Abstract
Predation of bacteria by phagocytic cells was first developed during evolution by environmental amoebae. Many of the core mechanisms used by amoebae to sense, ingest and kill bacteria have also been conserved in specialized phagocytic cells in mammalian organisms. Here we focus on recent results revealing how Dictyostelium discoideum senses and kills non-pathogenic bacteria. In this model, genetic analysis of intracellular killing of bacteria has revealed a surprisingly complex array of specialized mechanisms. These results raise new questions on these processes, and challenge current models based largely on studies in mammalian phagocytes. In addition, recent studies suggest one additional level on complexity by revealing how Dictyostelium recognizes specifically various bacterial species and strains, and adapts its metabolism to process them. It remains to be seen to what extent mechanisms uncovered in Dictyostelium are also used in mammalian phagocytic cells.
Introduction
Phagocytosis appeared during evolution of unicellular eukaryotic organisms essentially as a way to acquire food by predating other microorganisms. In higher multicellular eukaryotes, phagocytosis allows specialized immune phagocytic cells to ingest and destroy potential pathogens. Professional mammalian phagocytes (e.g. macrophages and neutrophils) share with unicellular phagocytes (e.g. Dictyostelium amoebae) the ability to ingest and kill a large number of microorganisms (Steinert, 2011). They also frequently face the same virulence traits developed by bacteria in the course of evolution: bacteria largely make use of the same mechanisms to resist predation by Dictyostelium and by mammalian phagocytes (Cosson and Soldati, 2008).
There have been a number of excellent recent reviews dealing with the manner in which pathogenic bacteria avoid killing by Dictyostelium cells and mammalian phagocytes (Clarke, 2010; Bozzaro and Eichinger, 2011; Steinert, 2011; Soldati and Neyrolles, 2012). This review is focused on the situation in which bacteria show little or no pathogenicity, and succumb easily to phagocytic cells. The distinction is somewhat arbitrary: even the most innocuous bacteria can exceptionally infect and kill some individuals [e.g. fatal Lactobacillus infections (Kalima et al., 1996)]. We designate here as ‘non-pathogenic’ bacteria that have a very low ability to infect mammals, and upon which Dictyostelium amoebae can efficiently feed. With this perspective, we are examining two emerging themes in the field of Dictyostelium research: which are the molecular mechanisms employed by amoebae to kill bacteria? How do amoebae recognize bacteria and adapt their physiology to optimize their feeding strategy?
Educated guesses on intracellular killing
A large number of mechanisms have been proposed to play a role in intracellular killing, based mostly on studies of mammalian phagocytic cells (Haas, 2007). These include production of toxic free radicals, control of the ionic environment, and lytic enzymes. Dictyostelium provides the opportunity to test how well we understand the molecular mechanisms ensuring intracellular bacterial killing. One way to address the question is to try to predict which gene products should be important for efficient intracellular killing of bacteria. It is then relatively easy to specifically inactivate the selected genes of interest in Dictyostelium, and to measure the ability of the resulting mutant amoebae to kill ingested bacteria. From the current knowledge of intracellular killing in mammalian cells, one may try to guess the gene products most likely to be essential for intracellular killing in Dictyostelium. Our best guesses initially were: free radicals, NRAMP1, and lysosomal hydrolases.
Conveniently, the ability of Dictyostelium cells to kill various bacteria can be tentatively inferred from the ability of mutant cells to feed and grow upon various bacteria, a growth assay that allows for the testing of thousands of mutants in a simple and inexpensive way (Fig. 1A) (Froquet et al., 2009). However, a defective growth invites further characterization since not only intracellular killing is necessary for efficient feeding of Dictyostelium on bacteria, but also phagocytosis, motility, and probably bacterial sensing and metabolic adaptation (see below). A defect in intracellular killing can be characterized more specifically by measuring the survival of bacteria inside phagosomes (Fig. 1B). A non-virulent isolate of Klebsiella pneumoniae has been used historically to feed and grow Dictyostelium amoebae, and several studies have focused on the mechanisms ensuring intracellular killing of this Klebsiella strain, but several other non-pathogenic bacterial species are equally amenable to this type of analysis, in particular Gram-positive Bacillus subtilis and Micrococcus luteus, or Gram-negative Escherichia coli and Pseudomonas aeruginosa strains.
Figure 1.
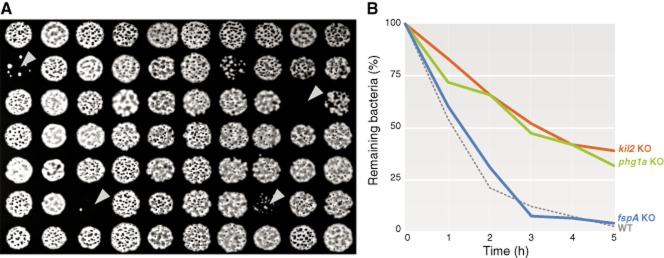
Surrogate methods for measuring intracellular killing in Dictyostelium.A. Growth of amoebae on a lawn of bacteria can be scaled up to allow for the screen of thousands of mutants at a time. Dictyostelium colonies able to feed on a lawn of Klebsiella bacteria (in black) form phagocytic plaques (white circles). On the contrary, mutants unable to feed on bacteria do not form such plaques (arrowheads).B. Intracellular killing of bacteria can be more specifically measured by mixing Dictyostelium cells and bacteria, and assessing the number of remaining live bacteria after different times. WT and fspA KO cells are able to efficiently eliminate Klebsiella (less than 10% of bacteria remaining after 3 h), while killing-deficient mutants (as kil2 and phg1a KO cells) are not (around 50% of bacteria remaining after 3 h).
The protein most clearly implicated in intracellular killing of bacteria in mammals is Nox2 (or gp91phox), a component of the NADPH-oxidase expressed in phagocytic cells. Nox2 is essential for the oxidative burst of phagocytic cells (e.g. neutrophils and monocytes), which is believed to play a key role in bacterial killing by free radicals (Winterbourn and Kettle, 2013). This hypothesis is based first on the observed bactericidal effect of free radicals, and second on the observation that genetic alterations of Nox2 lead to chronic granulomatous disease (CGD), a severe disorder in which patients suffer from recurrent bacterial and fungal infections (Goldblatt and Thrasher, 2000). In addition, neutrophils from mice with defective NADPH-oxidase activity kill inefficiently ingested Staphylococcus aureus both in vitro and in vivo (Ellson et al., 2006), and inhibiting the production of reactive oxygen species in human neutrophils (e.g. in hypoxic conditions) impairs intracellular killing of S. aureus (McGovern et al., 2011). More complex scenarios may be envisaged, since for example neutrophils from CGD patients are also defective for extracellular bacterial killing, as they do not produce Neutrophil Extracellular Traps (NETs), involved in binding and killing of a variety of microbes (Papayannopoulos and Zychlinsky, 2009).
The Dictyostelium genome contains three putative orthologues of Nox2 (NoxA, B and C), although only NoxA is expressed in vegetative cells (the other two isoforms are expressed during developmental stages) (Lardy et al., 2005; Bedard et al., 2007). While genetic inactivation of noxA, noxB or noxC causes altered multicellular development of Dictyostelium (Lardy et al., 2005), neither a noxA-null nor a noxA/noxB double null mutant show any defect in their ability to feed upon a wide variety of bacterial species, or in their ability to kill ingested Klebsiella bacteria (Lardy et al., 2005; Benghezal et al., 2006). Apparently, in Dictyostelium generation of superoxide by the NADPH oxidases NoxA and B plays mostly a role in signalling and is dispensable for efficient killing of Klebsiella. It remains to be seen if this result would hold true in different conditions (as in a triple noxABC KO), or when the killing of other bacterial species is considered.
Another protein potentially involved in intracellular bacterial killing is NRAMP1, a metal ion transporter present in the phagosomal membrane of mammalian macrophages (Nevo and Nelson, 2006) and of Dictyostelium (Peracino et al., 2006). It has been shown that NRAMP1 is essential for the ability of mice to efficiently kill Mycobacterium bovis and Salmonella typhimurium (Vidal et al., 1995; White et al., 2005). NRAMP1 uses the proton gradient generated by the activity of the vacuolar H+-ATPase to transport manganese and iron out of the phagosome, generating a metal-ion-depleted environment unfavourable for survival and replication of bacteria (Soldati and Neyrolles, 2012). Indeed, nramp1 KO Dictyostelium cells allow more efficient intracellular replication of Mycobacterium avium and Legionella pneumophila. These cells do not however exhibit any defect in their ability to feed upon or to kill non-pathogenic Klebsiella bacteria (Lelong et al., 2011).
Lysosomal hydrolases may also be responsible for the degradation and killing of different microbial species (Kornfeld and Mellman, 1989). For example, genetic inactivation of cathepsin G in mice renders them more sensitive to S. aureus infections, and this is paralleled by a decrease in the ability of neutrophils to kill these pathogens (Reeves et al., 2002). In mammals, other peptidases, as elastase and proteinase 3, are involved in the conversion to the active form of antimicrobial peptides (as cathelicidins), which are able to efficiently kill many bacteria in vitro and in vivo (Zanetti, 2005).
The Dictyostelium genome exhibits a large array of enzymes expected to hydrolyse carbohydrates (such as lysozymes and β-hexosaminidases) and proteins (as cathepsins) (Table 1), and many of them have been localized by proteomics to the phagosomal compartment (Gotthardt et al., 2002; 2006,; Boulais et al., 2010; Journet et al., 2012). To date the best-characterized Dictyostelium lytic enzyme is AlyA, which belongs to a new family of amoeba lysozymes with low levels of similarity to metazoan lysozymes. AlyA is responsible for almost 50% of the total cellular lysozyme activity, and alyA KO cells cannot grow efficiently upon non-pathogenic bacteria (E. coli, Klebsiella and Micrococcus). However, with successive passages in bacterial lawns, mutant cells recover their ability to grow upon bacteria, an effect linked to increased phagocytosis (Muller et al., 2005). Altogether, these observations provide no direct evidence for a role of AlyA in killing, and instead uncover an unexpected link between lysozyme activity and the regulation of phagocytosis. Clearly, cells with a decreased lysozyme activity are eventually able to compensate for this loss, pointing to the existence of other killing mechanisms. It is possible that combining alyA knockout with other mutations will reveal better its specific role in bacterial killing.
Table 1.
Role of various gene products in bacterial sensing and killing in Dictyostelium
| Gene | Dictybase ID | Molecular identity | KO phenotype – mammaliana | KO phenotype – Dictyostelium | Reference | |
|---|---|---|---|---|---|---|
| Candidates by analogy with mammalian system | alyA | DDB_G0275123 | Lysozyme | NAb |
|
Muller et al. (2005) |
| catD | DDB_G0279411 | Cathepsin D |
|
|
Journet et al. (1999) | |
| lvsB | DDB_G0271504 | LYSosomal Trafficking regulator (LYST) homologue |
|
|
Cornillon et al. (2002); Harris et al. (2002) | |
| nox2 | DDB_G0289653 | NADPH oxidase, large subunit |
|
|
Lardy et al. (2005) | |
| nramp | DDB_G0276973 | Fe3+/Mn2+ transporter |
|
|
Peracino et al. (2006) | |
| wshA | DDB_G0292878 | WASP and SCAR homologue |
|
|
Carnell et al. (2011) | |
| Candidates by random mutagenesis | fspA | DDB_G0277237 | Putative GPCR-like protein | NA |
|
Lima et al. (2014) |
| kil1 | DDB_G0267630 | Sulfotransferase | NAc |
|
Benghezal et al. (2006) | |
| kil2 | DDB_G0279183 | Type V Mg2+ P-ATPase |
|
|
Lelong et al. (2011) | |
| phg1a | DDB_G0267444 | TM9-family protein |
|
|
Cornillon et al. (2000); Lozupone et al. (2009) | |
| tirA | DDB_G0289237 | TIR domain-containing protein | NAd |
|
Chen et al. (2007) |
NA: no KO available for the homologous gene in mammalian systems.
Lysozymes have primarily a bacteriolytic function, and are involved in bacterial killing and immune response in general.
In mammalian cells, sulfation reactions play a role on epitope generation for ligands of cell adhesion receptors, although no role on bacterial sensing and killing has been described to date.
TIR domains in mammalian proteins (as MyD88, interleukin-1 receptor and TLRs) are involved in interactions between the TLRs and signal-transduction components.
Cathepsins are another important group of lysosomal hydrolases involved in protein degradation and recycling, and playing a role in several physiological processes in mammalian organisms, as antigen presentation, bone remodelling and hormone processing (Reiser et al., 2010), besides their well-established role in degrading bacterial wall components and virulence factors (Thorne et al., 1976; Carrasco-Marin et al., 2009; Flannagan et al., 2009). Cathepsin D is one of the most abundant proteases in mammalian lysosomes (Kato et al., 1972), and it is involved in killing of Streptococcus, Mycobacterium and Listeria (del Cerro-Vadillo et al., 2006; Bewley et al., 2011). Dictyostelium genome possesses more than 30 genes annotated as cathepsins or cysteine proteases. Cathepsin D is also a major marker for Dictyostelium lysosome maturation, but disruption of the gene does not impair the ability of cells to feed upon Klebsiella (Journet et al., 1999).
Affecting proteins responsible for phagosome maturation may also be expected to impair bacterial killing. Dictyostelium WASH protein, as other metazoan orthologues, has been implicated in vesicular trafficking and phagosome maturation (Carnell et al., 2011; King et al., 2013). Cells lacking WASH exhibit impaired phagosomal proteolysis and reduced amounts of lysosomal hydrolases (such as cathepsins and lysozymes), yet their growth on non-pathogenic Klebsiella and Bacillus is not affected, suggesting that their killing activity is not significantly reduced. In contrast, they grow inefficiently on several other bacterial strains or species [e.g. a pathogenic encapsulated KP52145 Klebsiella, or an attenuated quorum-sensing deficient P. aeruginosa strain (King et al., 2013)]. The ability of the wash mutant cells to kill ingested bacteria was not directly measured.
Another Dictyostelium protein directly involved in lysosome maturation is LvsB, an orthologue of the mammalian lysosomal trafficking regulator LYST. In Dictyostelium, as in mammalian cells, this protein regulates lysosome biogenesis, acidification and secretion (Cornillon et al., 2002; Harris et al., 2002; Charette and Cosson, 2007; Kypri et al., 2007). As seen for wash KO cells, lvsB KO cells also have specific growth defects: they are unable to feed upon M. luteus and some pathogenic Klebsiella strains, but can grow as efficiently as WT cells on other bacteria, as Bacillus and E. coli.
Overall, the general conclusion of these various attempts is that no single gene product targeted so far played a crucial role in intracellular killing of ingested non-pathogenic bacteria. This may be due to the presence of redundant killing mechanisms, or to the fact that the most important gene products were not tested. Dictyostelium provides a convenient model to inactivate specific genes and assess their role in intracellular killing.
Obscure words from the slime
Another way to test the depth of our knowledge of intracellular killing mechanisms, and to simultaneously test if functional redundancy prevents Dictyostelium mutants from exhibiting killing defects, is to isolate randomly killing-deficient mutants, and try to make sense of the gene products identified in this manner. The first killing-deficient mutant was identified serendipitously: phg1a KO cells, initially characterized as defective in adhesion to and ingestion of latex beads (Cornillon et al., 2000), were later found to also kill inefficiently ingested Klebsiella bacteria (Benghezal et al., 2006). This defect presumably accounts for the inability of phg1a KO cells to feed and grow upon Klebsiella bacteria. In the same study, Kil1 was identified as a high-copy suppressor of the killing defect phg1a KO cells, and kil1 KO cells were shown to kill inefficiently Klebsiella bacteria. The role of Phg1a in intracellular killing is probably due to the fact that it controls intracellular transport and stability of membrane proteins (Froquet et al., 2012), and that in its absence the Kil1 protein is unstable and virtually depleted from cells (Le Coadic et al., 2013) (Fig. 2 ). Kil1 is a sulfotransferase, and no direct link has previously been established between sulfation of host proteins and host–pathogen interactions in metazoans. Sulfation has been described to play a role in receptor–ligand interactions (Hemmerich and Rosen, 2000; Park et al., 2010), but its role in intracellular killing remains to be determined.
Figure 2.
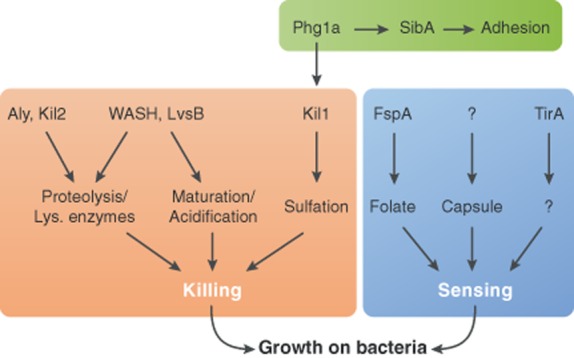
Molecular mechanisms involved in Dictyostelium sensing and killing of bacteria. Sensing of Klebsiella bacteria involves different players, notably FspA for bacteria-secreted folate, and a yet-unknown receptor of capsule components. TirA may also play a regulatory role in sensing. Mechanisms related to intracellular killing have been more extensively unravelled. Lysosomal activity (as denoted by the proteolytic efficiency inside the phagosome) and phagosomal biogenesis (including proper acidification and maturation) are major factors implicated in efficient killing. Proper regulation of adhesion and sulfation processes has also been implicated in successful killing.
A random screen for Dictyostelium mutants identified Kil2 as another essential gene for efficient growth on Klebsiella. Kil2 is a putative magnesium transporter in the phagosomal membrane, and its absence leads to diminished activity of phagosomal proteases, and to inefficient intracellular killing of Klebsiella (Lelong et al., 2011). Both killing and proteolytic activity are restored by supplementation with magnesium, indicating that ionic homeostasis inside the phagosome is essential for proper hydrolytic activity and efficient bacteria processing. The role of the ionic composition of phagosomes in bacterial killing is discussed nicely in a recent review (Soldati and Neyrolles, 2012). Kil2 belongs to the group V Ptype ATPase family, and no member of this family has been previously implicated in host–pathogen interactions, nor has magnesium been previously linked to intraphagosomal killing mechanisms.
It is remarkable how little we can comment about these findings at this stage, or try to delineate mechanistic relationships among them and with mammalian gene products implicated in killing. Phg1, Kil1 and Kil2 have potential orthologues in human (Table 1), but none of them have been linked previously to host–pathogen interactions. Another important observation is that although these three gene products are necessary for intracellular killing of Klebsiella, they are dispensable for efficient killing of non-pathogenic P. aeruginosa or of Gram-positive B. subtilis. This suggests that Dictyostelium cells may use several independent mechanisms to kill different bacterial species or strains.
The taste of bugs
In animals, the recognition of invading microorganisms is essential to trigger an adequate antibacterial response and a successful defence against infections. Several proteins involved in the recognition machinery have been identified, most notably Toll receptors in Drosophila fruit flies (Lemaitre et al., 1996). The discovery and characterization of these receptors was the first step in the elucidation of the different molecular pathways involved in the recognition of distinct bacterial MAMPs (microbial-associated molecular patterns) (Brennan and Anderson, 2004). Today, the Toll-like receptor (TLR) family has been extensively characterized in mammals, and each member has been linked to recognition of specific microbial components, such as lipopolysaccharides, peptidoglycans or nucleic acids. TLRs are the paradigm for differential pathogen recognition in metazoans (Hoffmann and Reichhart, 2002; Akira et al., 2006), together with several other membrane and cytoplasmic receptors. Cytosolic NOD receptors have been implicated in the recognition of peptidoglycan and flagellin; C-type lectin receptors can recognize lipopolysaccharides, capsule polysaccharides and glycolipids; and scavenger receptors can detect lipopolysaccharides, lipoteichoic acid and several bacterial proteins (Pluddemann et al., 2011).
In multicellular hosts, recognition of potentially harmful microorganisms can easily be distinguished from response to other physiological signals. Ideally it should elicit danger signals that ultimately allow the host to eliminate invading pathogens. The distinction may be subtler in amoebae, for which microorganisms are both a source of food and potential pathogens. Theoretically, to successfully feed upon bacteria, amoebae may need to sense bacterial factors and migrate towards them, and to adapt their physiology to optimize ingestion, killing and digestion of the available bacteria. Ideally, they should also be able to recognize pathogenic bacteria and avoid them. These largely speculative considerations have essentially not been tested so far, most probably due to the reduced number of studies addressing these or related issues. The Dictyostelium genome exhibits no clear Toll-like receptors, but several putative orthologues to other bacterial-sensing receptors (Cosson and Soldati, 2008), the function of which remains to be established. We describe below this nascent field of research, stemming from a few recently published studies.
Several large-scale transcriptional studies have been conducted to analyse changes in gene expression when Dictyostelium cells are exposed to different bacteria, e.g. E. coli, Pseudomonas and Legionella (Benghezal et al., 2006; Farbrother et al., 2006; Carilla-Latorre et al., 2008; Sillo et al., 2008; Nasser et al., 2013). These studies clearly demonstrate that Dictyostelium cells confronted to different bacterial species exhibit very different gene expression profiles. However, it is not clear whether these differences are caused by specific recognition of bacteria by amoebae, or result from a long-term metabolic adaptation of Dictyostelium to various food sources.
One founding study on bacterial sensing identified TirA, a cytosolic TIR-domain containing protein whose eukaryotic orthologues are involved in specific antimicrobial response, and showed that its genetic inactivation rendered Dictyostelium cells unable to feed upon Klebsiella (Chen et al., 2007). However, up to now it is not clear if and how this protein is involved in bacterial recognition or subsequent downstream pathways.
Another study (Nasser et al., 2013) also identified by random mutagenesis genes essential for growth on various bacteria. The ability of amoeba cells to feed upon Gram(+) bacteria appears to be dependent on proteins involved in bacterial cell wall breakdown and N-glycosylation (probably by modulating the activity of yet-unknown membrane receptors), while response to Gram(−) bacteria appears to involve AlyL lysozyme activity, in conjunction with the Spc3 signal peptidase (which has been proposed to act as a regulator of lysozyme biogenesis). The exact role of these proteins in bacterial sensing remains to be established. It is likely that the vast majority of gene products involved in bacterial recognition by amoebae, in particular specific receptors, are still not identified today.
In a recent study we made use of the observation that exposure of Dictyostelium to bacteria increases their motility to show that Dictyostelium cells can recognize different types of bacteria, and that recognition of Klebsiella involves at least two distinct pathways: one sensing the folate produced by bacteria, the other one responding to bacterial capsule (Lima et al., 2014) (Fig. 2). Moreover, we also identified one molecular component of the folate-sensing pathway: FspA, a putative G-protein coupled receptor (GPCR) protein that may act as a receptor or a regulator in this specific sensing pathway.
Collectively, these studies support the idea that Dictyostelium amoebae do possess several specific bacterial-sensing mechanisms, and that these mechanisms are essential to ensure efficient growth in the presence of bacteria.
Conclusion
Dictyostelium discoideum has been used as a powerful model to study cell motility and phagocytosis, and the underlying mechanisms have proven largely similar to those identified in mammalian cells. There is no clear evidence today that Dictyostelium and mammalian cells use similar mechanisms to kill ingested microorganisms. Mostly, this ambiguity results from the fact that our understanding of killing mechanisms is very incomplete in these two different cell types. Recognition of bacteria by phagocytic cells may also be a prerequisite for efficient intracellular killing, and only very few studies have analysed this function in Dictyostelium. It is likely that, as our knowledge progresses, we will be able to distinguish which sensing and killing mechanisms are conserved between amoebae and mammalian phagocytes, and which ones are specific for each system.
References
- Akira S, Uematsu S. Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bedard K, Lardy B. Krause KH. NOX family NADPH oxidases: not just in mammals. Biochimie. 2007;89:1107–1112. doi: 10.1016/j.biochi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- Bewley MA, Marriott HM, Tulone C, Francis SE, Mitchell TJ, Read RC, et al. A cardinal role for cathepsin d in co-ordinating the host-mediated apoptosis of macrophages and killing of pneumococci. PLoS Pathog. 2011;7:e1001262. doi: 10.1371/journal.ppat.1001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulais J, Trost M, Landry CR, Dieckmann R, Levy ED, Soldati T, et al. Molecular characterization of the evolution of phagosomes. Mol Syst Biol. 2010;6:423. doi: 10.1038/msb.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzaro S. Eichinger L. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets. 2011;12:942–954. doi: 10.2174/138945011795677782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA. Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- Carilla-Latorre S, Calvo-Garrido J, Bloomfield G, Skelton J, Kay RR, Ivens A, et al. Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 2008;8:109. doi: 10.1186/1471-2180-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, et al. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J Cell Biol. 2011;193:831–839. doi: 10.1083/jcb.201009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Marin E, Madrazo-Toca F, de los Toyos JR, Cacho-Alonso E, Tobes R, Pareja E, et al. The innate immunity role of cathepsin-D is linked to Trp-491 and Trp-492 residues of listeriolysin O. Mol Microbiol. 2009;72:668–682. doi: 10.1111/j.1365-2958.2009.06673.x. [DOI] [PubMed] [Google Scholar]
- del Cerro-Vadillo E, Madrazo-Toca F, Carrasco-Marin E, Fernandez-Prieto L, Beck C, Leyva-Cobian F, et al. Cutting edge: a novel nonoxidative phagosomal mechanism exerted by cathepsin-D controls Listeria monocytogenes intracellular growth. J Immunol. 2006;176:1321–1325. doi: 10.4049/jimmunol.176.3.1321. [DOI] [PubMed] [Google Scholar]
- Charette SJ. Cosson P. A LYST/beige homolog is involved in biogenesis of Dictyostelium secretory lysosomes. J Cell Sci. 2007;120:2338–2343. doi: 10.1242/jcs.009001. [DOI] [PubMed] [Google Scholar]
- Chen G, Zhuchenko O. Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. Recent insights into host–pathogen interactions from Dictyostelium. Cell Microbiol. 2010;12:283–291. doi: 10.1111/j.1462-5822.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, et al. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem. 2000;275:34287–34292. doi: 10.1074/jbc.M006725200. [DOI] [PubMed] [Google Scholar]
- Cornillon S, Dubois A, Bruckert F, Lefkir Y, Marchetti A, Benghezal M, et al. Two members of the beige/CHS (BEACH) family are involved at different stages in the organization of the endocytic pathway in Dictyostelium. J Cell Sci. 2002;115:737–744. doi: 10.1242/jcs.115.4.737. [DOI] [PubMed] [Google Scholar]
- Cosson P. Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Ellson CD, Davidson K, Ferguson GJ, O'Connor R, Stephens LR. Hawkins PT. Neutrophils from p40phox−/− mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing. J Exp Med. 2006;203:1927–1937. doi: 10.1084/jem.20052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbrother P, Wagner C, Na J, Tunggal B, Morio T, Urushihara H, et al. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell Microbiol. 2006;8:438–456. doi: 10.1111/j.1462-5822.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- Flannagan RS, Cosio G. Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Froquet R, Lelong E, Marchetti A. Cosson P. Dictyostelium discoideum: a model host to measure bacterial virulence. Nat Protoc. 2009;4:25–30. doi: 10.1038/nprot.2008.212. [DOI] [PubMed] [Google Scholar]
- Froquet R, le Coadic M, Perrin J, Cherix N, Cornillon S. Cosson P. TM9/Phg1 and SadA proteins control surface expression and stability of SibA adhesion molecules in Dictyostelium. Mol Biol Cell. 2012;23:679–686. doi: 10.1091/mbc.E11-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblatt D. Thrasher AJ. Chronic granulomatous disease. Clin Exp Immunol. 2000;122:1–9. doi: 10.1046/j.1365-2249.2000.01314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D, Warnatz HJ, Henschel O, Bruckert F, Schleicher M. Soldati T. High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol Biol Cell. 2002;13:3508–3520. doi: 10.1091/mbc.E02-04-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M. Soldati T. Proteomics fingerprinting of phagosome maturation and evidence for the role of a Galpha during uptake. Mol Cell Proteomics. 2006;5:2228–2243. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- Haas A. The phagosome: compartment with a license to kill. Traffic. 2007;8:311–330. doi: 10.1111/j.1600-0854.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- Harris E, Wang N, Wu Wl WL, Weatherford A, De Lozanne A. Cardelli J. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol Biol Cell. 2002;13:656–669. doi: 10.1091/mbc.01-09-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich S. Rosen SD. Carbohydrate sulfotransferases in lymphocyte homing. Glycobiology. 2000;10:849–856. doi: 10.1093/glycob/10.9.849. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Journet A, Chapel A, Jehan S, Adessi C, Freeze H, Klein G. Garin J. Characterization of Dictyostelium discoideum cathepsin D. J Cell Sci. 1999;112:3833–3843. doi: 10.1242/jcs.112.21.3833. [DOI] [PubMed] [Google Scholar]
- Journet A, Klein G, Brugiere S, Vandenbrouck Y, Chapel A, Kieffer S, et al. Investigating the macropinocytic proteome of Dictyostelium amoebae by high-resolution mass spectrometry. Proteomics. 2012;12:241–245. doi: 10.1002/pmic.201100313. [DOI] [PubMed] [Google Scholar]
- Kalima P, Masterton RG, Roddie PH. Thomas AE. Lactobacillus rhamnosus infection in a child following bone marrow transplant. J Infect. 1996;32:165–167. doi: 10.1016/s0163-4453(96)91622-9. [DOI] [PubMed] [Google Scholar]
- Kato T, Kojima K. Murachi T. Proteases of macrophages in rat peritoneal exudate, with special reference to the effects of actinomycete protease inhibitors. Biochim Biophys Acta. 1972;289:187–193. doi: 10.1016/0005-2744(72)90121-0. [DOI] [PubMed] [Google Scholar]
- King JS, Gueho A, Hagedorn M, Gopaldass N, Leuba F, Soldati T. Insall RH. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol Biol Cell. 2013;24:2714–2726. doi: 10.1091/mbc.E13-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S. Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Kypri E, Schmauch C, Maniak M. De Lozanne A. The BEACH protein LvsB is localized on lysosomes and postlysosomes and limits their fusion with early endosomes. Traffic. 2007;8:774–783. doi: 10.1111/j.1600-0854.2007.00567.x. [DOI] [PubMed] [Google Scholar]
- Lardy B, Bof M, Aubry L, Paclet MH, Morel F, Satre M. Klein G. NADPH oxidase homologs are required for normal cell differentiation and morphogenesis in Dictyostelium discoideum. Biochim Biophys Acta. 2005;1744:199–212. doi: 10.1016/j.bbamcr.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Le Coadic M, Froquet R, Lima WC, Dias M, Marchetti A. Cosson P. Phg1/TM9 proteins control intracellular killing of bacteria by determining cellular levels of the Kil1 sulfotransferase in Dictyostelium. PLoS ONE. 2013;8:e53259. doi: 10.1371/journal.pone.0053259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelong E, Marchetti A, Gueho A, Lima WC, Sattler N, Molmeret M, et al. Role of magnesium and a phagosomal P-type ATPase in intracellular bacterial killing. Cell Microbiol. 2011;13:246–258. doi: 10.1111/j.1462-5822.2010.01532.x. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM. Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Lima WC, Balestrino D, Forestier C. Cosson P. Two distinct sensing pathways allow recognition of Klebsiella pneumoniae by Dictyostelium amoebae. Cell Microbiol. 2014;16:311–323. doi: 10.1111/cmi.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone F, Perdicchio M, Brambilla D, Borghi M, Meschini S, Barca S, et al. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Rep. 2009;10:1348–1354. doi: 10.1038/embor.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern NN, Cowburn AS, Porter L, Walmsley SR, Summers C, Thompson AA, et al. Hypoxia selectively inhibits respiratory burst activity and killing of Staphylococcus aureus in human neutrophils. J Immunol. 2011;186:453–463. doi: 10.4049/jimmunol.1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller I, Subert N, Otto H, Herbst R, Ruhling H, Maniak M. Leippe M. A Dictyostelium mutant with reduced lysozyme levels compensates by increased phagocytic activity. J Biol Chem. 2005;280:10435–10443. doi: 10.1074/jbc.M411445200. [DOI] [PubMed] [Google Scholar]
- Nasser W, Santhanam B, Miranda ER, Parikh A, Juneja K, Rot G, et al. Bacterial discrimination by dictyostelid amoebae reveals the complexity of ancient interspecies interactions. Curr Biol. 2013;23:862–872. doi: 10.1016/j.cub.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo Y. Nelson N. The NRAMP family of metal-ion transporters. Biochim Biophys Acta. 2006;1763:609–620. doi: 10.1016/j.bbamcr.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V. Zychlinsky A. NETs: a new strategy for using old weapons. Trends Immunol. 2009;30:513–521. doi: 10.1016/j.it.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Park CJ, Han SW, Chen X. Ronald PC. Elucidation of XA21-mediated innate immunity. Cell Microbiol. 2010;12:1017–1025. doi: 10.1111/j.1462-5822.2010.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracino B, Wagner C, Balest A, Balbo A, Pergolizzi B, Noegel AA, et al. Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic. 2006;7:22–38. doi: 10.1111/j.1600-0854.2005.00356.x. [DOI] [PubMed] [Google Scholar]
- Pluddemann A, Mukhopadhyay S. Gordon S. Innate immunity to intracellular pathogens: macrophage receptors and responses to microbial entry. Immunol Rev. 2011;240:11–24. doi: 10.1111/j.1600-065X.2010.00989.x. [DOI] [PubMed] [Google Scholar]
- Reeves EP, Lu H, Jacobs HL, Messina CG, Bolsover S, Gabella G, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- Reiser J, Adair B. Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillo A, Bloomfield G, Balest A, Balbo A, Pergolizzi B, Peracino B, et al. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics. 2008;9:291. doi: 10.1186/1471-2164-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T. Neyrolles O. Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic. 2012;13:1042–1052. doi: 10.1111/j.1600-0854.2012.01358.x. [DOI] [PubMed] [Google Scholar]
- Steinert M. Pathogen-host interactions in DictyosteliumLegionellaMycobacterium and other pathogens. Semin Cell Dev Biol. 2011;22:70–76. doi: 10.1016/j.semcdb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Thorne KJ, Oliver RC. Barrett AJ. Lysis and killing of bacteria by lysosomal proteinases. Infect Immun. 1976;14:555–563. doi: 10.1128/iai.14.2.555-563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S, Gros P. Skamene E. Natural resistance to infection with intracellular parasites: molecular genetics identifies Nramp1 as the Bcg/Ity/Lsh locus. J Leukoc Biol. 1995;58:382–390. doi: 10.1002/jlb.58.4.382. [DOI] [PubMed] [Google Scholar]
- White JK, Mastroeni P, Popoff JF, Evans CA. Blackwell JM. Slc11a1-mediated resistance to Salmonella enterica serovar Typhimurium and Leishmania donovani infections does not require functional inducible nitric oxide synthase or phagocyte oxidase activity. J Leukoc Biol. 2005;77:311–320. doi: 10.1189/jlb.0904546. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- Zanetti M. The role of cathelicidins in the innate host defenses of mammals. Curr Issues Mol Biol. 2005;7:179–196. [PubMed] [Google Scholar]


