SUMMARY
Engineering clinically relevant cells in vitro holds promise for regenerative medicine, but most protocols fail to faithfully recapitulate target cell properties. To address this, we developed CellNet, a network biology platform that determines whether engineered cells are equivalent to their target tissues, diagnoses aberrant gene regulatory networks, and prioritizes candidate transcriptional regulators to enhance engineered conversions. Using CellNet, we improved B cell to macrophage conversion, transcriptionally and functionally, by knocking down predicted B cell regulators. Analyzing conversion of fibroblasts to induced hepatocytes (iHeps), CellNet revealed an unexpected intestinal program regulated by the master regulator Cdx2. We observed long-term functional engraftment of mouse colon by iHeps, thereby establishing their broader potential as endoderm progenitors and demonstrating direct conversion of fibroblasts into intestinal epithelium. Our studies illustrate how CellNet can be employed to improve direct conversion and to uncover unappreciated properties of engineered cells.
INTRODUCTION
The in vitro manufacture of clinically relevant cells offers a potential strategy for regenerative therapy and permits disease modeling, toxicology testing and drug discovery. Current approaches aim to engineer cell identity by means of directed differentiation from a pluripotent state or by transcription factor-driven conversion between differentiated states (Morris and Daley, 2013; Vierbuchen and Wernig, 2011). Directed differentiation typically comprises multiple steps, is time-consuming and inefficient, and commonly yields immature cells corresponding to embryonic counterparts rather than mature adult cells (Cohen and Melton, 2011). By comparison, direct conversion is relatively straightforward and rapid but there is evidence for incomplete conversion, especially between divergent cell types (Morris and Daley, 2013; Willenbring, 2011).
Many examples of direct conversion between differentiated states have been reported in mouse and human, for example: from fibroblasts to cardiomyocytes, hepatocytes, and neurons (Huang et al., 2011; Ieda et al., 2010; Sekiya and Suzuki, 2011; Son et al., 2011; Vierbuchen et al., 2010). More recently, several groups have described direct conversion to progenitor states, including hematopoietic, neuronal and hepatic progenitors (Lujan et al., 2012; Pereira et al., 2013; Yu et al., 2013). These engineering strategies predominantly employ transcription factor overexpression as a means to drive fate conversion.
Current conversion strategies are often unable to fully specify a defined cell fate. For example, hepatic gene expression is not fully extinguished in neural cells derived from hepatocytes, and macrophages derived from fibroblasts harbor the originating cell signature and are prone to de-differentiation (Feng et al., 2008; Marro et al., 2011). Furthermore, conversion of fibroblasts to cardiomyocytes yields cells that do not fully recapitulate the profile of neonatal cardiomyocytes (Ieda et al., 2010). These observations are concerning since the extent to which an in vitro engineered cell population resembles its in vivo correlate transcriptionally and functionally is seldom assessed in a comprehensive or standardized manner. Measuring functional engraftment via transplantation into animal models lacks rigorous quantitation and the transcriptional similarity of engineered cell populations is commonly assessed by expression-profiling followed by simple hierarchical clustering analysis. Such global analyses do not provide a quantitative means for assessing deficiencies of engineered cells, nor do they provide a systematic approach to prioritize interventions to improve fate specification.
To address this, we developed a computational platform, CellNet, which reconstructs gene regulatory networks (GRNs) using publically available gene expression data for a range of cell types and tissues, and then classifies engineered cells according to establishment of GRNs for particular target cells, providing a precise metric of cell similarity. CellNet also identifies regulatory nodes at which engineered cells are distinct from target cells, and provides a ranked list of transcription factors whose manipulation is predicted to bring the engineered cell closer to the target. In an accompanying study, we have analyzed expression data for over 200 derived cell populations from 56 published reports and found that cells generated through directed differentiation more closely resemble their in vivo correlates compared to cells engineered via direct conversion, mainly due to failure of the converted cells to extinguish the expression programs of the starting cell type. Unexpectedly, we discovered that the establishment of GRNs associated with alternate fate was common to nearly all engineering strategies (Cahan et al.).
Here we apply CellNet to two distinct cell fate engineering paradigms: conversion of B cells to macrophages, and fibroblasts to hepatocyte-like cells (iHeps). CellNet revealed that neither strategy generated fully-converted cells; B cell identity was not extinguished in induced macrophages, whereas a progenitor state was transiently and partially established. Engineering the conversion to macrophages by knocking down CellNet-prioritized candidates improved target cell fate and function. iHeps, unlike primary hepatocytes, demonstrate impaired hepatocyte function, are immortalized and exhibit progenitor marker expression which is extinguished following transplantation (Sekiya and Suzuki, 2011). In agreement with this, CellNet revealed that iHeps manifest minimal liver identity. Surprisingly, CellNet unveiled considerable hindgut identity harbored by iHeps, regulated by Cdx2. We were able to demonstrate their long-term functional colon engraftment, indicating that iHeps in fact represent an endoderm progenitor rather than a differentiated cell type as previously thought. CellNet is thus a potent tool for reassessing and refining established conversion protocols.
RESULTS
Application of the CellNet network biology platform to assess direct conversion
Gene regulatory networks (GRNs) govern the steady-state expression program of a particular cell type and thus act as major molecular determinants of cell identity (Davidson and Erwin, 2006). Measuring the establishment of cell and tissue-specific GRNs in engineered populations serves as both a robust metric of cellular identity and as a tool to detect the establishment of alternate GRNs. We designed CellNet to classify engineered cell populations by their similarity to target cells and tissues, to assess the extent to which target GRNs are established, and to score transcriptional regulators according to their likelihood of improving the engineered population (Figure 1A). The CellNet platform is fully detailed in our accompanying paper and is publically available (Cahan et al., http://cellnet.hms.harvard.edu).
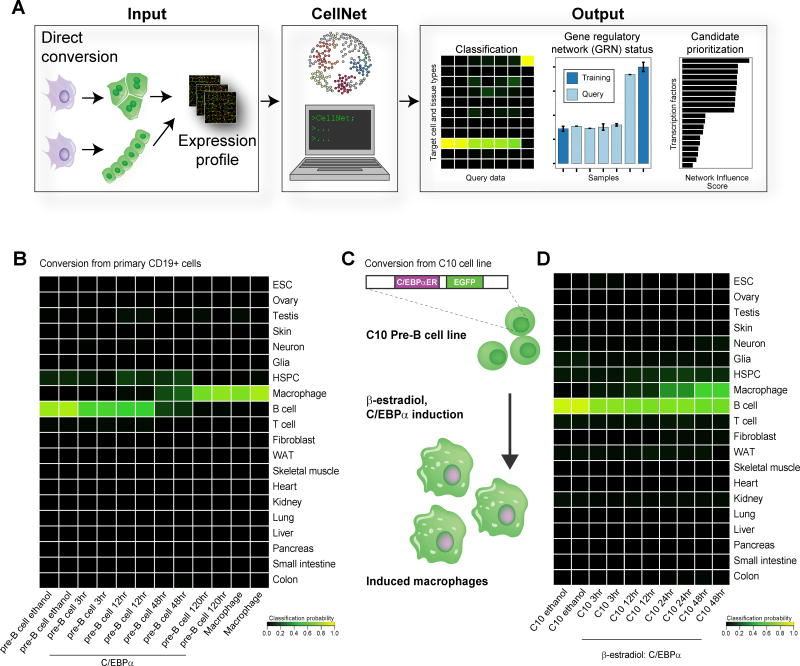
Figure 1. Application of CellNet classification to the direct conversion of B cells to macrophages.
(A) Adapted from Cahan et al. We designed CellNet to query gene expression profiles of engineered cell populations to classify input samples by their similarity to target cells and tissues, in order to assess the extent to which cell type and tissue GRNs are established and to score transcriptional regulators according to their likelihood of improving the engineered population. GRNs were used to construct a cell type classifier which was initially used to assess C/EBPα-mediated conversion of B cells to macrophages. (B) Each row in the classifier represents one cell or tissue type, and each column represents one array. Higher classification scores indicate a higher probability that a query sample expresses the target GRN genes at a level indistinguishable from the same cell or tissue in the training data; classification heatmap of primary B cell to macrophage conversion showing that converted cells classify exclusively as macrophages (Di Tullio et al., 2011). (C) C10 estradiol-inducible B cell to macrophage conversion (Bussmann et al., 2009). (D) Cell and tissue classification heatmap showing that conversion is not complete with induced macrophages maintaining B cell classification and only partially classifying as macrophages. See also Figure S1.
B cell to macrophage conversion is a major paradigm for understanding principles of direct cell fate conversion (Bussmann et al., 2009). Application of CellNet revealed that enforced expression of the transcription factor C/EBPα converts freshly harvested B cells into macrophages, following a time-dependent extinction of B cell identity and acquisition of a strong, exclusive macrophage classification score (Di Tullio et al., 2011; Mean scores, B cell: 0.012, Macrophage: 0.73; Figure 1B). In contrast to this, conversion of a cultured pre-B cell line engineered with estradiol-inducible C/EBPα (C10 cells, Bussmann et al., 2009; Figure 1C) fails to fully extinguish B cell identity, yielding cells that only partially classify as macrophages 48 hours post-conversion (Mean scores: B cell: 0.65, Macrophage: 0.54; Figure 1D). Interestingly, no significant changes in DNA methylation have been observed during this conversion, which may indicate that the C10 cell line harbors conversion barriers unique to in vitro cultured cell lines (Rodríguez-Ubreva et al., 2012). Despite this limitation, the inducible C10 cell line represents a robust system to interrogate the molecular mechanisms of direct conversion, and a valuable model for applying CellNet to enhance cell fate conversion.
To enable identification of GRNs that fail to extinguish or are incompletely or aberrantly established in engineered cells, we devised a metric of GRN status. We assessed the expression of each gene in a GRN relative to its expected value, and then weighted each gene by its importance to the network and to the associated cell and tissue classifier (Figure 2A). To prioritize transcriptional regulator interventions that might improve engineering strategies, we computed a network influence score that integrates the expression level of the regulator in the target cell or tissue, the extent of dysregulation of the regulator and its predicted targets in the query sample, and the number of predicted targets (Figure 2B). We applied these functions of CellNet to the estradiol-inducible C10 B cell to macrophage conversion. Assessment of GRN status revealed that the macrophage GRN was gradually but not completely established over 48 hours, whereas the B cell GRN remained intact (Figure 2C). These data were in contrast to the almost complete conversion of native B cells (Figure 1B, S1A). Interestingly, CellNet also exposed the partial and transient establishment of a hematopoietic stem and progenitor cell (HSPC) GRN during conversion of C10 cells (Figure 2C), and similarly in the conversion of native B cells (Figure 1B; Di Tullio et al., 2011), raising the provocative possibility that expression of C/EBPα induces a transient wave of progenitor GRN reactivation from which macrophages ultimately emerge.
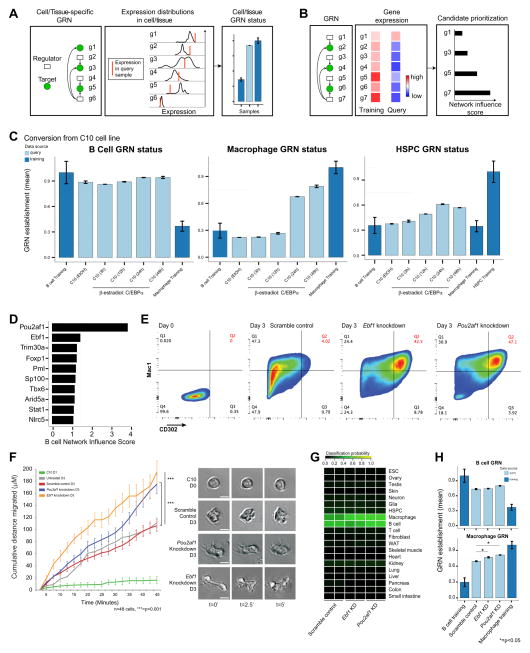
Figure 2. Application of the CellNet GRN status metric and candidate prioritization functions to B cell to macrophage conversion.
(A) Adapted from Cahan et al. To identify cell and tissue specific GRN establishment, we devised a precise metric of GRN status. We defined this metric as the closeness of the expression of each gene in a GRN to its expected value, and then weighted each gene (g1–g6) by its importance to the network and its importance in the associated classifier. (B) To prioritize transcriptional regulator interventions, we devised a Network Influence Score integrating the target expression level of the regulator, the extent of dysregulation of the regulator and its predicted targets in the query sample, and the number of predicted targets. We applied these functions of CellNet to the estradiol-inducible B cell to macrophage conversion: (C) The B cell GRN is not extinguished and the macrophage GRN not fully established following conversion. The HPSC GRN is transiently and partially established during conversion. Data are represented as mean +/− SD. (D) CellNet prioritization of B cell transcriptional regulators whose expression is maintained in induced macrophages. (E) C/EBPα induced direct conversion of C10 B cells to macrophages following shRNA knockdown of Ebf1 and Pou2af1: flow cytometry plots of Mac1 and CD302 expression three days after conversion. The Mac1BrightCD302+ population is expanded following Ebf1 and Pou2af1 knockdown. (F) Cumulative distance migrated by induced macrophages increases following knockdowns: still images of cell morphology from timelapse imaging. Scale bars, 10μM. (G) Cell and tissue classification heatmap following knockdowns and (H) corresponding B cell and macrophage GRN establishment. *=p<0.05, t-Test. Macrophage identity is fortified following knockdown. Data are represented as mean +/− SEM. See also Figure S1, S2 and Table 1 and 2.
Engineering cell fate
In an attempt to improve conversion of C10 induced macrophages, we extended the duration of C/EBPα expression to four days, but neither B cell nor macrophage statuses improved, suggesting that the partial conversion was not due to insufficient duration of C/EBPα expression (Figure S1B). B cell GRN persistence was striking in the converted cells, thus we consulted CellNet to score transcriptional regulators of B cell identity. CellNet prioritized Pou2af1 (Pou2 associating factor 1) and Ebf1 (Early B cell factor 1) as transcriptional regulators associated with B cell identity that remained expressed at inappropriately high levels in the converted cells (Figure 2D). Ebf1 promotes B cell fate and blocks alternate fates (Nechanitzky et al., 2013) whereas Pou2af1 co-activates Oct1 and Oct2 on B cell specific promoters and is important for B cell activation and maturation (Lins et al., 2003).
In our experiments, conversion of the C10 B cell line was highly reproducible, with the majority of cells gaining Mac1 positivity and losing CD19 expression within 48 hours (Figure S1C). To facilitate investigation of CellNet-enhanced conversion, we identified CD302 as a cell surface antigen that was inadequately expressed on induced relative to native macrophages (Figure S1D). We then knocked down Ebf1 or Pou2af1 using lentivirally delivered shRNAs, resulting in an ~80% reduction in target gene expression (Figure S2A). Next, we initiated conversion with β-estradiol, and after 72 hours analyzed expression of Mac1 and CD302 by flow cytometry. Knockdown of either Ebf1 or Pou2af1 significantly expanded the Mac1brightCD302+ population relative to scramble shRNA treated cells (Figure 2E). We then tested the macrophage-associated functions of Ebf1 and Pou2af1 depleted cells. Cell migration, formation of lamellipodia and filopodia, and phagocytosis were all significantly enhanced relative to untreated and scramble-treated cells (Figure 2F; S2B,C).
Quantitative PCR analysis showed a decrease in expression of some B cell specific genes whereas others were paradoxically upregulated; however, there was a more consistent increase in macrophage specific gene expression (Figure S2D). We re-applied CellNet to the engineered cells, and found that knockdown of Ebf1 or Pou2af1 fortified their macrophage classification, corroborating their improved in vitro function (Figure 2G; Mean scores: Scramble: 0.39, Ebf1 knockdown: 0.52, Pou2af1 knockdown: 0.54). However, although macrophage GRN status significantly improved (Figure 2H: Ebf1 knockdown: p=0.029, Pou2af1 knockdown: p=0.011), detailed analysis revealed that the B cell GRN was not extinguished following knockdown of these factors, perhaps due to the existence of feedback loops reinforcing B cell identity. To explore this, we searched CellNet GRNs for regulatory loops in which Pou2af1 loss is predicted to increase B-cell factor expression. Indeed we found several such loops, including one connecting Pou2af1 downregulation to the up-regulation of the master B cell regulator Pax5 and Ebf1 itself through Runx2 and Runx3 (Figure S2D, E). These data suggest that residual expression of Ebf1 and Pou2af1 in the converted cells serves primarily to repress macrophage fate.
We explored the possibility that Ebf1 and Pou2af1 mutually repress myeloid-associated GRNs, as previously reported for Ebf1 (Nechanitzky et al., 2013). Examining targets directly linked to Ebf1/Pou2af1 in the overall GRN, as well as targets of transcription factors that Ebf1/Pou2af1 target themselves, we found that the set of macrophage genes up-regulated by B cell factor knockdown were significantly enriched in genes that CellNet predicted to be repressed by Pou2af1 or Ebf1 (p=<0.005 and 0.05, respectively, Table 1, 2; Figure S2F). Thus, the improved in vitro function of Ebf1/Pou2af1-depleted cells following C/EBPα-induced conversion of C10 cells is at least in part due to the repressive actions of these B cell regulators on macrophage gene expression programs.
B cell to macrophage conversion demonstrates the utility of CellNet to dissect cell identity and prioritize candidates to improve fate specification iteratively. Conversion of B cells to macrophages represents a conversion between closely related cells. To investigate a dramatic change in fate between more disparate germ layers, we focused on conversion of mesoderm-derived fibroblasts to endoderm-derived hepatocytes.
CellNet: induced hepatocytes fail to classify as liver
Mouse embryonic fibroblast (MEF) to hepatocyte-like cell conversion has been reported by two independent groups: Sekiya and Suzuki employed Hnf4α, with either Foxa1, 2 or 3, while Huang and colleagues utilized Gata4, Hnf1α, Foxa3 in concert with inactivation of p19(Arf) (Huang et al., 2011; Sekiya and Suzuki, 2011). In both reports, the resulting induced hepatocytes (iHeps) were capable of engrafting fumarylacetoacetate hydrolase-deficient (Fah−/−) mice, a tyrosinemia model of liver failure. However, unlike primary hepatocytes, iHeps demonstrated impaired hepatocyte function and were unable to fully repopulate Fah−/ − mice. Liver function remained abnormal, resulting in reduced survival rates. Curiously, iHeps were immortalized and expressed progenitor markers such as Alphafetoprotein (Afp) whose expression ceased following transplantation (Sekiya and Suzuki, 2011).
We reproduced the Sekiya and Suzuki MEF to iHep conversion (Figure 3A). Careful analysis of the derived cells showed that we attained comparable derivation efficiency, and that the cells expressed E-cadherin and albumin, stored glycogen, took up low-density lipoprotein (LDL), secreted albumin and produced urea (Figure S3A–F). As previously reported, we observed emergence of epithelial-like iHep colonies three weeks following transduction and culture on collagen-coated plates. Moreover, these cells were immortalized, in accordance with the previous report, and we have been able to continuously passage established iHep lines for over a year. These analyses confirm that our iHeps appear equivalent to those previously reported (Sekiya and Suzuki, 2011). Ex vivo cultured primary hepatocytes are known to de-differentiate (Strain, 1994). qPCR analysis of key liver markers in iHeps showed very weak hepatic gene expression relative to both freshly isolated primary and de-differentiated hepatocytes, and much higher levels of Afp and E-cadherin (Figure S3G). The failure of iHeps to reach expression levels of even de-differentiated primary hepatocytes suggests that their inadequacy is not due to culture conditions alone.
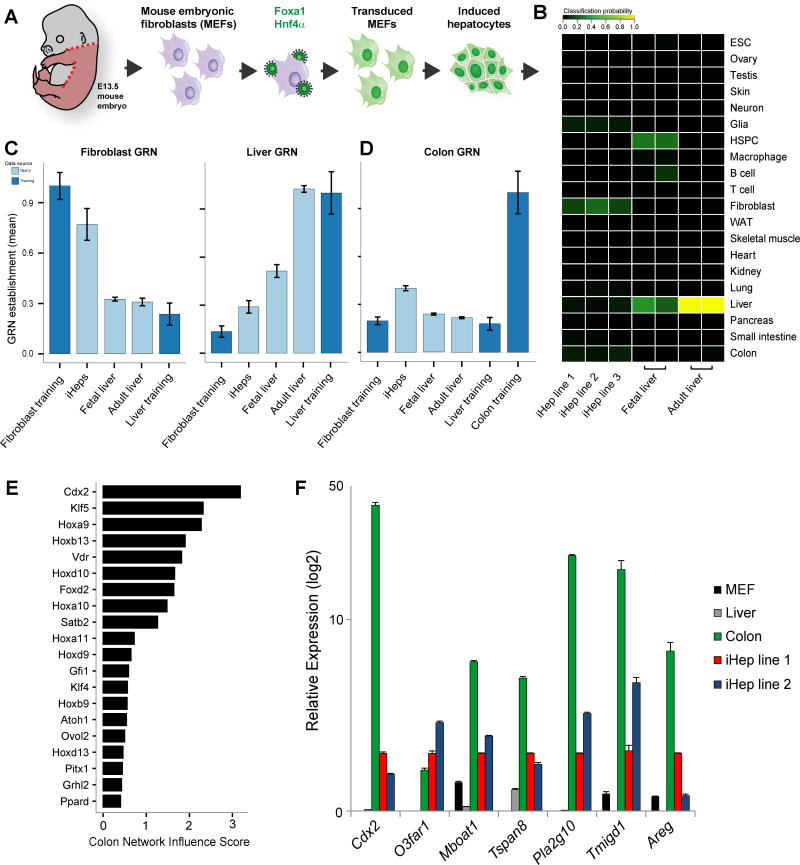
Figure 3. Application of CellNet to direct conversion of fibroblasts to hepatocytes.
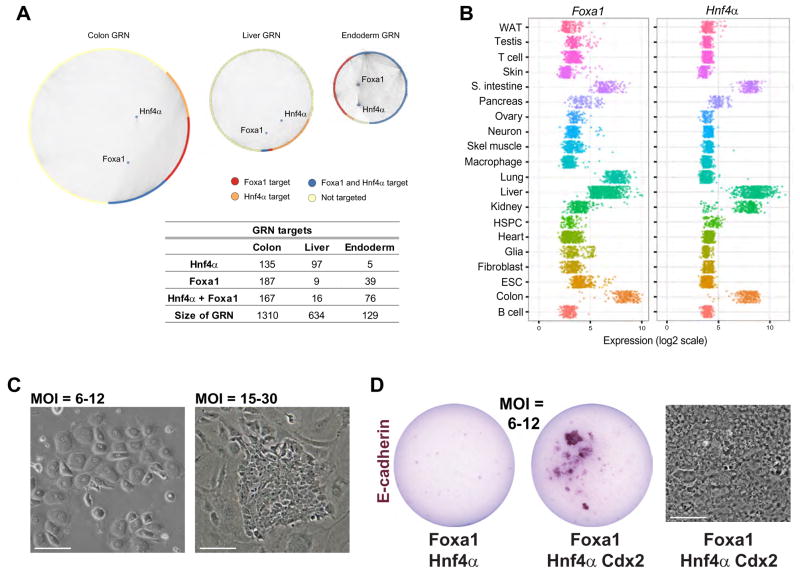
(A). (B) Cell and tissue classification heatmap of three independently derived iHep lines, fetal liver and adult liver: iHeps poorly classify as liver. (C) Liver and fibroblast GRN status in iHeps, fetal liver, and adult liver: the fibroblast GRN is not extinguished and liver GRN not fully established in iHeps. HSPC classification of the fetal liver reflects its role as a hematopoietic organ at this stage. (D) Partial establishment of a colon sub-GRN (424 genes) in iHeps. (E) Prioritization of transcriptional regulators of the colon GRN overexpressed in iHeps relative to native liver, led by the master intestinal regulator, Cdx2. (F) Confirmation of intestine-specific gene expression in iHeps relative to native fibroblasts, liver, and colon. Data are represented as mean +/− SEM. See also Figure S3.
We next transcriptionally profiled our iHeps and applied the classification function of CellNet. iHeps classified predominantly as fibroblasts and barely manifested any liver classification (Figure 3B; Mean scores: 0.186 and 0.049, respectively). CellNet analysis of GRN status in these cells confirmed that fibroblast status was not extinguished while liver identity was only weakly established (Figure 3C). This classification was not accounted for by incomplete conversion nor the persistence of fibroblasts, as the rapidly dividing iHeps dominate the culture. Considering our accompanying findings that alternate cell fates are established in most fate engineering protocols, we assessed all alternate identities suggested by CellNet (Cahan et al.). Unexpectedly, CellNet detected the partial establishment of an intestinal GRN, relating to colon (Figure 3D).
Intestinal fate specification in iHeps
During endoderm development, liver is derived from foregut and intestine from hindgut (Sheaffer and Kaestner, 2012). Considering our findings that knockdown of B cell transcriptional regulators could de-repress macrophage fate, we reasoned that factors responsible for a hindgut program in iHeps may act to repress specification of foregut fate. We consulted CellNet and found that the homeobox transcription factor Cdx2 emerged as the top prioritized regulator of colon fate in iHeps (Figure 3E). Cdx2 is a regulator of Hox gene expression that is required for intestinal cell lineage specification in early endoderm (Gao et al., 2009). Interestingly, CellNet also revealed several Hox genes as prioritized regulators of the colon GRN, suggesting that iHeps were aberrantly patterned with respect to the Hox code. Whereas freshly-isolated adult hepatocytes did not express Cdx2, we confirmed expression of Cdx2 and additional CellNet-nominated intestine-specific marker genes in three independent iHep lines, consistent with expression data previously reported for iHeps (Sekiya and Suzuki, 2011; Figure 3F; S4A, B).
Cdx2 is required for the generation of iHeps
Cdx2 expression in posterior gut epithelium antagonizes foregut differentiation (Gao et al., 2009). Thus we proceeded to knock down Cdx2 in iHeps in an attempt to fortify their liver identity. As predicted, we observed significant loss of CellNet-appointed intestine-specific gene expression, with more efficient Cdx2 knockdown resulting in reduced intestinal marker expression (Figure 4A; S4C: Cdx2low, ~85% knockdown). Moreover, knockdown of Cdx2 resulted in moderate increases in expression of the liver markers Albumin, Afp and alpha-1-antitrypsin (α1AT) (Figure 4B). Functionally, following Cdx2 knockdown, albumin expression and urea secretion significantly increased (Figure 4C, D). Additionally, Cdx2 knockdown resulted in higher gene expression associated with liver functions and appeared to alter the Hox code to favor anterior gene expression (Figure S4D). However, neither liver classification nor liver GRN status improved following Cdx2 knockdown (Figure S4E). Interestingly, these gains in liver-specific gene expression and function were more evident for the weaker Cdx2 knockdown: Cdx2mod (Figure S4C: Cdx2mod, ~60% knockdown). Cdx2 disruption has been shown to disrupt Hnf4α occupancy at Cdx2-bound enhancers (Verzi et al., 2013) and analysis of our iHep microarray data revealed that expression of endogenous Hnf4a was lost as Cdx2 expression decreased, suggesting coordinate regulation of these two factors during iHep induction (Figure 4E).
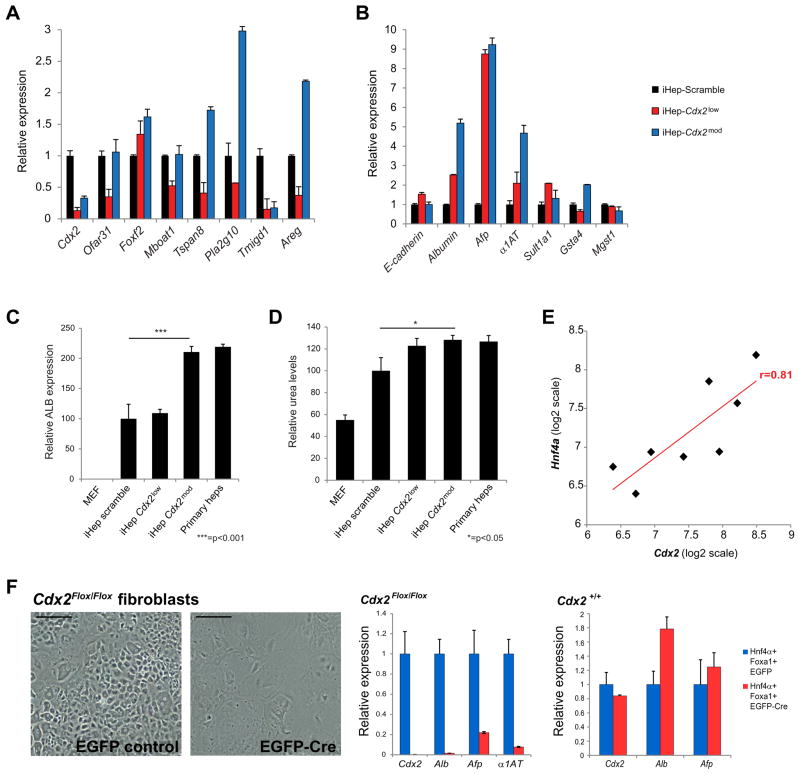
Figure 4. Requirement of Cdx2 for function and generation of iHeps.
(A) qPCR analysis of intestine-specific and liver-specific (B) gene expression following Cdx2 knockdown. Albumin expression (C) and urea production (D) in iHeps following Cdx2 knockdown. *=p<0.05, ***=p<0.001, t-Test. Cdx2 knockdown in iHeps reduces intestinal gene expression while moderately increasing liver gene expression and function, although these gains diminish with stronger Cdx2 knockdown. (E) Correlation between endogenous Hnf4α and Cdx2 expression in several iHep lines. (F) Fibroblasts fail to be converted to iHeps in the absence of Cdx2: transduction of conditional Cdx2 knockout fibroblasts. Far right: Cre expression does not impair generation of iHeps. Scale bars, 50μM. Data are represented as mean +/− SEM. See also Figure S4 and S5.
To further probe the role of Cdx2 in fibroblast to iHep conversion, we attempted to derive iHeps from MEFs carrying exon 2 of the Cdx2 gene flanked by loxP sites (Verzi et al., 2010; Figure S5A). Following transduction with Cre-EGFP to knock out Cdx2, we initiated direct conversion to iHeps with Foxa1 and Hnf4α. In the GFP control, iHep colonies began to emerge after 7 days, whereas no colonies were detected following Cdx2 knockout, demonstrating that Cdx2 is necessary for the establishment of iHeps (Figure 4F).
Levels of Foxa1 and Hnf4α are important for hepatic or intestinal fate specification
To understand how intestinal fate might be specified in iHeps, and to identify candidate factors to further engineer the cells, we consulted CellNet to score transcriptional regulators of the endoderm, liver and colon GRNs. Interestingly, we found the transcription factors used to generate iHeps to be putative regulators of a broad endoderm GRN, and Foxa1 in particular to be more heavily biased toward targets in the endoderm and colon GRNs than the liver GRN (Figure 5A). Surveying Foxa1 and Hnf4α expression over all CellNet target cells and tissues reveals their expression across a range of endoderm-derived tissues and restriction of co-expression to liver, colon, and small intestine. Indeed, the highest levels of Foxa1 expression are found in the colon (Figure 5B). These data highlight the roles of these transcription factors in both liver and intestinal tissues (Garrison et al., 2006; Parviz et al., 2003). Together, this suggests that the conversion factors employed in this protocol fail to establish a unique liver fate but instead specify endoderm tissues more broadly. This indicates that higher levels of Foxa1 and Hnf4α may target a much broader endoderm GRN, resulting in the establishment of colon GRNs in addition to liver GRNs.
Figure 5. Foxa1 and Hnf4α, employed in the generation of iHeps are putative regulators of a broad endoderm GRN, as predicted by CellNet.
(A) Foxa1 in particular is more heavily biased toward targets in the endoderm and colon GRNs than the liver GRN. (B) Foxa1 and Hnf4α expression over all datasets from the 20 CellNet target cells and tissues reveals their expression across a range of endoderm-derived tissues and restriction of co-expression to liver, colon, and small intestine. (C) Morphology of Foxa1-Hnf4α converted fibroblasts at MOI=6–12 (left) and MOI=15–30 (right). (D) Alkaline phosphatase staining of E-cadherin expressing iHep colonies with Foxa1-Hnf4α transduction at MOI=6–12 in the presence and absence of Cdx2. Cdx2 expression enables iHep establishment from low levels of Foxa1 and Hnf4α. Scale bars, 50μM. See also Figure S5.
To further investigate the effect of variable levels of reprogramming factor expression, we employed retroviral infection at low (6–12) and high (15–30) multiplicities of infection (MOI) for Foxa1 and Hnf4α to drive conversion to iHeps. Low MOI promoted conversion to cells resembling native hepatocytes—binucleate, possessing an orthogonal morphology distinct from the epithelial morphology reported for iHeps (Sekiya and Suzuki, 2011; Figure 5C). In contrast, high MOI conditions, equivalent to the conditions employed by Sekiya and Suzuki, generated colonies with characteristic iHep morphology that were capable of expanding indefinitely (Figure 5C). Monitoring the behavior of low MOI iHeps over the course of six days revealed that these cells became quiescent after limited cell division, corresponding to ex vivo-cultured primary hepatocytes. Moreover, qPCR analysis of low MOI iHeps revealed reduced Cdx2 expression and enhanced Alb expression relative to high MOI iHeps (Figure S5B). Supplementing low MOI Foxa1 and Hnf4α infection with exogenous Cdx2 permitted the establishment of large iHep colonies, whereas in the absence of Cdx2 no E-cadherin-positive epithelial colonies were generated (Figure 5D; S5C).
These data further corroborate the importance of Cdx2 expression for the conversion of MEFs to iHeps, and suggests that different levels of Foxa1, Hnf4α, and Cdx2 might establish alternate endodermal cell fates. Furthermore, our analysis shows a greater number of Foxa1 and Hnf4α targets in the endoderm and colon, relative to liver. Given the close association of these transcription factors to hindgut fate, we decided to further investigate the intestinal potential of iHeps.
Intestinal fate and function of iHeps
To explore the potential for iHeps to form epithelial structures in vitro, we cultured iHeps as 3D-organoids, which could be expanded from single cells in matrigel to form large spherical structures possessing epithelial polarity and whose formation was dependent on Cdx2 expression (Figure 6A; S6A). However, neither prolonged culture nor the addition of Wnt3a resulted in budding structures characteristic of organoids grown from adult intestinal stem cells (Yui et al., 2012; Figure S6A). Moreover, Lgr5 positive cells, indicative of adult intestinal stem cells, were only rarely detected in iHeps cultured under these conditions, and their culture could be maintained in the absence of R-spondin and Wnt3a (Figure S6B). Although distinct from intestinal stem cells, the morphology and behavior of iHeps was reminiscent of Fetal Enterospheres, which may represent transient developmental intestinal progenitors (Fordham et al., 2013).
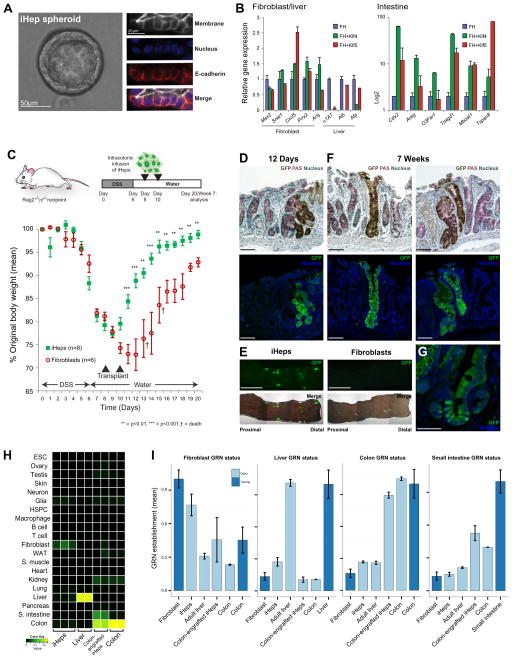
Figure 6. iHeps are an endoderm progenitor harboring intestinal identity and can functionally engraft mouse colon.
(A) Culture of iHeps as spheroids and determination of cell polarity by immunofluorescence staining of E-cadherin. (B) qPCR analysis of fibroblast, liver and intestinal gene expression following three-factor driven conversion to ‘Klf-iEPs’ with Hnf4α, Foxa1 and Klf4 or Klf5 (C) Functional engraftment of GFP-expressing iHeps into mice with DSS-induced colitis. Weight gain following DSS withdrawal in GFP-iHep/iEP recipient animals: the latter regain weight significantly faster than recipients of GFP-fibroblasts, **=p<0.01, ***=p<0.001, t-Test. (D) Immunohistochemistry and immunofluorescence of short-term engrafted GFP-iHeps/iEPs in serial sections of colon 12 days after transplantation. (E) Whole-mounts of engrafted GFP-iHeps/iEPs. Engraftment of GFP-fibroblasts is not observed. Scale bars, 0.5cm. (F) Long-term engrafted GFP-iHeps/iEPs in the colon 7 weeks after transplantation. Goblet cells are visualized by PAS staining. Scale bars, 100μm. (G) Detail of long-term engrafted crypt. (H) CellNet classification of engrafted iHeps/iEPs recovered 12 days following transplantation. Colon classification is strongly fortified in engrafted iHeps/iEPs, comparable to native colon, relative to in vitro cultured iHeps/iEPs. (I) Fibroblast, liver and broad colon GRN status in colon-engrafted iHeps/iEPs: Fibroblast and liver GRNs are extinguished where colon GRN status is fortified following maturation in vivo. Data are represented as mean +/− SEM. Scale bars as indicated. See also Figure S6.
iHeps functionally engraft colon
iHeps can functionally engraft liver, and over time within this in vivo niche, they cease to divide and their hepatocyte gene expression signature matures (Sekiya and Suzuki, 2011). Considering that our iHeps formed spheroids and resembled intestinal progenitors in 3D matrigel culture, we wondered if they might differentiate further toward mature intestine. We again consulted CellNet for candidate transcriptional regulators to drive iHeps closer to intestine, and identified Klf4 and Klf5, whose functions are known in the differentiated epithelium and dividing crypt cells, respectively (McConnell et al., 2007). We then drove the conversion of MEFs to an intestinal fate using high-MOI Hnf4α and Foxa1 supplemented with either Klf4 or Klf5. Addition of either Klf transcription factor dampened liver marker expression and strongly fortified intestinal identity, without altering fibroblast-specific genes significantly (Figure 6B).
Taken together, these data show that iHeps generated by the published protocol of Sekiya and Suzuki can differentiate toward both liver and intestine, suggesting that iHeps are actually induced endoderm progenitors, ‘iEPs’. To test the potential for iHeps/iEPs and Klf-matured cells (termed ‘Klf-iEPs’) to differentiate toward functional intestinal cells in vivo, we transplanted GFP-labeled iHeps/iEPs, Klf-iEPs, and fibroblasts into superficially damaged mouse colon, a model recently employed to assess the functional engraftment of ESC-derived intestinal stem cells (Yui et al., 2012; Figure 6C). Following six days of DSS-induced colitis and 48 hours of recovery, 5 million cells were delivered into the lumen of the colon, twice over a two-day period, and the colon was examined for cellular engraftment 12 days later. During this period control animals receiving fibroblasts had not returned to their original weight, demonstrated ongoing colitis resulting in death of 2/6 mice, and showed no engraftment. In contrast, iHep/iEP recipient animals returned to their original weight significantly faster, showed no sign of colitis after 12 days, and instead revealed the presence of numerous GFP+ donor-derived cells integrated into the colonic epithelium (Figure 6C–E). Klf-iEPs did not engraft the colon (Figure S6C), probably because mature intestinal epithelial cells cannot fulfill this role (Zhou et al., 2012). At 12 days, crypts contained a mix of GFP+ and GFP- colonocytes, as the known cell turnover would predict (Figure 6D). By 7 weeks after transplantation, however, 5 of 8 recipient mice carried patches of GFP+ colonic crypts (Figure S6D), which signifies competent reconstitution of the damaged mucosa by iHep/iEPs. The patchy pattern of GFP+ crypts strongly indicates clonal repopulation by cells with intestinal stem-like self-renewal (Figure 6F, G), and the lack of Lgr5 expression in iHeps in vitro is compatible with low Lgr5 gene activity in native mouse colonic crypts, distinct from the small intestine (Barker et al., 2007).
To evaluate if iHeps/iEPs could mature in vivo, we harvested colonic crypts from recipient animals 12 days post-transplantation, dissociated the colonic epithelium, and profiled gene expression in GFP+ cells purified by flow cytometry (Figure S6E). By CellNet analysis, colon-engrafted iHeps/iEPs received a high colon classification score, significantly stronger than iHeps cultured in vitro prior to engraftment (average colon scores 0.79 and 0.07, respectively) and a weaker small intestine classification score of 0.33 (Figure 6H). Both the fibroblast GRN persistent in iHeps/iEPs and their weak liver GRN were significantly silenced, with an accompanying small increase in small intestine GRN status and striking fortification of the colon GRN to 90% of the native colon status (Figure 6I). To rule out fusion of iHeps/iEPs with native colonic cells, we transplanted Cre-EGFP transduced iHeps/iEPs into Rosa26-Lox-STOP-Lox-Tomato transgenic mouse colons: engrafted cells did not show tomato fluorescence, dismissing this formal possibility (Figure S6F). The long-term functional engraftment of iHeps/iEPs into superficially damaged mouse colon, along with the previously reported functional engraftment within mouse liver (Sekiya and Suzuki, 2011), establishes that these cells represent progenitors with broad endodermal rather than hepatocyte-restricted potential. Thus we demonstrate how the CellNet network biology platform guided a direct conversion strategy to generate endoderm progenitors capable of functional intestinal repopulation.
DISCUSSION
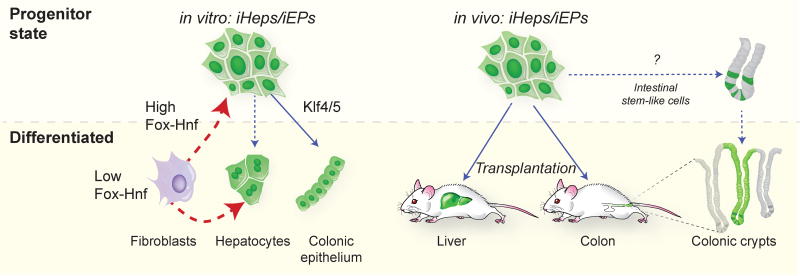
Here, in a practical application of CellNet to two distinct cell fate conversion protocols, we document the incomplete specification of macrophage fate in a B cell line, and the unexpected establishment of intestinal identity in fibroblasts reportedly converted into liver. CellNet analysis yielded the surprising discovery that iHeps in fact represent an induced endoderm progenitor (‘iEP’), which is capable of long-term functional engraftment of the mouse colon. Additional manipulation of CellNet-prioritized candidates, Klf4 and Klf5, promoted intestinal maturation in vitro, but these differentiated ‘Klf-iEP’ cells failed to engraft damaged colon, indicating that cell populations engineered in vitro must be transplanted at a specific stage that is amenable to functional end organ engraftment (Figure 7).
Figure 7. Model: Foxa1 and Hnf4α specify an endoderm progenitor state.
Left: In this study we find that low levels of Foxa1 and Hnf4α expression in fibroblasts in vitro specifies cells bearing the hallmarks of differentiated hepatocytes. High levels of Foxa1 and Hnf4α specifies a progenitor state, ‘iHeps/iEPs’ from which differentiation toward an intestinal fate, ‘Klf-iEPs’, can be coaxed by Klf4/5 expression. Right: Transplantation of iHeps/iEPs into mouse liver (as in Sekiya and Suzuki, 2011) or mouse colon leads to differentiation of progenitors to support long-term engraftment of the colon suggesting that iHeps/iEPs possess intestinal stem-like properties.
Our findings establish CellNet as a potent tool for assessing the fidelity of cell fate conversions, and for revealing unexpected properties of cells engineered in vitro. CellNet analysis calls into question the underlying premise that direct conversion bypasses progenitor or stem cell intermediates. First, in B cell to macrophage conversion, we found that progenitor GRNs were partially and transiently engaged, suggesting that C/EBPα has the capacity to establish a progenitor-like program from which macrophage fate emerges. Second, we demonstrate that iHeps are endoderm progenitors, iEPs, rather than mature hepatocyte-like cells as previously reported (Sekiya and Suzuki, 2011). Additionally, there have been several reports describing the generation of progenitors from direct conversion of fibroblasts: hematopoietic, neural and hepatic progenitors (Lujan et al., 2012; Pereira et al., 2013; Yu et al., 2013). The capture of a progenitor state in vitro may be facilitated by selection for these expandable populations over non-dividing differentiated cells.
The existence of iHeps as endoderm progenitors rather than differentiated cells explains their failure to exclusively classify as a unique cell or tissue until they mature within the colon niche. Our accompanying study shows that cells derived via directed differentiation more faithfully establish the desired target GRNs relative to direct conversion (Cahan et al.). It is possible that cells generated via direct conversion classify poorly because this approach frequently specifies progenitors which must be subsequently differentiated. The resolution and power of CellNet to accurately identify these intermediates will continue to improve as sufficient training datasets are collected for embryonic lineages. At present we can assert that in vitro cultured iHeps/iEPs are neither mature liver nor colon but behave as a progenitor, although their in vivo equivalent is unclear.
Pancreas and lung GRNs were not detected in iHeps/iEPs, suggesting that their potential may be restricted to liver and intestine. Hnf4α is required for development of the liver (Parviz et al., 2003) and colon (Garrison et al., 2006). Similarly, Foxa1 is involved in liver and intestinal development and function (Bernardo and Keri, 2012). Foxa1 is a pioneer factor, capable of remodeling chromatin and facilitating binding of other transcription factors, such as Hnf4α (Zaret and Carroll, 2011). In a variation to the protocol of Sekiya and Suzuki, we found that lower expression of Foxa1 and Hnf4α generates cells resembling native hepatocytes. In contrast, high levels of Foxa1 may facilitate a permissive chromatin state accessible to Hnf4α thus establishing an embryonic endoderm progenitor state (Figure 7).
CellNet prioritized Cdx2 as an intestinal fate regulator in iHeps/iEPs, leading to the finding that Cdx2 is essential to establish the endoderm progenitor state in these cells. Cdx2 specifies embryonic intestinal epithelium and is required for intestinal gene expression, including Hnf4α (Gao et al., 2009). By E9.0 emergence of Cdx2 in the hindgut and Sox2 in the foregut demarcates the endoderm (Sherwood et al., 2009) suggesting that iHeps/iEPs may more resemble a hindgut progenitor. iHep/iEP behavior in vitro is reminiscent of recently reported Fetal Enterospheres (Fordham et al., 2013), a transient intestinal progenitor population, rather than adult intestinal stem cells, further supporting the notion that high levels of Foxa1 and Hnf4α install a hindgut progenitor state. The capacity of iHeps to engraft liver raises the possibility that a foregut progenitor is concomitantly specified, although our failure to isolate any expandable progenitor population in the absence of Cdx2 expression does not support this. Likewise, any low MOI hepatocyte-like cells are unlikely to engraft liver due to their scarcity and selection against in vitro. It must also be emphasized that direct conversion is an artificial situation where high levels of pioneer factor expression may open inaccessible chromatin domains to endow cells with enhanced plasticity – thus iHeps/iEPs may not possess a true in vivo correlate.
In addition to assigning a metric of cell identity and thereby providing a measure of the fidelity of cell fate conversions, CellNet prioritizes candidates for interventions to help improve cell fate specification. At present, predicting gene expression in GRNs remains challenging and is limited to simplified networks. Here, knockdown of prioritized B cell candidates, Pou2af1 or Ebf1 improves the classification and function of induced macrophages. Moreover, CellNet prioritization of regulators of the colon GRN established in iHeps led to the discovery that Cdx2 is required for the generation of iHeps, an instrumental finding helping reveal their full potential. Furthermore, CellNet assisted in the identification of Klf4 and Klf5 as factors to differentiate iHeps further toward mature colon in vitro. These results demonstrate the utility of CellNet to dissect and enhance cell engineering strategies. However, it is clear from the outcome of our interventions that refining cell identity is not trivial, as illustrated by the persistence and fortification of B cell GRNs following knockdown of prioritized B cell regulators. Due to this complex feedback regulation within gene regulatory networks, attempting to refine engineering strategies will require a painstaking, rational, iterative approach.
Undoubtedly, the best strategy to capture a desired fate harnesses the in vivo niche. In our accompanying study, our analysis shows that cardiomyocyte-like cells generated from cardiac fibroblasts in situ show a markedly higher heart classification than those converted in vitro (Cahan et al.). Similarly, here we have shown that following engraftment into colon, iHeps exhibited stem cell behavior to reconstitute the intestinal epithelium, maturing in vivo to increase their colon classification, associated with 90% colon GRN establishment and silencing of liver and fibroblast GRNs. Achieving such a high degree of tissue identity in vivo may be ideal for therapeutic applications of engineered cells, but for in vitro applications in toxicology testing and disease modeling, it will be essential to develop more robust cell fate conversions. Understanding how in vivo cues mature fate will assist in this endeavor, as will bioengineering approaches designed to recapitulate the niche in vitro. CellNet analysis will be of great assistance in developing these strategies.
This study establishes CellNet as a powerful platform for assessing and refining engineered cell populations. In addition to providing a metric for the establishment of target cell identity, CellNet can reveal the unanticipated hybrid or intermediate identity of cells, which can be easily overlooked with current methods of broad transcriptional profiling and functional assays. This approach has revealed the unrealized potential of iHeps as an endoderm progenitor and establishment of intestinal fate from a direct conversion strategy. In the future, we anticipate continued refinement of CellNet and greater potential to marshal cellular engineering towards more precise and impactful applications in regenerative medicine.
EXPERIMENTAL PROCEDURES
Extended Experimental Procedures are available online.
Mice
Strains used in this study: C57BL/6J (The Jackson laboratory: 000664), C;129S4-Rag2tm1.1Flv Il2rgtm1.1Flv/J (The Jackson laboratory: 014593), B6.129P2-Lgr5tm1(cre/ERT2)Cle/J (The Jackson laboratory: 008875), B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (The Jackson laboratory: 007905), Cdx2Flox/Flox (Verzi et al., 2010). All animal procedures were based on animal care guidelines approved by the Institutional Animal Care and Use Committee.
Direct conversion of B cells to macrophages
The C10 pre-B cell line was converted to induced macrophages, as in (Bussmann et al., 2009). Briefly, C10 cells were grown in RPMI 1640 without phenol red (Lonza) supplemented with 10% charcoal/dextran-treated FBS (Hyclone) and 50mM 2-mercaptoethanol (GIBCO). They were induced by the addition 100nM of β-estradiol (Sigma) and grown with 10nM IL-3 and CSF-1 (Peprotech).
Direct conversion of MEFs to iHeps
MEFs were converted to iHeps as in (Sekiya and Suzuki, 2011). Briefly, MEFs were prepared from E13.5 embryos and serially transduced with Foxa1 and Hnf4α or Hnf4α-2ta-Foxa1 retroviruses over a five-day period, followed by culture on gelatin for two weeks in hepato-medium (DMEM:F-12, supplemented with 10% FBS, 1mg/ml insulin (Sigma), 10−27M dexamethasone (Sigma-Aldrich), 10mM nicotinamide (Sigma-Aldrich), 2mM L-glutamine, 50mM β mercaptoethanol (Life Technologies) and penicillin/streptomycin, containing 20ng/ml hepatocyte growth factor (Sigma-Aldrich) and 20ng/ml epidermal growth factor (Sigma-Aldrich), after which the emerging iHeps were cultured on collagen. iHeps generated with mono- and bicistronic constructs were equivalent.
Lenti- and retrovirus production
Open reading frames were cloned into the lentiviral vector, pSMAL-GFP or the retroviral vector, pGCDNsam. Lentiviral particles were produced by transfecting 293T-17 cells (ATCC: CRL-11268) with pCMV-dR8.2 dvpr (Addgene plasmid 8455) and pCMV-VSVG (Addgene plasmid 8454). Virus was harvested 48 and 72 hours after transfection and PEG concentrated. Constructs were titered by serial dilution on 293T cells. Foxa1, Hnf4α and Hnf4α-t2a-Foxa1 retroviruses were packaged with pCL-Eco (Imgenex), titered on MEFs and cells transduced according to Sekiya and Suzuki, 2011.
Gene expression microarray
The RNAeasy Microkit (QIAGEN) was used to collect and prepare total RNA for microarray analysis. The Ovation Picokit (Nugen) was used for pre-amplification, as required. Gene expression profiling was performed on Affymetrix 430 2.0 gene chips per standard protocol. Microarray data from this study has been deposited to the GEO database under accession number GSE59037.
Conditional Cdx2 knockout
E13.5 MEFs were derived from Cdx2Flox/Flox embryos (Verzi et al., 2010), transduced with either EGFP or Cre-EGFP retrovirus (Addgene plasmid 24064), cultured for 48 hours and then GFP+ cells were flow sorted and cultured on gelatin. The cells were then transduced with Foxa1 and Hnf4α retroviruses.
Immunohistochemistry and Immunostaining
Cultured cells were fixed with 4% PFA, permeabilized, blocked in 10% FCS and incubated with the following antibodies: mouse anti-E-cadherin (BD Biosciences) and rabbit anti-Albumin (Biogenesis). After washing, samples were incubated with Alexa 568-conjugated secondary antibody (1:500; Molecular Probes) with DAPI and far-red phalloidin (Molecular Probes) for immunofluorescence staining. Colon tissue was fixed with 10% formalin and paraffin sectioned. After rehydration and blocking in 10% goat serum, the sections were incubated with rabbit anti-GFP antibody (Abcam: ab6556). For immunofluorescence analysis, the sections were incubated with Alexa 488-conjugated secondary antibody (1:500; Molecular Probes) and DAPI and imaged with a Zeiss LSM 710 confocal microscope. For Immunohistochemistry, samples were processed using the Vectastain ABC system and DAB kit (Vector Laboratories), and PAS staining was performed with a kit (Sigma).
Intracolonic transplantation of iHeps
Colitis was induced in Rag2γc−/− mice by administration of 2.5% Dextran Sodium Sulfate (DSS, MP Biomedicals) in the drinking water for six days, followed by two days of recovery. On day 8, animals losing between 15–20% of their original body weight were selected at random to receive delivery of 5 million cells (GFP-labeled iHeps, fibroblasts or Klf4-iEPs) into the lumen of the colon. In a modification to the procedure reported in Yui et al., 2012, the cells were resuspended in Matrigel (BD Biosciences) and 100–150μl of the suspension was delivered via a 22G angiocatheter (BD Biosciences). Transplantation of cells was repeated on day 10 and the colons were examined for engraftment on day 20 and week 7 post-initiation of DSS treatment. For recovery of GFP-expressing iHeps, transplanted colons were cut into small pieces and incubated in HBSS with 30mM EDTA at 37°C to liberate crypts. The crypts were then dissociated with 0.25% Trypsin and the resulting single cell suspension was flow sorted to recover engrafted iHeps.
Supplementary Material
HIGHLIGHTS.
The CellNet network biology platform evaluates cell fate engineering strategies
CellNet reveals incomplete conversion of engineered macrophages and hepatocytes
Conversion to induced hepatocytes specifies intestinal fate, regulated by Cdx2
Induced hepatocytes are endoderm progenitors that can functionally engraft colon
Acknowledgments
We would like to thank members of the Daley laboratory for critical discussion, the Dana Farber Cancer Institute Hematological Neoplasia flow facility for assistance with cell sorting, and Thomas Graf for the C10 and RAW264.7 cells. G.Q.D. is supported by grants from the NIH (Progenitor Cell Biology Consortium UO1-HL100001 and R24DK092760), and the Harvard Stem Cell Institute. G.Q.D. is an affiliate member of the Broad Institute, and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research. S.A.M. is supported by a Young Investigator Award from Alex’s Lemonade Stand Foundation and P50HG005550. P.C is supported by 1K01DK096013, 2T32HL066987, and 5T32HL007623. H.L. is supported by Mayo Clinic Center for Individualized Medicine and Mayo Clinic Center for Regenerative Medicine. J.J.C. is supported by NIH grant R24DK092760 and the HHMI. R.A.S. is supported by NIH grant R01DK082889.
Footnotes
AUTHOR CONTRIBUTIONS
S.A.M., P.C., and H.L. designed, performed, and interpreted experiments and wrote the paper. A.M.Z. assisted with experiments. A.K.S. and R.A.S. designed, interpreted and A.K.S. assisted in Cdx2 knockout and colon engraftment experiments. J.J.C. assisted in design and interpretation of experiments. G.Q.D. designed and interpreted experiments and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: Network Biology Applied to Stem Cell Engineering. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DE, Melton D. Turning straw into gold: directing cell fate for regenerative medicine. Nat Rev Genet. 2011;12:243–252. doi: 10.1038/nrg2938. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham RP, Yui S, Hannan NRF, Soendergaard C, Madgwick A, Schweiger PJ, Nielsen OH, Vallier L, Pedersen RA, Nakamura T, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem Cell Reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins K, Reményi A, Tomilin A, Massa S, Wilmanns M, Matthias P, Schöler HR. OBF1 enhances transcriptional potential of Oct1. EMBO J. 2003;22:2188–2198. doi: 10.1093/emboj/cdg199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Südhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Daley GQ. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell Res. 2013;23:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechanitzky R, Akbas D, Scherer S, Györy I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma’ayan A, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ubreva J, Ciudad L, Gómez-Cabrero D, Parra M, Bussmann LH, di Tullio A, Kallin EM, Tegnér J, Graf T, Ballestar E. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res. 2012;40:1954–1968. doi: 10.1093/nar/gkr1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Sheaffer KL, Kaestner KH. Transcriptional networks in liver and intestinal development. Cold Spring Harb Perspect Biol. 2012;4:a008284. doi: 10.1101/cshperspect.a008284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen TYA, Melton DA. Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain AJ. Isolated hepatocytes: use in experimental and clinical hepatology. Gut. 1994;35:433–436. doi: 10.1136/gut.35.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tullio A, Vu Manh TP, Schubert A, Castellano G, Månsson R, Graf T. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc Natl Acad Sci U S A. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Shin H, San Roman AK, Liu XS, Shivdasani RA. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol Cell Biol. 2013;33:281–292. doi: 10.1128/MCB.01185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbring H. A simple code for installing hepatocyte function. Cell Stem Cell. 2011;9:89–91. doi: 10.1016/j.stem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Yu B, He ZY, You P, Han QW, Xiang D, Chen F, Wang MJ, Liu CC, Lin XW, Borjigin U, et al. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell. 2013;13:328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Price DD, Dreher KL, Pronold B, Callam CS, Sharma J, Verne GN. Localized colonic stem cell transplantation enhances tissue regeneration in murine colitis. J Cell Mol Med. 2012;16:1900–1915. doi: 10.1111/j.1582-4934.2011.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.