or more TPA concentrations compared with untreated control. GAPDH was used as a loading control.
TPA treatment uniformly led to an escalation in p53, and p21WAF1 protein levels in both HRBEC cell lines and finite-life primary HRBECs (quantitation relative to untreated control shown in Supplementary Figure 2 is available at Carcinogenesis Online). This observation is in striking contrast to BPA-induced downregulation of the G1-checkpoint proteins—p53 and p21WAF1 [with concomitant Cyclin A/CDK2 induction (13)]. Simultaneously, induction or activation of the G1/S transition proteins—pRB and Cyclin D1/CDK4 was also observed over the range of TPA concentrations. Nevertheless, on balance, the accompanying repression of the S-phase CDK complex (Cyclin A/CDK2) was the dominant factor leading to TPA-induced HRBEC growth arrest (Figure 2C). Altogether, 6/6 non-malignant HRBEC donor samples displayed a G1-arrest response due to TPA exposure, accompanied by the characteristic profile of perturbations in cell cycle regulatory proteins shown here.
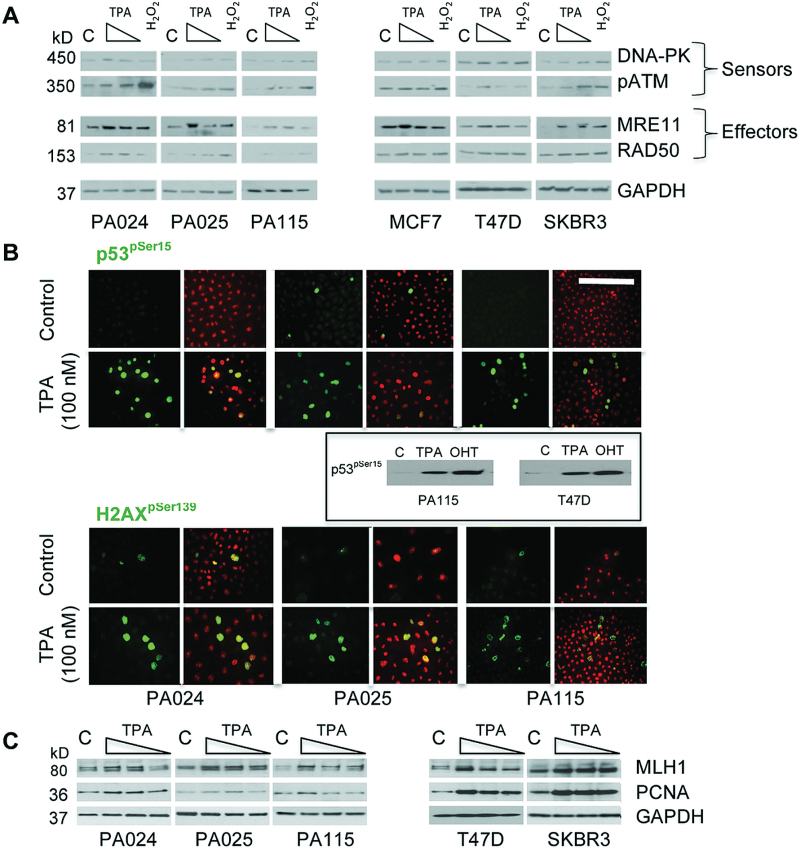
TPA activates DNA-damage response pathways
To determine the biological basis for the unexpected cell cycle arrest in TPA-treated HRBECs, lysates were analyzed to gauge the induction of DNA damage pathway proteins. A consistent increase was observed across all test samples in the expression of DNA-PK—a DNA repair protein required for rejoining DNA double-strand breaks (DSBs) and in the activation of ATM—a serine/threonine protein kinase recruited by DSBs. Additionally, TPA treatment led to the induction or activation of members of the MRN complex (MRE11 and RAD50) known to play an important role in the processing of DSBs prior to repair (Figure 3A). Such perturbations were less apparent in breast cancer cell lines, probably due to preexisting intrinsic genomic instability characteristic of cancer cell lines propagated in vitro for several decades.
Fig. 3.
TPA activates DNA-damage signaling and response pathways. (A) Western blot analysis of DNA-damage and repair checkpoints in HRBEC and breast cancer cell lines. Each set of four lanes represents untreated control (C), 10 μM, or 1nM TPA and a positive control treated with 100 μM H2O2 for DNA damage induction. Increased expression of DNA-PK, MRE11 and RAD50 proteins, and phosphorylation of ATM is comparable in TPA or H2O2-treated lysates of HRBEC and cancer cell lines. GAPDH was used as a loading control. (B). Recruitment of repair proteins to DNA-damage sites within HRBEC lines treated with 100nM TPA for 7 days and visualized by FITC immunofluorescence with anti-p53pSer15 (top panel) or anti-H2AXpSer139 (bottom panel). Nuclei counterstained with PI. In each grouping of four images, left panels show FITC alone and right panels show merged FITC and PI signal. Bar in top right corner = 50 µm. Note induction of activated proteins (green or yellow nuclei) in all HRBEC lines. Boxed insert shows comparable levels of p53Ser15 phosphorylation (ATM and DNA-PK binding site) in the presence of TPA or by OHT in HRBEC (PA115) and T47D protein lysates. (C) Western blot analysis of DNA repair proteins in the DNA-bound fraction of TPA-treated cell lysates. Each set of four lanes represents untreated control (C), 10 μM, 100nM or 1nM TPA-treated HRBECs and breast cancer cell lines. Note increase in DNA-bound MLH1 and PCNA proteins at one or more TPA concentration in all cell lines. GAPDH was used as a loading control.
Reflecting the consequences of the presence of DSBs within the DNA of TPA-treated HRBECs, foci of activated p53 and of γ-H2AX were observed by immunofluorescence (Figure 3B). The induction of p53pSer15 by TPA or by the known DNA-damaging drug—OHT, was comparable as shown by western blot analysis. Representing shifts in upstream molecular signaling leading to the nuclear foci of p53pSer15 and γ-H2AX pSer139, we observed the induction of MLH1—a DNA mismatch repair protein, as well as PCNA—a DNA processivity factor, within the DNA-bound fraction of protein lysates of HRBECs and cancer cell lines treated with increasing concentrations of TPA (Figure 3C). Altogether, our findings consistently implicate activation of DNA damage response signaling, and consequently the involvement of the mismatch repair pathway in the cascade of molecular events following TPA exposure of non-malignant HRBECs.
Cells with DNA damage re-enter the cell cycle upon TPA removal
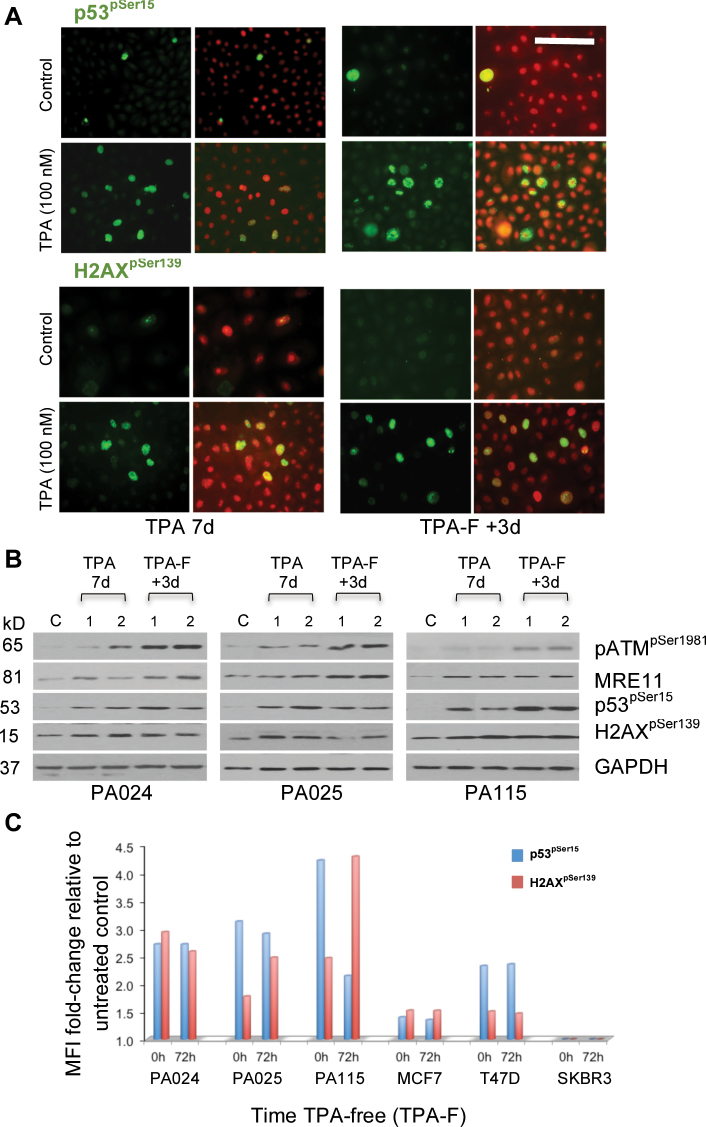
Finally, a TPA-free (designated TPA-F) recovery period was allowed for spontaneous reversal of functional perturbations detected in HRBECs:
A. TPA-induced DNA damage persists: Immunofluorescence staining of HRBECs maintained TPA-free for 72h post-TPA treatment continued to display increased levels of p53pSer15 compared with untreated controls suggesting that unrepaired DNA damage persisted in these cells (Figure 4A). Confirming this observation, western blot analysis of protein lysates of such HRBECs demonstrated sustained overexpression of MRE11, and phosphorylation of ATM at Ser198, p53 at Ser15 and γ-H2AX at Ser139 (Figure 4B). Evidence of the activated status of p53 and γ-H2AX proteins in TPA-F cultures was also obtained by FACS-based quantitation. Expression levels of p53pSer15 and γ-H2AXpSer139 after a 72h TPA-free period were generally similar to those maintained within 7 days TPA-treated cells confirming that DNA damage from prior TPA exposure was retained by most if not all exposed cells. A range of MFI in six test cell lines showed quantitative variation in DNA damage signaling proteins induced by TPA (Figure 4C).
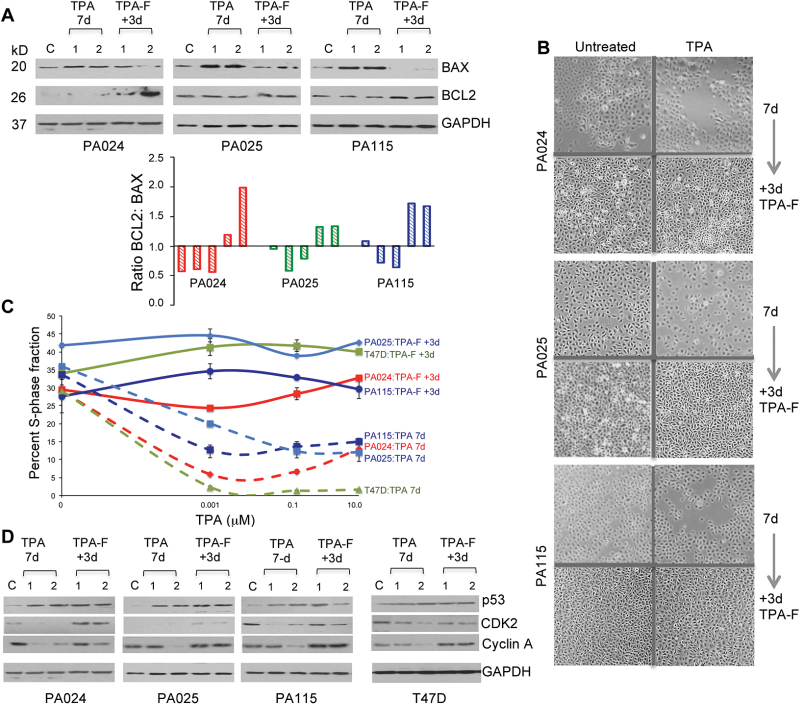
B. TPA-damaged cells are sustained: The un-induced or baseline apoptotic fraction within untreated, TPA-treated (100nM) and post-TPA treated HRBEC cultures was found to be closely similar (Supplementary Figure 3 is available at Carcinogenesis Online). Since increased accumulation of Annexin V-positive cells was not observed during the post-TPA recovery period, our data suggest that TPA-damaged cells that were arrested in G1 were not targeted for elimination by apoptosis. Quantitation of apoptosis associated proteins demonstrated that while TPA-treated HRBECs displayed increased levels of the proapoptotic protein—BAX, its expression in 3 days TPA-F cells was reduced to the level of untreated control cultures, or lower. Simultaneously however, expression of the antiapoptotic protein—BCL2 was visibly elevated in TPA-F cultures (Figure 5A). Consequently, the BCL2:BAX ratio shifted to favor antiapoptotic signaling in post-TPA exposed HRBECs harboring DNA damage.
C. TPA-induced growth arrest is reversible: Despite TPA-induced DNA damage and growth arrest, the intact cellular morphology displayed by HRBEC cultures indicated that they were viable. As our data demonstrate, TPA-F HRBEC cultures re-entered the cell cycle within 3 days of TPA removal from the growth medium, and attained confluence through robust proliferation (Figure 5B). Normally, the G1/S checkpoint prevents the initiation of DNA replication until any DNA damage incurred is repaired. However, cell cycle measurements for BrdU-labeled TPA-F HRBECs showed a rapid and consistent induction of S-phase at all doses of prior TPA treatment. The highest average percent S-phase increase occurred in PA025—from 2.3±0.5 to 46.3±1.8. Comparable increases were observed for other test cell lines: PA024—from 5.9±0.2 to 24.3±1.2, PA115—from 12.6±0.2 to 34.6±1.2 and T47D—from 20±1 to 44.5±1.4 (Figure 5C). Quantitation of cell cycle proteins within TPA-F lysates of all test cell lines demonstrated a reversal of TPA-induced suppression of CDK2 and Cyclin A providing a plausible mechanism for re-entry into the cell cycle despite elevated p53—an indicator of persistent DNA damage (Figure 5D). Taken together, these results confirm that TPA-damaged HRBEC cultures remain viable, resist cell death and are proliferation-competent.
Fig. 4.
TPA-mediated DNA damage is persistent. Damage from 7 days exposure to TPA persists in HRBEC lines maintained TPA-free (TPA-F) for 3 days. (A) Microscopic evaluation of representative immunostained TPA-F HRBECs showed sustained phosphorylation of p53 and H2AX proteins above control levels in paraformaldehyde-fixed cells. Nuclei counterstained with PI. In each grouping of four images, left panels show FITC alone and right panels show merged FITC and PI fluorescence. Scale bar is 50 µm. Note continued expression of DNA damage related proteins (green or yellow nuclei). (B) Quantitation of DNA damage and repair proteins in TPA-treated (10 μM—noted as 1 and 100 nM—noted as 2) and TPA-F HRBECs by western blot analysis illustrates sustained activation of ATM, p53, H2AX and CHK1 by phosphorylation in all TPA-F samples. Increased MRE11 expression levels were also observed. GAPDH served as a loading control. (C) Quantitation of p53pSer15 and H2AXpSer139 expression in immunostained HRBECs and breast cancer cell lines by FACS. While differences in the MFI values between untreated control and 7 days TPA (100nM) treatment were significant for all cell lines (two-tailed P value < 0.005), with the exception of SKBR3, the minor shift in the values derived from TPA-treated cells without recovery and with 72h TPA-free recovery, was not statistically significant.
Fig. 5.
Reversal of cell cycle arrest is facilitated by TPA-induced apoptosis evasion. (A) Top panel: Western blot quantitation of apoptosis associated proteins—BAX and BCL2 in TPA-treated HRBECs (10 μM—noted as 1 and 100 nM—noted as 2). The proapoptotic protein—BAX is increased by TPA treatment whereas expression of the antiapoptotic protein—BCL2 is relatively unchanged. TPA removal from the cultures rapidly shifts the ratio of these proteins. Bottom panel: Ratio of BCL2:BAX proteins (antiapoptotic:proapoptotic) derived from the densitometric scan of the blot shown above. GAPDH expression was used for normalization. (B) Increased cell density in micrographs of cultures 72h post-TPA treatment illustrates growth resumption in HRBECs (Brightfield images—×4 objective). (C) Cell cycle progression measured by FACS in BrdU pulse-labeled TPA-F cultures at 0 and 72h post-treatment with TPA dose ranging from 0.001 to 10 μM. There is a rapid increase in S-phase in 3 days TPA-F (solid lines) compared with paired cultures at the end of 7 days exposure to TPA (dashed lines). Differences between TPA-treated cells without recovery and with 3 days recovery are significant at all doses for all cell lines (two-tailed P value < 0.05). (D) Western blot analysis of 72h TPA-F lysates (10 μM—noted as 1 and 100 nM—noted as 2) shows induction of cell cycle proteins accompanying the increase in culture density and S-phase fraction relative to 7 days TPA treatment.
Discussion
The biological effects of TPA observed here in non-malignant breast epithelial cells present a complex scenario compared with those of the xenoestrogen—BPA. Representing an overall agonistic effect at human exposure levels, BPA closely mimics natural estrogen in terms of the induction of ERα, apoptosis evasion, p53 suppression, induction of cell cycle proteins and increased growth rate in non-malignant human breast epithelium (12,13). In sharp contrast, TPA exposure induces an unexpected agonistic–antagonistic duality of responses. Similar to BPA and E2, TPA increases ERα levels and induces apoptosis evasion but unlike the former, it activates cell cycle arrest associated with evidence of DNA damage. We demonstrate further that despite persistent TPA-mediated DNA damage, such cells are not targeted for elimination. Instead they resume proliferation when returned to TPA-free conditions, suggesting that the net balance of opposing effects of TPA may lead to proliferation of cells harboring DNA damage.
The dichotomy of TPA-induced perturbations demonstrates that detecting the adverse effects of chemicals requires careful consideration of assay endpoints and experimental models. Relying entirely upon cell proliferation based tests, TPA might be classified as a safe chemical without recognition of its potential to induce DNA damage. Such critical issues remain relatively unaddressed despite the strategic regulatory intent to augment long-term animal studies with rapid in vitro tests (16). The integration of immortalized cancer cell lines into the high-throughput screening pipeline (17) has undoubtedly expedited chemical testing over a wide dose range. However, to evaluate underlying cause and effect related to carcinogenesis requires the use of non-cancerous cells as an experimental target, isolated from tissues of chemical vulnerability. Since the effects of estrogenic chemicals in estrogen-sensitive tissues such as the breast, is largely mediated by interaction with ER, therefore including ER-positive test models is critical for XE screening. As demonstrated, non-malignant HRBECs employed here uniquely maintain ER expression. Moreover, interindividual variability in ERα:ERβ ratios resulting from genetic, dietary or other factors, is detectable as well (12,13). Incorporating assays with donor-derived non-malignant breast cell models into the design of future breast carcinogen identification approaches could add an important dimension in assessing the biologically relevant range of concentrations, and thereby lend greater accuracy to chemical screening data.
The duality of TPA action at the molecular and functional level within HRBECs is conceptually intriguing. On one hand, by the induction of ERα, it stimulates cells to enter the cell cycle, as evident by downstream phosphorylation of pRB, and increased G1-cyclins, Cyclin D/CDK4. On the other hand, due to the induction of genotoxic stress and DNA damage with consequent p53 protein expression, TPA treatment inhibits HRBECs from exiting G1 and entering S-phase, as evident by a decline in the expression of the S-cyclins, Cyclin A/CDK2, known to be negatively regulated by p53 (18). Another aspect of the contrasting effects of TPA on HRBECs is the observation that despite p53 activation and DNA damage-induced cell cycle arrest, such cells are not targeted for apoptotic elimination, suggesting that other perturbations play a role in overriding normal cell regulation. These data are analogous to the repression of p53-mediated apoptotic response to cancer therapy by the recruitment of ER to sites within proapoptotic p53 target genes resulting in the lack of p53 regulation of such genes (19,20). Similarly, TPA-treated HRBECs when returned to TPA-free conditions displayed BCL2 induction, thereby sustaining cell survival, and enabling S-phase entry of cells with restored Cyclin A/CDK2 levels. Moreover, since BCL2 overexpression interferes with DNA replication dynamics (21), it might contribute to further genetic instability within cells harboring unrepaired DNA damage. Exposure of ER-positive breast cancer cells to E2 leads to DNA DSBs evident as γ-H2AX foci in Cyclin A expressing cells mainly during progression through S-phase (22). However, DSB foci induced by TPA exposure occurred in cell populations with decreased Cyclin A/CDK2 expression, arrested in G1 due to the onset of an earlier checkpoint response characteristic of the activation of the p53 cascade by DNA-damaging agents. Our data derived from TPA-treated HRBECs are consistent with the role for the mismatch repair proteins—MLH1 and PMS2 as sensors of structural DNA aberrations and as signal transducers for p53 activation following chemical alterations of the helix (23). The striking overexpression of p21WAF1 in TPA-treated HRBECs also sheds light on the persistence of damaged cells that evade apoptotic cell death. Aside from its role as a negative regulator of cell proliferation, p21 is well known for apoptosis-inhibiting (procancer) signaling particularly in cells with wild-type p53 (24) such as the HRBECs employed here, which display concordant induction of p53 and its downstream effector—p21 (13). For example, p21 binding to Cyclin/CDK complexes is known to induce cell cycle arrest, which in turn prevents cell death from catastrophic nuclear division (25). Oncogenic properties attributed to this protein through its interaction with proliferating cell nuclear antigen, PCNA—a DNA replication and repair protein (26,27) include inhibition of mismatch repair activity (28), as well as long patch base excision repair (29). Additionally, induction of p21 is associated with the inhibition of apoptosis-inducing signals of natural peptides (30–32), as well as cancer drugs (33). It is not surprising therefore that overexpression of p21 is common in many human cancers, and generally correlated with poor prognosis (34,35). Consequently, apoptosis evasion is a key hallmark of cancer (14), including breast cancer. In the light of the cascade of above-mentioned deleterious effects involving intersecting pathways within cells, it is evident that limited or inappropriately selected in vitro tests could underestimate the true consequences of chemical exposure. Instead of adopting a single functional assay, or gene expression endpoint based on a priori assumptions, a broad mechanistic understanding of the cellular mode of action particularly for high-volume chemicals must therefore be a priority.
PET is a polymer synthesized with few additives, and used most extensively for bottling water. Migration tests of the chemical components of PET into bottled water are conducted routinely to meet regulatory requirements, and test reports suggest levels low enough to pose no major concerns (9). Nevertheless, PET packaging is considered a possible source of leached estrogenic activity in some test assays (36–38). Expectedly, such activity is not detected in the E-screen bioassay (9), which specifically measures growth induction, since our data regarding the PET monomer—TPA, demonstrates induction of growth arrest but accompanied by other estrogen mimicking phenotypes described above. Studies of the genotoxicity of PET bottled water have included plant species, such as Allium cepa (39) and Tradescantia (40) making extrapolation to human cells difficult. The human donor derived non-malignant cellular models used here are well suited to examine the effects of potential endocrine disruptors such as TPA because they express levels of ERα and β characteristic of normal breast epithelium. In contrast, cancer cell lines deviate significantly from the norm with regard to ER expression due to microenvironmental selection pressures during malignant progression. Our pairwise analysis of non-malignant cells with and without TPA at a range of biologically and environmentally relevant concentrations of exposure reproducibly show an emergence of characteristic hallmarks of cancer across independent samples. In the light of our findings, it is important to consider that cellular perturbations evident as induction of DNA damage, cell cycle arrest and subsequent escape from programmed cell death during in vitro TPA exposure likely reflect in vivo potency. Thus, it is critical to ascertain the safety of long-term, cumulative exposure to this chemical, particularly in young children due to their lower body weight and susceptibility during maturation.
Supplementary material
Supplementary Western Blot Data and Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
The Cancer League, The Clarence E Heller Charitable Foundation and the California Breast Cancer Research Program (17UB-8702).
Supplementary Material
Acknowledgements
The authors appreciate the review of RPFNA cytopathology by Dr Ian Jaffee, Department of Pathology, California Pacific Medical Center, San Francisco. M.G.L.-T. designed and performed laboratory experiments. D.H.M. performed statistical analysis. W.H.G. collaborated in study design and acquisition of clinical resources. S.H.D. developed and directed overall research. All authors contributed toward data analysis and manuscript preparation and approved the final version. This study was presented in part as a poster at the San Antonio Breast Cancer Symposium, December 2013.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- BPA
bisphenol A
- BrdU
bromodeoxyuridine
- DSB
double-strand break
- E2
17β-estradiol
- ER
estrogen receptor
- FACS
fluorescence-activated cell sorting
- HRBEC
high-risk donor breast epithelial cell
- OHT
hydroxytamoxifen
- PET
polyethylene terephthalate
- RPFNA
random periareolar fine needle aspirate
- TPA
terephthalic acid.
References
- 1. Köpnick H., et al. (2000). Polyesters. In Ullmann’s Encyclopedia of Industrial Chemistry. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, pp. 21–227. [Google Scholar]
- 2. De Meulenaer B., et al. (2004). Packaging and other food contact material residues. In Nollet L.M.L. (ed.) Handbook of Food Analysis. Vol. 2 Marcel Dekker, New York, NY, pp. 1297–1330. [Google Scholar]
- 3. Begley T., et al. (2005). Evaluation of migration models that might be used in support of regulations for food-contact plastics. Food Addit. Contam., 22, 73–90. [DOI] [PubMed] [Google Scholar]
- 4. Monarca S., et al. (1994). Studies of migration of potentially genotoxic compounds into water stored in pet bottles. Food Chem. Toxicol., 32, 783–788. [DOI] [PubMed] [Google Scholar]
- 5. Cui L., et al. (2006). Modification of N-Methyl-N-Nitrosourea initiated bladder carcinogenesis in Wistar rats by terephthalic acid. Toxicol. Appl. Pharmacol., 210, 24–31. [DOI] [PubMed] [Google Scholar]
- 6. Shi Y., et al. (2006). Elevated prostaglandin E2 level via cPLA2–COX-2–mPGES-1 pathway involved in bladder carcinogenesis induced by terephthalic acid-calculi in Wistar rats. Prostaglandins. Leukot. Essent. Fatty Acids, 74, 309–315. [DOI] [PubMed] [Google Scholar]
- 7. Muncke J. (2009). Exposure to endocrine disrupting compounds via the food chain: is packaging a relevant source? Sci. Total Environ., 407, 4549–4559. [DOI] [PubMed] [Google Scholar]
- 8. Yang C.Z., et al. (2011). Most plastic products release estrogenic chemicals: a potential health problem that can be solved. Environ. Health Perspect., 119, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bach C., et al. (2012). Chemical compounds and toxicological assessments of drinking water stored in polyethylene terephthalate (PET) bottles: a source of controversy reviewed. Water Res., 46, 571–583. [DOI] [PubMed] [Google Scholar]
- 10. Osimitz T.G., et al. (2012). Lack of androgenicity and estrogenicity of the three monomers used in Eastman’s Tritan™ copolyesters. Food Chem. Toxicol., 50, 2196–2205. [DOI] [PubMed] [Google Scholar]
- 11. Dairkee S.H., et al. (2008). Bisphenol A induces a profile of tumor aggressiveness in high-risk cells from breast cancer patients. Cancer Res., 68, 2076–2080. [DOI] [PubMed] [Google Scholar]
- 12. Goodson W.H.III et al. (2011). Activation of the mTOR pathway by low levels of xenoestrogens in breast epithelial cells from high-risk women. Carcinogenesis, 32, 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dairkee S.H., et al. (2013). Bisphenol-A-induced inactivation of the p53 axis underlying deregulation of proliferation kinetics, and cell death in non-malignant human breast epithelial cells. Carcinogenesis, 34, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanahan D., et al. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 15. Hoshi A., et al. (1968). Distribution of terephthalic acid in tissues. Chem. Pharm. Bull. (Tokyo)., 16, 131–135. [DOI] [PubMed] [Google Scholar]
- 16. National Research Council. (2007). Toxicity Testing in the 21st Century: A Vision and a Strategy. Committee on Toxicity Testing and Assessment of Environmental Agents, National Research Council/National Academies Press, Washington, DC. [Google Scholar]
- 17. Tice R.R., et al. (2013). Improving the human hazard characterization of chemicals: a Tox21 update. Environ. Health Perspect., 121, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamamoto M., et al. (1994). Effect of tumor suppressors on cell cycle-regulatory genes: RB suppresses p34cdc2 expression and normal p53 suppresses cyclin A expression. Exp. Cell Res., 210, 94–101. [DOI] [PubMed] [Google Scholar]
- 19. Sayeed A., et al. (2007). Estrogen receptor alpha inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res., 67, 7746–7755. [DOI] [PubMed] [Google Scholar]
- 20. Bailey S.T., et al. (2012). Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc. Natl Acad. Sci. USA, 109, 18060–18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie M., et al. (2014). Bcl2 induces DNA replication stress by inhibiting ribonucleotide reductase. Cancer Res., 74, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williamson L.M., et al. (2011). Estrogen receptor α-mediated transcription induces cell cycle-dependent DNA double-strand breaks. Carcinogenesis, 32, 279–285. [DOI] [PubMed] [Google Scholar]
- 23. Duckett D.R., et al. (1999). hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl Acad. Sci. USA, 96, 12384–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kokontis J.M., et al. (2001). A transcriptional activation function of p53 is dispensable for and inhibitory of its apoptotic function. Oncogene, 20, 659–668. [DOI] [PubMed] [Google Scholar]
- 25. Vogelstein B., et al. (2001). Achilles’ heel of cancer? Nature, 412, 865–866. [DOI] [PubMed] [Google Scholar]
- 26. Xiong Y., et al. (1992). D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell, 71, 505–514. [DOI] [PubMed] [Google Scholar]
- 27. Zhang H., et al. (1993). Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol. Biol. Cell, 4, 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Umar A., et al. (1996). Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell, 87, 65–73. [DOI] [PubMed] [Google Scholar]
- 29. Tom S., et al. (2001). Regulatory roles of p21 and apurinic/apyrimidinic endonuclease 1 in base excision repair. J. Biol. Chem., 276, 48781–48789. [DOI] [PubMed] [Google Scholar]
- 30. Yan Q., et al. (1998). Transforming growth factor-beta1 induces apoptotic cell death in cultured retinal endothelial cells but not pericytes: association with decreased expression of p21waf1/cip1. J. Cell. Biochem., 70, 70–83. [PubMed] [Google Scholar]
- 31. Zhu J., et al. (1998). The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res., 58, 5061–5065. [PubMed] [Google Scholar]
- 32. Hobeika A.C, et al. (1999). IFN-gamma induction of p21(WAF1) is required for cell cycle inhibition and suppression of apoptosis. J. Interferon Cytokine Res., 19, 1351–1361. [DOI] [PubMed] [Google Scholar]
- 33. Li W, et al. (1999). Overexpression of p21(waf1) decreases G2-M arrest and apoptosis induced by paclitaxel in human sarcoma cells lacking both p53 and functional Rb protein. Mol. Pharmacol., 55, 1088–1093. [DOI] [PubMed] [Google Scholar]
- 34. Abbas T, et al. (2009). p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer, 9, 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Musgrove E.A., et al. (2011). Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer, 11, 558–572. [DOI] [PubMed] [Google Scholar]
- 36. Pinto B., et al. (2009). Screening of estrogen-like activity of mineral water stored in PET bottles. Int. J. Hyg. Environ. Health, 212, 228–232. [DOI] [PubMed] [Google Scholar]
- 37. Sax L. (2010). Polyethylene terephthalate may yield endocrine disruptors. Environ. Health Perspect., 118, 445–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wagner M., et al. (2011). Endocrine disruptors in bottled mineral water: estrogenic activity in the E-Screen. J. Steroid Biochem. Mol. Biol., 127, 128–135. [DOI] [PubMed] [Google Scholar]
- 39. Evandri M.G., et al. (2000). Toxicological evaluation of commercial mineral water bottled in polyethylene terephthalate: a cytogenetic approach with Allium cepa. Food Addit. Contam., 17, 1037–1045. [DOI] [PubMed] [Google Scholar]
- 40. Biscardi D., et al. (2003). Evaluation of the migration of mutagens/carcinogens from PET bottles into mineral water by Tradescantia/micronuclei test, Comet assay on leukocytes and GC/MS. Sci. Total Environ., 302, 101–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.