SUMMARY
Homeotic (HOX) genes are dysregulated in multiple malignancies including several AML subtypes. We demonstrate that H3K79 dimethylation (H3K79me2) is converted to mono-methylation (H3K79me1) at HOX loci as hematopoietic cells mature thus coinciding with a decrease in HOX gene expression. We show that H3K79 methyltransferase activity as well as H3K79me1 to H3K79me2 conversion is regulated by the DOT1L co-factor AF10. AF10 inactivation reverses leukemia-associated epigenetic profiles, precludes abnormal HOXA gene expression and impairs the transforming ability of MLL-AF9, MLL-AF6 or NUP98-NSD1 fusions – mechanistically distinct HOX-activating oncogenes. Furthermore, NUP98-NSD1 transformed cells are sensitive to small-molecule inhibition of DOT1L. Our findings demonstrate that pharmacological inhibition of the DOT1L/AF10 complex may provide therapeutic benefit in an array of malignancies with abnormal HOXA gene expression.
INTRODUCTION
The clustered homeobox (HOX) genes are conserved throughout evolution and play important roles in development (Lappin et al., 2006). Multiple human malignancies, including approximately 50% of acute myeloid leukemia (AML), show dysregulated expression of the clustered homeobox genes (Argiropoulos and Humphries, 2007). Specific members of the HOX cluster are critical for hematopoietic stem cell (HSC) self-renewal and the expression of a particular sub-set of HOXA gene cluster, specifically the Hoxa7-10 genes, is sharply down-regulated when murine HSCs differentiate to granulocyte macrophage progenitors (GMPs) (Krivtsov et al., 2006). HOXA gene repression in myeloid progenitors such as GMPs is critical, since aberrant HOXA gene expression is a central component of a leukemogenic gene expression program driven by diverse oncogenes (Argiropoulos and Humphries, 2007).
One of the classic examples of a malignancy with aberrant HOXA activation is leukemia with rearrangements of the Mixed Lineage Leukemia gene (MLL), an aggressive disease with limited treatment options and poor survival rates. In these leukemias, MLL is fused to one of more than 60 different fusion partners leading to the formation of dominantly acting oncogenic fusion proteins (Krivtsov and Armstrong, 2007). Expression of the MLL-fusion oncoprotein perpetuates inappropriate high level HOXA gene expression in GMPs and induces leukemias in mice that are arrested at a GMP-like differentiation stage (Krivtsov et al., 2006; Somervaille and Cleary, 2006). Some of the recurrent C-terminal fusion partners of MLL such as AF9, ENL, AF17, and AF10 normally interact with the histone methyl-transferase DOT1L (reviewed in Deshpande et al., 2012). DOT1L is the sole histone methyltransferase that catalyzes histone 3 lysine 79 monomethylation (H3K79me1), dimethylation (H3K79me2) and trimethylation (H3K79me3) (reviewed in Nguyen and Zhang, 2011), chromatin modifications that are widely associated with highly expressed genes in mammalian cells (Steger et al., 2008). MLL-fusions with DOT1L interacting proteins are believed to misdirect DOT1L to the promoters of HOXA genes, leading to H3K79 methylation and constitutive activation of these genes in MLL-rearranged leukemia. However, recent studies have reported that MLL-fusions with no apparent DOT1L recruiting activity are also dependent on the H3K79 methylating activity of DOT1L for transformation (Chang et al., 2010; Deshpande et al., 2013). These studies offer an alternative, though not mutually exclusive possibility: H3K79 methylation may be broadly important for the epigenetic regulation of specific HOXA genes and HOXA gene mediated oncogenesis. The most interesting implication of this alternative hypothesis is that the transforming activity of other HOX-activating oncogenes may also depend on H3K79 methylation.
There is tremendous interest in DOT1L as a potential therapeutic target in MLL-rearranged leukemia and clinical trials with a DOT1L small molecule inhibitor are currently ongoing. However, little is known about potential co-factors that mediate the chromatin modifying activity of DOT1L. DOT1L has been identified as a component of large multi-protein complexes that are associated with transcribed genes (Mohan et al., 2010; Mueller et al., 2009) although critical constituents of the DOT1L complex are not well defined. DOT1L binds to the PHD and leucine zipper containing protein AF10, an interaction that is highly conserved throughout evolution. Despite the fact that AF10 is involved in recurrent chromosomal translocations that fuse it to one of at least five different fusion partners apart from MLL in human leukemia (Brandimarte et al., 2013; Dreyling et al., 1996; Soler et al., 2013; Zhang et al., 2012), there have been few studies aimed at understanding the functional and physiological role of AF10 in mammalian cells (Chamorro-Garcia et al., 2012; Linder et al., 2000). AF10-rearranged (AF10-R) leukemias show two recurrent features: consistent retention of the leucine rich DOT1L-interacting AF10 domain in the fusion and the aberrant upregulation of HOXA cluster genes. Strikingly, these two features span all AF10-R leukemias regardless of the N-terminal fusion partner. These observations, taken together with the fact that DOT1L as well as AF10 orthologs regulate the expression of the bithorax gene cluster in Drosophila (Perrin et al., 2003; Shanower et al., 2005) strongly suggest that DOT1L and AF10 may be generally involved in HOXA gene regulation.
RESULTS
Normal and malignant HOXA gene expression correlates with higher H3K79 methylation states
We wanted to test a potential role for DOT1L in the regulation of HOXA gene expression in murine hematopoietic stem and progenitor cells. We sorted lineage−Sca-1+c-Kit+ (LSK) cells, which are enriched for HSCs, from bone marrow (BM) prepared from Dot1lfl/fl mice crossed with interferon inducible Mx1-Cre transgenic mice in which Dot1l deletion was induced upon administration of polyinosinic-polycytidylic acid (pIpC) and compared these LSKs to their pIpC injected Dot1l wild-type counterparts. Gene expression analysis showed that Hoxa9 and Meis1 were significantly down-regulated in LSKs with homozygous Dot1l deletion (Figure 1A) demonstrating a role for DOT1L in the normal regulation of these homeobox genes.
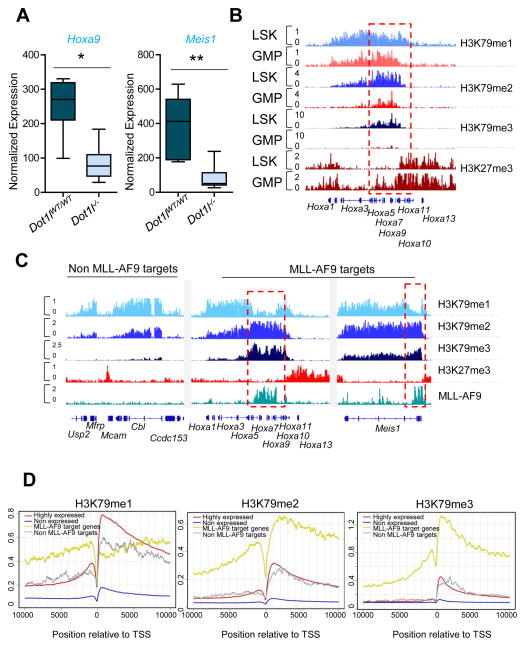
Figure 1. Histone methylation profiles in normal cells and MLL-AF9 leukemia.
(A) Expression of Hoxa9 and Meis1 in LSKs sorted from Dot1lfl/fl or Dot1lWT/WT mice crossed with Mx1-Cre mice treated with pIpC 10 days before. The box plot shows normalized expression values from 6 independent replicates. Whiskers represent the upper and lower limits of the range. Boxes represent the first and third quartile, and the line represents the median. (B) Representative profiles for ChIP-seq using anti-H3K79me1, H3K79me2, H3K79me3 and H3K27me3 antibodies in LSK and GMP cells at the HOXA cluster. (C) Representative profiles for ChIP-seq using anti-H3K79me1, H3K79me2, H3K79me3 and H3K27me3 antibodies at various genomic loci in MLL-AF9 leukemia cells. Binding of MLL-AF9 fusion gene is shown at the bottom in green. In both B and C, the y-axis scales represent read density per million sequenced reads and the Hoxa5-10 genomic region is marked by a dotted red rectangle. (D) Meta-analysis of averaged ChIP-seq signal at four sets of genes across a +10 kb to −10 kb genomic region around the transcriptional start site. See also Figure S1 and Table S1.
Since the state of H3K79 methylation could have different functions, we assessed H3K79me1, H3K79me2 and H3K79me3 in LSKs, which express high levels of Hoxa7-10 and Meis1 genes, and compared them to GMPs, which express much reduced levels of these genes (Krivtsov et al., 2006). We performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) using well-characterized antibodies that predominantly recognize H3K79me1, H3K79me2, or H3K79me3 (Figure S1A). Each of the three H3K79 methylation states directly correlated with gene expression levels and showed broadly overlapping patterns across the genome (Figure S1B). H3K79me3 demonstrated a more contracted pattern surrounding the transcription start sites (TSS) (Figure S1B). Interestingly, this analysis showed the sharp decrease in Hoxa7-10 expression in GMPs compared to LSKs was accompanied by a diminution in H3K79me2 and H3K79me3 at the HOXA genes with minimal changes in H3K79me1 (Figure 1B). In fact some genes such as Hoxa9 showed higher H3K79me1 in GMPs than in LSKs. Furthermore, the LSK-GMP transition was associated with an encroachment of H3K27me3 into the Hoxa7-10 gene cluster (Figure 1B), consistent with the observed down-regulation of these transcripts. These results demonstrate that HOXA gene expression is correlated with higher H3K79 methylation states during hematopoietic differentiation and suggests that H3K79me1 may not be sufficient to maintain high-level HOXA gene expression.
MLL-rearranged leukemias show constitutive expression of the Hoxa7-10 gene cluster and Meis1, a process that is dependent on H3K79 methylation. We therefore assessed H3K79 methylation states in LSK-derived MLL-AF9 leukemias by ChIP-seq. Similar to LSKs and GMPs, all three states of H3K79 methylation showed overlapping profiles with H3K79me3 particularly enriched at genes where the MLL-AF9 fusion protein binds (Bernt et al., 2011) (Figure S1C and Table S1). Strikingly, in contrast to the genome wide profiles of H3K79 methylation, promoters of HOXA genes were characterized by low levels of H3K79me1 and high levels of H3K79me2/3 (Figure 1C and S1D). The increased conversion of H3K79 methylation to higher methylation states at the Hoxa7-10 genes was accompanied by the absence of the repressive H3K27me3 mark as expected from the high level expression of these genes in MLL-AF9 leukemia cells (Figure 1C). We then conducted a meta-analysis of H3K79me1/2/3 relationships across MLL-AF9 target genes. Strikingly, MLL-AF9 target genes showed low level H3K79me1 and high level H3K79me2/3 profile observed on the Hoxa7-10 and Meis1 genomic loci (Figure 1D). Meta-analysis of the averaged H3K79me1 and H3K79me2 profiles across the body of MLL-AF9 target genes showed that the H3K79me1 dip was especially pronounced near the promoter proximal regions of MLL-AF9 target genes (Figure S1D). In contrast, control sets of non MLL-AF9 target genes showed similar profiles of H3K79me1 and H3K79me2 across the gene body (Figure S1D). These results demonstrate that similar to normal hematopoietic differentiation, higher degrees of H3K79me2/3, rather than H3K79me1, correlate with high HOXA gene expression as well as exclusion of the repressive H3K27me3 from the Hoxa7-10 locus. This suggests that the expression of HOXA, and perhaps other genes, is critically dependent on the conversion of H3K79me1 to H3K79me2.
AF10 is critical for higher H3K79 methylated states
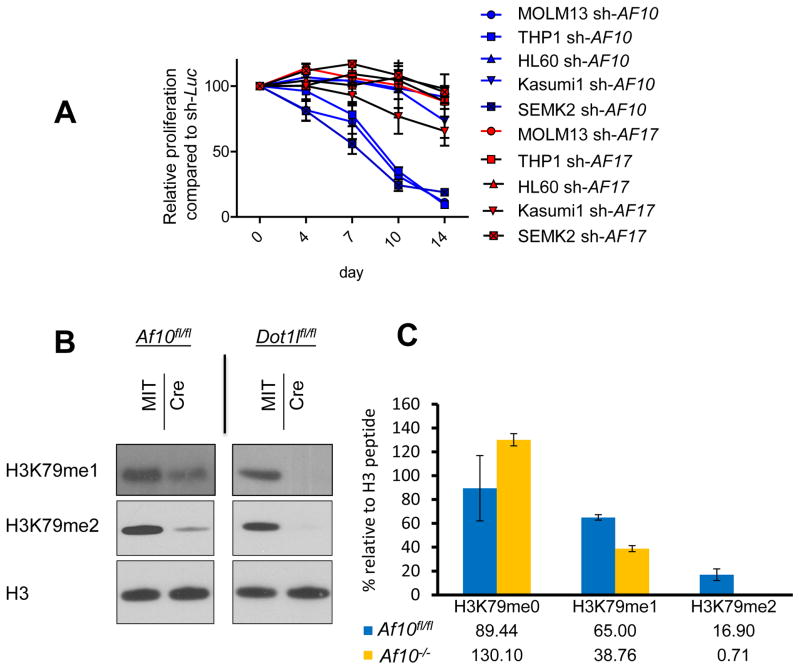
Given that H3K79me2, but not H3K79me1, correlated with high level expression of select genes, we wanted to determine how the conversion of H3K79me1 to H3K79me2 is regulated. We found that the expression of Dot1l transcripts do not correlate with Hoxa5-10 gene expression in LSK compared to GMP cells (Figure S2A) suggesting that Dot1l transcript levels do not explain the HOXA locus associated decrease in H3K79me2 and H3K79me3 in GMPs. There is increasing appreciation of the role of co-factor proteins in the modulation of specific methylation states by chromatin modifying enzymes (Dou et al., 2006; Pasini et al., 2004). We therefore wondered whether a co-factor of DOT1L could be responsible for enhancing the ability of DOT1L to catalyze increased H3K79 di/tri-methylation at the HOXA genes. Even though several DOT1L interacting proteins have been reported (Mohan et al., 2010; Park et al., 2010), the composition of the DOT1L complex in human leukemia cells remains undefined. We purified the DOT1L complex from a human MLL-rearranged and a MLL-germline cell line using previously described protocols (Kim et al., 2009) and identified a number of DOT1L interacting proteins, notably AF10, AF17, ILF2, MEF2C, HDAC1, and CDK9 (Figure S2B and Table S2). Of note, other than AF10 and AF17, most other proteins were either found in low abundance or were occasionally also retrieved from control cells (Table S2). We focused our attention on the PHD and octapeptide-motif leucine zipper containing paralogous proteins AF10 and AF17 for further studies. In order to assess a potential role for AF10 or AF17 in the conversion from H3K79me1 to H3K79me2, we performed shRNA knockdown experiments in human leukemia cell lines. We found that AF10 suppression produced a clear reduction in H3K79me2, but AF17 suppression had no observable effect on H3K79 dimethylation (Figure S2C). In contrast to DOT1L suppression, we observed that AF10 suppression led to the reduction of H3K79me2 but not H3K79me1 (Figure S2C and S2D). Next we performed proliferation assays with shRNAs targeting AF10 or AF17 in human MLL-rearranged or MLL-germline leukemia cell lines. We found that AF10 knockdown significantly impaired the proliferation of MLL-rearranged cell lines MOLM13, SEMK2 and THP1 but not the MLL-germline leukemia cell lines HL60 and Kasumi1 (Figure 2A and Fig. S2E). In contrast, none of the cell lines were sensitive to AF17 knockdown suggesting that AF10 plays a more prominent role in H3K79 methylation in MLL-rearranged leukemia (Figure 2A). Moreover, the levels of Af10 but not Af17 correlated with the decrease in HOXA gene expression in murine LSKs compared to GMP cells (Figure S2A) marking AF10 as a prime candidate for further studies.
Figure 2. The impact of genomic Af10 deletion on H3K79 methylation.
(A) Growth of human leukemia cell lines MOLM13 (MLL-AF9), THP1 (MLL-AF9), SEMK2 (MLL-AF4), HL60 (non-MLL-rearranged), and Kasumi1 (non-MLL-rearranged) after transduction with anti-AF10 and anti-AF17 shRNAs. Viable cells were counted every 3 to 4 days, and cell numbers after transduction with an anti-luciferase shRNA (sh-Luc) were set as 100%. (B) Western blots showing H3K79 methylation in HOXA9-MEIS1 transformed clones with indicated floxed allele following empty vector (MIT) or Cre expression. (C) Quantification of integrated signals for area under the curve values of indicated clones. Error bars represent SD. See also Figure S2 and Table S2.
Given the results above, we interrogated AF10 in more detail. We generated a conditional Af10 knockout mouse (Figure S2F). We first assessed the effect of AF10 loss on cells insensitive to the loss of H3K79 methylation to avoid the confounding effects of apoptosis or differentiation that might be seen in DOT1L dependent cells. For this, we used bone marrow cells transformed with HOXA9 and MEIS1 or immortalized fibroblast cell lines generated from Af10fl/fl mice. We confirmed the absence of AF10 both at the genomic and the protein level after Cre mediated excision of Af10 (Figure S2F and Figure S2G). Even though our knockout strategy targeted the 3′ exons of Af10 encoding the OM-LZ domains, we did not observe a truncated AF10 protein with an anti-AF10 N-terminus antibody suggesting that our model generates a more extensive AF10 loss of function (Figure S2G). We did not observe any changes in the DOT1L or AF17 protein level in AF10 deficient cells, suggesting that these complex members are not adversely affected upon AF10 loss (Figure S2G). Moreover, Af10 excision had no significant effect on the colony forming ability of HOXA9-MEIS1 transformed cells (Figure S2H) and Af10 excised single cell clones could be propagated indefinitely (data not shown). Interestingly, AF10 inactivation produced a dramatic decrease in H3K79me2 levels in the HOXA9-MEIS1 transformed cells as well as in the AF10 deficient fibroblasts as assessed by Western blotting, and this loss of H3K79me2 could be rescued by ectopic AF10 expression (Figure 2B and Figure S2I). Notably, we observed that the effect of Af10 deletion on H3K79me1 was less severe than that of Dot1l deletion, which completely eliminated both H3K79me1 and H3K79me2, consistent with DOT1L as the sole H3K79 methyl-transferase (Figure 2B). We next determined H3K79 methylation levels in histones purified from Af10 deleted HOXA9-MEIS1 transformed single cell clones using Mass Spectrometry (MS) and used Dot1l wild-type or deleted clones for comparison. While Dot1l deleted clones harbored essentially undetectable levels of H3K79me1 and H3K79me2, Af10 deleted cells had an average of almost 20 fold reduction in H3K79me2 levels and a much less severe reduction in H3K79me1 levels, recapitulating the results from Western blotting (Figure 2C and Figures S2J and S2K). These results show that loss of AF10 impairs the conversion of H3K79me1 to H3K79me2 in hematopoietic cells.
Inactivation of AF10 impairs the transforming activity of MLL-AF9 and MLL-AF6
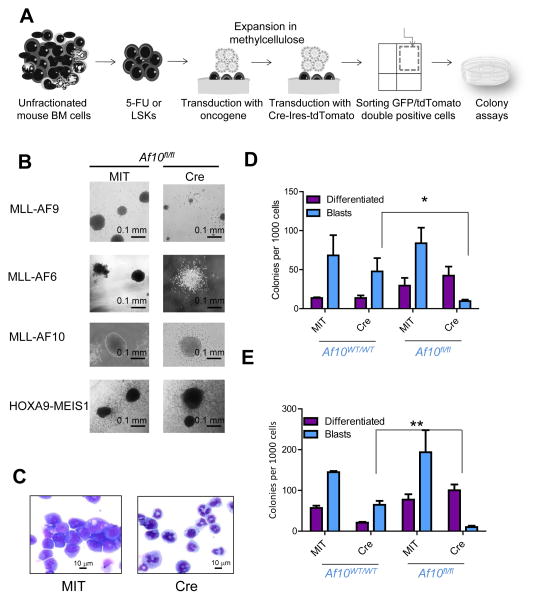
Leukemic transformation driven by different MLL-fusion oncogenes is dependent on DOT1L (Bernt et al., 2011; Chang et al., 2010; Chen et al., 2013; Daigle et al., 2013; Deshpande et al., 2013; Jo et al., 2011; Nguyen et al., 2011). We determined whether the transforming activity of MLL-fusions was dependent on AF10, since it is important for generation of H3K79me2. We used MLL-fusions with fusion partners that interact with DOT1L (MLL-AF9 and MLL-AF10) or an MLL-fusion that is believed to operate through a distinct mechanism (MLL-AF6). We used BM-derived LSKs from wild-type or Af10fl/fl mice to establish blast-colony forming cultures expressing the MLL-AF9, MLL-AF6 or MLL-AF10. We then deleted Af10 using a retrovirus expressing the Cre recombinase and performed colony-forming cell (CFC) assays (Figure 3A). In the first week, Af10 deletion profoundly impaired blast colony formation from MLL-AF9 transformed LSKs leading to increased myeloid differentiation (Figure 3B–D). Subsequent replatings showed the emergence of blast colonies that had escaped Af10 excision (data not shown). MLL-AF6 transformed cells showed a similar dependence on AF10 (Figure 3B and Figure 3E). In contrast, the blast colony forming ability of LSKs transformed with MLL-AF10 was unaffected by Af10 deletion (Figure 3B and Figure S3). These results show that endogenous AF10 is critical for MLL-fusion mediated transformation, and this dependence can be rescued when the AF10OM-LZ domain is present in the fusion protein as in the case of MLL-AF10.
Figure 3. The DOT1L-AF10 interaction is critical for MLL-transformation.
(A) Schematic representation of colony assays. (B) Pictures of representative colonies from cells transformed with different oncogenes 7 days after Af10 excision (Cre) compared to Af10 floxed colonies (MIT). (C) Wright-Giemsa stained cytospins of MLL-AF9 transformed cells 7 days after expression of the Cre recombinase (Cre) or the control vector (MIT). (D, E) Colony numbers obtained 7 days after expression of the Cre recombinase in wildtype or Af10 floxed MLL-AF9 (D) or MLL-AF6 (E) transformed cells. *p=0.013, **p=0.04. (D) and (E) represent mean +/− SEM values from 3 independent experiments. p values were calculated between the mean numbers of blast colonies in Cre expressing wildtype cells compared to Cre expressing Af10 floxed counterparts to account for Cre-mediated toxicity. See also Figure S3.
AF10 loss of function impairs the maintenance of diverse MLL-driven leukemias
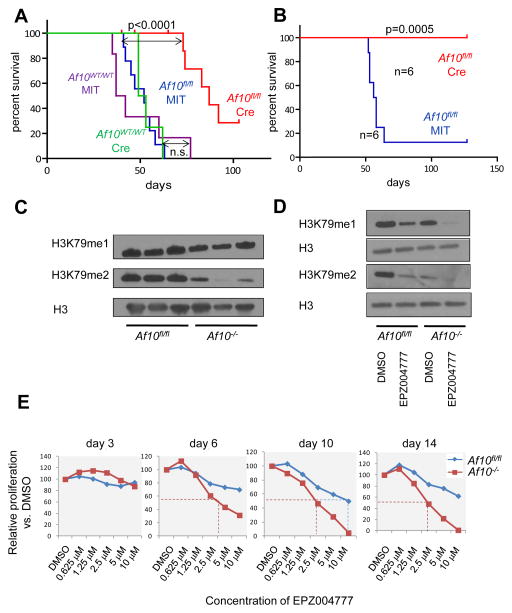
Next we investigated the effects of Af10 deletion on leukemia initiation from MLL-AF9 transformed cells. Af10 deletion significantly delayed leukemia initiation by MLL-AF9 (Figure S4A). We then deleted Af10 from established MLL-AF9 or MLL-AF6 driven leukemias and injected cells into secondary recipient mice to assess the impact on leukemia maintenance. AF10 inactivation significantly increased the latency of secondary MLL-AF9 leukemias (Figure 4A) and completely abrogated MLL-AF6 leukemia propagation (Figure 4B). Gene expression analysis by quantitative PCR (q-PCR) showed that expression of key HOXA cluster genes (Hoxa5-10) and Meis1 was dramatically reduced in AF10 deficient leukemias (Figure S4B). Next we assessed global H3K79 methylation in Af10 deleted MLL-AF9 leukemia cells. Similar to the HOXA9-MEIS1 transformed cells, we observed that AF10 inactivation drastically reduced H3K79me2, whereas H3K79me1 was affected to a lesser degree (Figure 4C and Figure S4C). We then investigated whether AF10 deficient MLL-AF9 leukemias were still dependent on DOT1L for their continued proliferation. We performed proliferation assays on wild type and AF10 deficient MLL-AF9 leukemias cells using the DOT1L inhibitor EPZ004777 (Daigle et al., 2011). Treatment of the AF10 deficient MLL-AF9 leukemias with EPZ004777 eliminated H3K79me2 and residual H3K79me1 and further reduced the expression of Hoxa7-10 and Meis1 genes (Figure 4D and Figure S4D). Strikingly, AF10 deficient MLL-AF9 leukemia cells showed an increased sensitivity to DOT1L inhibition compared to wild-type MLL-AF9 leukemia cells. AF10 deficiency substantially lowered both the time and the dose required to impair proliferation of MLL-AF9 leukemia cells (Figure 4E). These results demonstrate that AF10 may be a therapeutic target in MLL-rearranged leukemias and that targeted disruption of the DOT1L-AF10 interaction may also sensitize leukemias to small-molecule DOT1L inhibition.
Figure 4. Impact of AF10 inactivation on leukemia maintenance.
(A)Survival curves for secondary leukemia from MIT or Cre expressing primary MLL-AF9 leukemias in the Af10fl/fl background compared to MLL-AF9 leukemias from the Af10 wildtype background. (n=6–10 mice for each cohort). (B) Percent survival of mice injected with primary MLL-AF6 leukemia cells in the Af10fl/fl background after Cre or MIT expression (n=6 for each cohort). (C) Western blots showing H3K79me1 and H3K79me2 in the BM cells of 3 independent Af10fl/fl or Af10−/− MLL-AF9 leukemias. Total H3 is used as the loading control. (D) Western blots showing H3K79me1 and H3K79me2 in a representative Af10fl/fl or Af10−/− MLL-AF9 leukemia treated with 10 μM of EPZ004777 or the DMSO vehicle control. (E) Representative experiment showing relative proliferation as assessed by Trypan blue exclusion for Af10 floxed (MIT) or deleted (Cre) MLL-AF9 leukemia cells plotted as a percent of vehicle control (DMSO) in various concentrations of EPZ004777 at different time-points (Representative of 3 independent experiments). See also Figure S4.
AF10 inactivation reverses MLL-AF9 driven gene expression and H3K79 methylation profiles
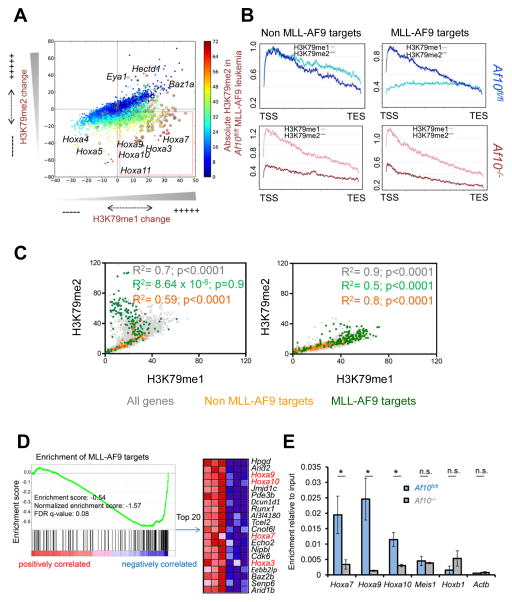
Next we performed a comprehensive investigation of histone methylation and gene expression in the Af10 deleted leukemias compared to wild-type counterparts. We found dramatic locus-specific changes in H3K79 methylation in the AF10 deficient leukemias by ChIP-seq. The most remarkable change was the marked decrease in H3K79me3 in the AF10 deficient cells (Figure S5A and S5B). Specifically at the HOXA gene cluster, we observed a progressive reduction in H3K79 methylation with the most profound reduction in H3K79me3, accompanied by a substantial decrease of H3K79me2 (Figure S5C). H3K79me1 however, appeared to be either retained or increased at the Hoxa7-10 genes (Figure S5C). This discrepancy between H3K79me1 and H3K79me2/3 prompted us to analyze the genome wide relationship between H3K79me1 and H3K79me2 in greater detail. Our analysis showed that thousands of genes, including most of the HOXA cluster genes, lost H3K79me2 (Figure 5A lower 2 quadrants) consistent with our previous assessments of global H3K79me2 levels. Interestingly, a few of these H3K79me2 depleted genes appeared to gain H3K79me1 (Figure 5A red box). We found that the genes with H3K79me2 loss accompanied by an apparent H3K79me1 gain were largely those with the highest absolute H3K79me2/3 levels in MLL-AF9 leukemia (Figure 5A, colored from yellow to red), and were highly enriched for MLL-AF9 target genes (Figure 5A, circled dots). We confirmed the decrease in H3K79me2/3 and retention of H3K79me1 at HOXA/MEIS1 promoters upon Af10 excision more quantitatively by ChIP-qPCR where the input sample was used to assess relative enrichment (Figure S5D). We then conducted a meta-analysis of all 129 MLL-AF9 target genes to assess changes in the relationships between H3K79me1 and H3K79me2 upon Af10 deletion across the gene body. Strikingly, we observed that Af10 deletion altered the relationship between H3K79me1 and H3K79me2 at the majority of MLL-AF9 target genes restoring it to a pattern resembling non MLL-AF9 target genes (Figure 5B and Figure 5C green dots vs. yellow dots). This reversal happened at all but a few MLL-AF9 target genes such as Eya1, Hectd1, and Baz1a (Figure S5E and data not shown). These results demonstrate that AF10 loss of function prevented the conversion of H3K79me1 to H3K79me2/3 at most MLL-AF9 target genes such as Hoxa7, Hoxa9, Hoxa10, Meis1, Mef2c and Runx2, resulting in the depletion of H3K79me2/3 and the preservation or accumulation of H3K79me1.
Figure 5. Genomic and epigenomic changes upon Af10 deletion.
(A) Scatter plot showing genome-wide changes in the values of promoter-proximal H3K79me1 (x-axis) or H3K79me2 (y-axis) in Af10fl/fl vs. Af10−/− leukemias. Each dot represents the difference in averaged methylation values around the promoter proximal regions (−2 kb to +2 kb around the TSS); positive and negative values reflect increases and decreases respectively in Af10−/− compared to Af10fl/fl leukemias. Methylation values are colored according to absolute H3K79me2 values in wild type MLL-AF9 leukemia (blue- least methylation, red-highest methylation). (B) A representative meta-analysis plot showing averaged profile across the gene body from the TSS to the transcription end site (TES) of MLL-AF9 target genes compared to control genes. Profiles of Af10fl/fl MLL-AF9 leukemias in (blue) compared to their Af10−/− counterparts (in red) are presented. Light colors represent H3K79me1 and dark colors represent H3K79me2. MLL-AF9 targets are genes bound by MLL-AF9 and non MLL-AF9 target genes are a size and expression matched set of genes that are not bound by MLL-AF9. (C) Scatter plot showing the genome-wide relative relationship between H3K79me1 and H3K79me2 values around promoter-proximal regions. Values are representative of 5 independent replicates for each group. (D) Enrichment of MLL-AF9 target genes in the Af10−/− compared to Af10fl/fl leukemias shown by GSEA. The top 20 MLL-AF9 targets downregulated in Af10−/− leukemia are shown on the right. (E) ChIP for DOT1L protein at the promoter proximal regions of indicated genes in MLL-AF9 leukemias. n=2, * p<0.05, n.s.= not significant.
Given that Af10 excision erased H3K79me2, but not H3K79me1, at critical target genes, we wanted to determine how this influenced the expression of MLL-fusion target genes. Microarray analysis of AF10 deficient leukemias showed significant reduction in the expression of key transcripts associated with MLL-rearranged leukemia (Table S3), and gene-set enrichment analysis (GSEA) showed that transcripts down-regulated upon Af10 deletion were significantly enriched for MLL-AF9 target genes (Figure 5D). Transcripts down-regulated in AF10 deficient leukemias were also significantly enriched for a previously reported gene-set of DOT1L regulated genes in MLL-AF9 leukemia (Figure S5F) (Bernt et al., 2011). Therefore, the lack of conversion of H3K79me1 to H3K79me2 corresponds with a decrease in gene expression for most of the critical MLL-AF9 target genes. However there are a few MLL-AF9 target genes such as Eya1, which show elevated H3K79me2 and increased expression after AF10 inactivation suggesting that these genes may be regulated by alternative DOT1L complexes.
Af10 deletion impairs Dot1L localization and promotes H3K27me3 at HOXA genes
Next, we assessed whether chromatin localization of MLL-AF9 or of DOT1L was influenced in AF10 deficient cells. For this, we made use of in vivo biotin-tagging to tag either MLL-AF9 or DOT1L and performed ChIP using Streptavidin-conjugated beads followed by qPCR. While there was no significant difference in MLL-AF9 occupancy in Af10−/− compared to Af10fl/fl cells, (Figure S5G) we observed significantly lower DOT1L occupancy on the promoter-proximal regions of the Hoxa7-10 genes in the Af10−/− cells (Figure 5E). In contrast to the Hoxa7-10 genes, we observed significantly increased DOT1L localization at the Eya1 promoter-associated region in Af10−/− compared to Af10fl/fl leukemia cells (Figure S5H), which corresponds with increased Eya1 promoter associated H3K79me2/3 and mRNA expression upon Af10 deletion.
We next assessed if there were changes in H3K27 methylation at MLL-AF9 target genes after Af10 deletion. Since the Drosophila AF10 ortholog plays a role in Polycomb repulsion from homeotic loci, we wanted to determine whether AF10 played a similar role in the spread of repressive PRC2 activity on HOX genes in MLL-AF9 leukemia. Our ChIP-qPCR analysis of H3K27me3 showed a significant increase in H3K27me3 at the promoters of Hoxa7-10 genes in Af10−/− leukemias (Figure S5I). We then performed ChIP-seq for H3K27me3 to assess whether H3K79 diminution was generally accompanied by H3K27 increase in other loci. Our analysis showed alterations of H3K27 on various genes in the Af10 deleted leukemias, but strikingly genes that lost H3K79me3 or H3K79me2 and concomitantly gained high levels of H3K27me3 were almost exclusively comprised of exactly those HOXA cluster genes that are highly expressed in LSKs and are associated with stem-cell self renewal (Krivtsov et al., 2006) (Figure S5J and Figure S5K). These results demonstrate that AF10 has specific chromatin modulatory activity at the self-renewal associated HOXA cluster genes in MLL-AF9 leukemia cells. Importantly, similar to our observation in normal GMPs, the continued presence of H3K79me1 was insufficient to prevent the increase in H3K27me3 at the Hoxa7-10 gene promoters in the Af10 deleted leukemias, thereby disrupting the mutually exclusive nature of H3K79 and H3K27 methylation normally observed across the genome (Figure S5L) (Bernt et al., 2011). These results show that inactivation of AF10 leads to impaired DOT1L association with HOXA cluster genes, loss of higher H3K79 methylated states, and the spread of H3K27me3, which coincides with a decrease in gene expression.
AF10 enhances the enzymatic activity of DOT1L
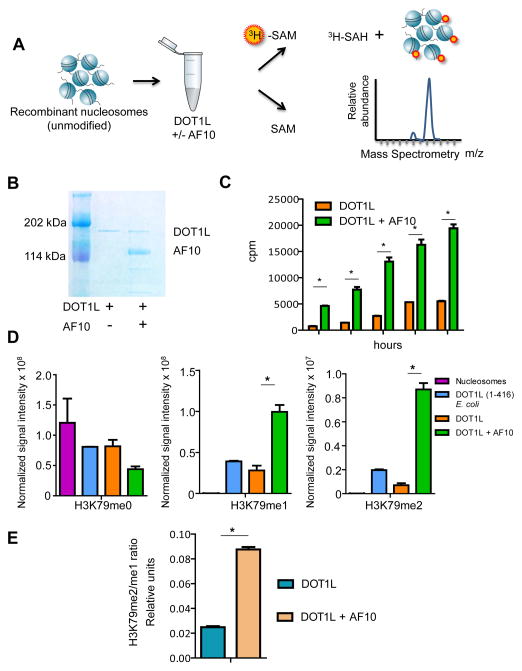
Next we tested whether AF10 can directly affect the histone methyl-transferase (HMTase) activity of DOT1L in cell-free HMTase assays using purified flag-tagged human DOT1L, alone or in a complex with full length human AF10, and reconstituted unmodified mononucleosomes and radioactive 3H-S-adenosyl-methionine (3H-SAM) substrates (Figure 6A and Figure 6B). We found that the DOT1L-AF10 complex had greater HMTase activity than DOT1L alone, as assessed by 3H incorporation into histones in filter binding assays (Figure 6C). Next, we extracted histones from the DOT1L or DOT1L-AF10 methylated mononucleosomes and investigated H3K79me0, H3K79me1 and H3K79me2 using MS, which confirmed that the DOT1L-AF10 complex had a significantly higher overall methylating activity as compared to DOT1L alone (Figure 6D and Figure S6A–C). In addition, histones methylated by the DOT1L-AF10 complex had a significantly higher H3K79me2/me1 ratio than those methylated by DOT1L alone (Figure 6E). These results demonstrate that AF10 plays a crucial role in enhancing the HMTase activity of DOT1L, influencing its ability to stimulate higher order H3K79 methylation.
Figure 6. Effect of AF10 on the HMTase activity of DOT1L.
(A) Schematic depiction of the experimental design. SAM: S-adenosylmethionine, SAH: S-Adenosylhomocysteine. (B) Flag-tagged DOT1L overexpressed either alone or in combination with untagged AF10 in insect cells and purified using Flag-affinity purification is shown by Coomassie staining. (C) Bar graph showing quantification of methylation as performed by filter binding followed by liquid scintillation counting at various time-points denoted on the x-axis. cpm: counts per million. n=3, *p<0.01 in each case. (D) Comparative levels of H3K79me0, H3K79me1 and H3K79me2 by MS in nucleosomes incubated with a human DOT1L fragment expressed in E. coli (positive control), full-length human DOT1L or DOT1L+AF10. (n=2; *p<0.05). Ion intensities of methylated peptides (residues 74–84) are scaled relative to an internal standard. (E) Ratio of H3K79me2/me1 normalized to internal standard in the different conditions is depicted. *p<0.05.
Error bars represent SD. See also Figure S6.
AF10 controls HOXA gene expression and proliferation of NUP98-NSD1 transformed cells
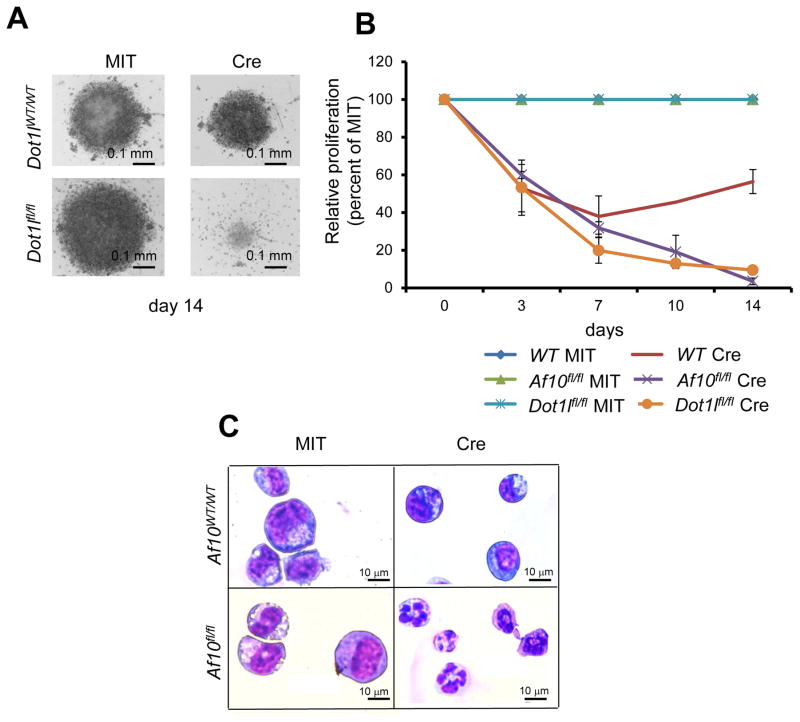
Our observations from LSKs, GMPs and different MLL-fusion leukemias suggest that higher degree H3K79 methylation mediated by the DOT1L-AF10 complex is critical for HOX gene expression irrespective of the cellular context. D. melanogaster studies have shown that both DOT1L and AF10 orthologs are required for normal HOX gene expression (Perrin et al., 2003; Shanower et al., 2005), suggesting an evolutionarily conserved function of the DOT1L-AF10 complex in homeotic gene expression. Since aberrant HOX gene expression is not limited to MLL-rearranged leukemia, we wondered whether the DOT1L-AF10 complex might also regulate HOX gene expression and consequently transforming activity of other AML inducing oncogenes that show aberrant HOX gene activation. The NUP98-NSD1 fusion resulting from a chromosomal translocation is observed in 16% of cytogenetically normal pediatric AML and in a small subset of adult AML (Fasan et al., 2013; Hollink et al., 2011; Thol et al., 2013). NUP98-NSD1 positive AMLs possess high-level HOXA gene expression and have a very poor prognosis (Hollink et al., 2011). We generated a retrovirus that expresses NUP98-NSD1 and transduced LSK cells from either wild-type or homozygous Af10 floxed mice. NUP98-NSD1 transduced cells proliferate indefinitely in culture and displayed features of differentiation arrest as assessed by immunophenotype and morphology (Figure S7A and S7B). Bi-allelic Af10 deletion led to a dramatic decrease in H3K79 dimethylation accompanied by a more subtle decrease in H3K79 monomethylation in the NUP98-NSD1 transformed cells (Figure S7C). Furthermore, AF10 inactivation led to a significantly reduced expression of the Hoxa7-10 cluster (Figure S7D). Importantly, deletion of either Af10 or Dot1l in NUP98-NSD1 transformed bone marrow cells dramatically impaired their proliferation and increased apoptosis (Figure 7A, Figure 7B and S7E) and cells growing after 14 days had incomplete deletion of the Dot1l or Af10 (data not shown). NUP98-NSD1 cells deleted of either Af10 or Dot1l demonstrated a cellular morphology consistent with differentiation (Figure 7C and data not shown). These findings suggest that, similar to MLL-rearranged leukemias, oncogenic transformation driven by NUP98-NSD1 is critically dependent on H3K79 methylation mediated by the DOT1L-AF10 complex.
Figure 7. Role of the DOT1L-AF10 complex beyond MLL-rearranged leukemia.
(A) Representative picture of a well with MIT or Cre expressing NUP98-NSD1 transformed cells in the Dot1lWT/WT and Dot1lfl/fl backgrounds at day 14 after culture. (B) Relative proliferation of NUP98-NSD1 cells after Af10 or Dot1l deletion (Cre) compared to wildtype controls are presented as a percentage of their Af10 or Dot1l non deleted (MIT) counterparts. n=3, * p< 0.05. Error bars represent SD. (C) Wright-Giemsa stained cytospins of NUP98-NSD1 transformed cells 7 days after MIT or Cre expression in the Af10WT/WT or Af10fl/fl backgrounds. See also Figure S7.
NUP98-NSD1 transformed cells are sensitive to small-molecule DOT1L inhibition
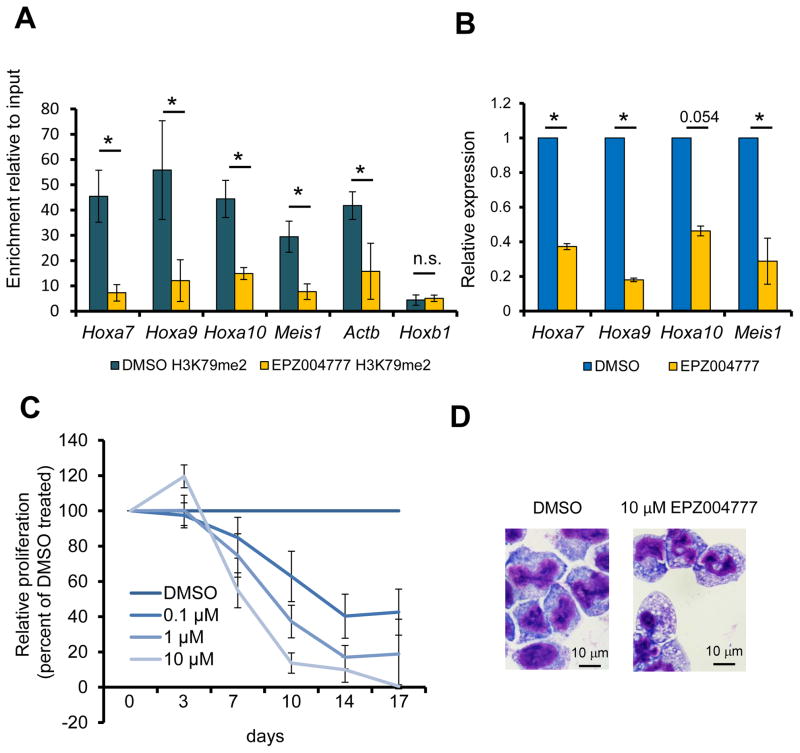
The observations above using Dot1l or Af10 conditional knockout mice indicated that cells transformed with HOX-activating oncogenes other than MLL-fusion proteins might show sensitivity to therapeutic targeting of DOT1L activity. Given the clear clinical implication of this hypothesis, we tested the sensitivity of NUP98-NSD1 transformed cells to pharmacological DOT1L inhibition. Treatment of the NUP98-NSD1 transformed cells with the DOT1L inhibitor EPZ004777 significantly reduced global and promoter associated H3K79me2, and a significant reduction in Hoxa7-10 gene expression (Figure S8A, Figure 8A and Figure 8B). Notably, the effect of EPZ00477 exposure on HOXA gene expression led to a dramatic dose-dependent decrease in proliferation starting from day 7 after exposure to the inhibitor (Figure 8C) accompanied by increased differentiation and apoptosis (Figure 8D and Figure S8B). The sensitivity of the NUP98-NSD1 cells to EPZ004777 could be completely rescued by retroviral overexpression of HOXA9-MEIS1 (Figure S8C) demonstrating that NUP98-NSD1 mediated transformation was critically dependent on DOT1L-mediated regulation of HOXA/MEIS1 gene expression. These results provide compelling evidence in support of a broader role for H3K79 methylation, mediated by the DOT1L-AF10 complex, in leukemogenic HOXA gene regulation and oncogenesis. These data also indicate that therapeutic targeting of the DOT1L activity may provide benefit in multiple leukemia subtypes showing aberrant HOXA gene activation.
Figure 8. Effect of pharmacological DOT1L inhibition on NUP98-NSD1 transformed cells.
(A) The H3K79me2 level at the promoters of the Hoxa7-10 and Meis1 genes upon DOT1L inhibition with 10 μM EPZ004777 for 7 days is assessed by ChIP with an H3K79me2 specific antibody followed by qPCR. *p< 0.05, n.s.= not significant. (B) Expression of HOXA/MEIS1 genes 7 days after treatment of NUP98-NSD1 transformed BM cells with 10 μM EPZ004777 is shown relative to DMSO vehicle carrier treated cells. *p< 0.05. (C) Relative proliferation of NUP98-NSD1 transformed cells during 17 days of incubation with varying concentrations of EPZ004777 is plotted as a percentage of the vehicle control treated cells. (D) Wright-Giemsa stained cytospins of NUP98-NSD1 transformed cells 10 days after treatment with DMSO or the EPZ004777 inhibitor. Error bars represent SD. See also Figure S8.
DISCUSSION
The biological responses associated with histone methylation are determined both by the amino acid residue and the degree of methylation (Kouzarides, 2007). H3K79 methylation is associated with actively transcribed genes and euchromatin in multiple species. Mono-, di-, and trimethylation of H3K79 are all solely catalyzed by DOT1L, in multiple organisms (reviewed in Nguyen and Zhang, 2011). In S. cerevisiae, mutation of H3K79 influences SIR2 binding and heterochromatin formation thus affecting gene activity. Whether H3K79 methylation influences similar mechanisms to control gene expression in mammalian cells is unclear. Our results suggest that different levels of H3K79 methylation serve as a “volume control” mechanism for gene expression during hematopoiesis and leukemia development, with higher degrees of H3K79 methylation correlating with elevated levels of gene expression. Of particular interest, genes decorated with H3K79me1 but not H3K79me2/3 are unable to maintain high-level expression either in developing hematopoietic cells or leukemia. Of note, H3K27me3 invaded HOX loci even though H3K79me1 was present. Therefore, the loss of one methyl group on H3K79 appears to have allowed gene repression and H3K27me3 accumulation. This suggests that during hematopoietic development the H3K79 methylation state decreases as myeloid development progresses thus ensuring a finely tuned decrease in gene expression to the point at which repressive mechanisms dominate. Such processes thus allow developing myeloid cells the potential to rapidly and faithfully modulate expression of genes that are highly expressed in less differentiated cells until cells are fully committed to a particular fate. However, this mechanism presents a risk of being coopted by leukemogenic oncoproteins, a mechanism that is taken advantage of by MLL or NUP98-fusions.
There is growing appreciation of aberrant chromatin modifications in cancer cells. One fascinating aspect is the apparent oncogenic impact of abnormal variations in the degree of histone methylation at particular histone residues. In MLL-AF9 leukemia cells we find a decrease in H3K79me1 and concomitant increase in H3K79me2/3 at MLL-AF9 target genes which suggests continuous conversion of H3K79 methylation to higher methylated states. The MLL-AF9 fusion protein prevents a differentiation associated decrease in higher degree H3K79 methylation at key MLL-AF9 target genes including the HOXA genes, leading to the perpetuation of a constantly self-renewing state. These findings underscore the critical roles that specific methylation states play in oncogenesis. Other examples of specific degrees of histone methylation contributing to oncogenic development are the recently described gain of function mutations in the H3K27 methyl-transferase EZH2 leading to hyper di and trimethylation of H3K27 in B-cell lymphoma (McCabe et al., 2012a; Sneeringer et al., 2010) as well as the accumulation of H3K36me2 in subsets of multiple myeloma and acute lymphoid leukemia (ALL) cells harboring chromosomal translocations or activating mutations of the H3K36 methyl-transferase MMSET/NSD2 (Martinez-Garcia et al., 2011).
The low H3K79me1/high H3K79me2 pattern that we observed on MLL-AF9 target genes could reflect enhanced DOT1L recruitment and/or enzymatic activity at these genes. A recent study demonstrated that inactivation of the C. elegans AF10 homolog abrogates chromatin localization of the C. elegans homolog of DOT1L (Cecere et al., 2013). In our study, we demonstrate that AF10 depletion reduces DOT1L localization to HOX genes indicating that AF10 plays a role in locus-specific DOT1L recruitment. Our observation that some genes, including some MLL-AF9 target genes such as Eya1, Hectd1 and Baz1a gain H3K79 methylation upon Af10 deletion suggests that AF10 is responsible for DOT1L localization on most, but not all, MLL-AF9 target genes, and in the absence of AF10, DOT1L levels and consequently H3K79 methylation may be relocated to other chromatin loci, potentially through other DOT1L co-factors. Although AF10 inactivation dramatically impairs leukemia initiation and maintenance, AF10 deficiency does not completely preclude MLL-AF9 leukemic outgrowth. In the case of MLL-AF6 however, AF10 depletion seems to completely inhibit leukemia development. The difference in the degree of AF10 dependence between these two leukemias may be attributable to the ability of MLL-AF9, but not MLL-AF6, to recruit DOT1L to HOX and other loci. It is important to mention that both leukemias show absolute dependence on H3K79 methylation, since DOT1L inhibition in AF10 deficient MLL-AF9 leukemias swiftly and completely eliminates those cells.
We demonstrate that AF10 augments the HMTase activity of DOT1L. AF10 is therefore important for both recruitment and enzymatic activity of DOT1L, analogous to co-factors of other chromatin modifying complexes (Dou et al., 2006; Wysocka et al., 2005). There is increasing interest in the development of small molecule inhibitors against chromatin modulators. Studies have shown that inhibition of chromatin modifying enzymes can have therapeutic benefits in pre-clinical models (Chen et al., 2013; Daigle et al., 2011; Deshpande et al., 2013; McCabe et al., 2012b). A small molecule DOT1L inhibitor is currently in phase I clinical trials for the therapy of MLL-rearranged leukemia (Daigle et al., 2013). It is worth noting that most drug-screening efforts aimed at therapeutically targeting the HMTase activity of DOT1L use the catalytically active DOT1L N-terminal 1-416 amino acid fragment. In our study, we have demonstrated that full-length DOT1L in complex with AF10 not only has a significantly enhanced HMTase activity but also is better able to produce H3K79 dimethylation than DOT1L alone. Our data not only provides an important starting point for further detailed analyses into the enzymology of H3K79 methylation, but also indicate that the DOT1L-AF10 complex might be a more physiologically relevant target for future drug development efforts.
Efforts to target chromatin-modifying proteins are increasingly gaining traction. One prime example of this is small molecule inhibition of the PRC2 complex in lymphoma cells for which clinical trials are ongoing (Knutson et al., 2012; McCabe et al., 2012b; Qi et al., 2012). These rapid advances predict an emerging wave of anti-cancer therapeutics based on the inhibition of chromatin regulatory factors. Our results demonstrate that in addition to histone methyltransferases themselves, the identification of oncogenic dependencies on chromatin co-factors may present additional avenues for therapeutic intervention in malignancies with aberrant changes in the epigenome. Targeting the DOT1L-AF10 interaction may synergize with and thus improve the efficacy of agents that inhibit DOT1L. These studies present a rationale for therapeutic targeting of the DOT1L-AF10 interaction and suggest that dimethylation and trimethylation of H3K79 via the DOT1L complex is a potential therapeutic target in an array of different leukemias, and perhaps other tumor types associated with elevated HOX gene expression.
EXPERIMENTAL PROCEDURES
ChIP-qPCR and ChIP-Seq
ChIP was performed as described previously (Bernt et al., 2011). Briefly, crosslinking was performed with 1% formalin, and the cells were lysed in SDS buffer. Sonication was used to fragment DNA. ChIP for H3K79me1, H3K79me2, H3K79me3, and H3K27me3 was performed using the antibodies ab2886, ab3534, ab2621 (Abcam) and 07-449 (Millipore) specific to the respective modifications. Eluted DNA fragments were analyzed by qPCR, or subjected to sequencing using next-generation Ilumina sequencing. More details can be obtained in the supplemental methods section.
Mice
A pFlexible-based targeting vector containing a ~2.7 kb genomic locus region that included exons 17 and 18 of Af10 (reference transcript: REFSEQ NM_010804) flanked by loxp sites were used to target CJ9 ES cells (129 background). Conditional mice were maintained on a B6/129 background (Taconic). The generation of Dot1l knockout mice was previously described (Bernt et al., 2011). Dot1lfl/fl mice were crossed to Mx1-Cre mice (Kuhn et al., 1995) (Jackson Labs, Bar Harbor, ME), and the Mx1-Cre allele was maintained as a heterozygous allele on a mixed B6/129 background. Genotyping strategies, primers and conditions are described in the supplemental experimental procedures. All animal experiments described in this study including the experiments with conditional knockout mice as well as mouse BM transplantation models were approved by, and adhered to guidelines of, the Memorial Sloan-Kettering Cancer Center and the Children’s Hospital Boston institutional animal care and use (IACUC) committees.
Small molecule inhibitor
EPZ004777 was synthesized by the laboratory of Jay Bradner (Dana Farber Cancer Institute, Boston, MA). 50 mM stock solutions were prepared in DMSO and stored at −20° C. Serial dilutions of stock solutions were carried out just prior to use in each experiment and final DMSO concentrations were kept at, or below 0.02%.
Accession information
ChIP and Microarray data is deposited at the NCBI gene expression omnibus (GEO) - accession number: GSE54500.
Data Analysis and Statistical Methods
Significance for all individual experiments described in the manuscript was calculated using a 2-tailed unpaired Student’s T-test in Microsoft Excel or GraphPad Prism. For mouse survival curves the Log-Rank test was used to calculate statistical significance using GraphPad Prism. Raw expression data was normalized using Genepattern software by RMA algorithm with quantile normalization and background correction. Probesets that had 50% or more present calls (using MAS5 algorithm) were marked as expressed and the remainder as not expressed. The expressed probe-sets were further divided into 3 groups of high, medium and low expression based on raw expression values. Gene set enrichment analysis of microarray was performed with GSEA (Subramanian et al., 2005), and relative changes in methylation values were plotted using Icanplot software (www.icanplot.org) (Sinha and Armstrong, 2012).
Supplementary Material
Figure S1 related to Figure 1: (A) ChIP-competition experiment to assess the cross-reactivity of H3K79me1, H3K79me2 and H3K79me3 antibodies by ChIP-qPCR at the Hoxa9 locus. Enrichment relative to input at the Hoxa9 promoter-proximal region after addition of different synthetic peptides described below are shown. Values are plotted as mean +/− SD. (B) Upper panel shows meta-analysis profiles of H3K79 mono, di and tri-methylation across +/−10 kb around the transcription start site (TSS) of LSK gene sets divided based on expression. Gene expression data from microarray experiments on LSKs was used to derive these four equally sized sets of genes. Bottom 25 % (non expressed) are shown by the blue line and the other three quartiles are divided into pink- low expressed genes, red- medium expressed genes, and top 25 % (dark red) highly expressed genes; Lower panel depicts heat maps of ChIP sequencing data across +/−10 kb around the transcription start site (TSS) of all genes sorted from highest methylation levels (top) to lowest (bottom); (C) Heat maps of ChIP sequencing data for H3K79me3 a the gene bodies of MLL-AF9 targets and control genes; (D) Meta- cross analysis of H3K79me2 (dark blue) and H3K79me1(light blue) across the gene body of MLL-AF9 target genes compared to controls. Control genes in both C and D are a size and expression matched set of non MLL-AF9 target genes.
Figure S2 related to Figure 2: (A) Normalized expression of Hoxa5-10, Actb (upper panel), and Dot1l, Af10 and Af17 (lower panel) from microarray profiling of LSK vs GMP cells in C57BL6 mice. Microarray data can be obtained from (Krivtsov et al., 2006) or from GEO accession ID GSE54500. The box plot shows normalized expression values from 6 independent replicates. Whiskers represent the upper and lower limits of the range. Boxes represent the first and third quartile, and the line represents the median. (B) Schematic representation of proteins identified in the human DOT1L complex purified from MLL-rearranged SEMK2 cells and non MLL-rearranged REH cells. VENN diagram shows the overlapping proteins DOT1L, AF10, AF17 and NPM1 found in both cell lines. NPM1, which was found at low representation, was also found in at least one empty vector control cell line. (C) Levels of AF10 or AF17 after knockdown with corresponding shRNAs relative to sh-Luc as a control are shown on the left and Western Blots for AF10 or AF17 protein after shRNA knockdown is shown on the right with LaminB1 (LMNB1) as a protein loading control. The bottom panel shows Western blots for H3K79me1 or H3K79me2 upon AF10 or AF17 knockdown in SEMK2 and MOLM13 cells with total H3 as a loading control. (D) Western blot showing H3K79me1 and H3K79me2 after DOT1L or AF10 knockdown in the MLL-rearranged MOLM13 cell line compared to cells expressing anti-Luciferase (Luc) of anti-GFP shRNAs. (E) Cell cycle changes in various cell lines upon AF10 knockdown. Error bars represent SD. (F) Mouse exons harboring the octapeptide-motif leucine zipper (OM-LZ) domain of mouse Af10 were flanked by loxp sites. Lower panel shows gel electrophoresis of representative PCR samples showing Af10 floxed and deleted amplicons. (G) Western blots for AF10, DOT1L, AF17 in nuclear lysates of HOXA9-MEIS1 transformed cells (left panel) or wh ole cell lysates of immortalized fibroblasts in the Af10fl/fl compared to the Af10−/− backgrounds. (right panel) Lamin B1 (LMNB1) or alpha-Tubulin TUBA1A are used as loading controls respectively. (H) Number of blast-like or differentiated colonies obtained from HOXA9-MEIS1 transformed cells 1 week after expression of the Cre recombinase or the empty vector in the Af10WT/WT or Af10fl/fl background are plotted; p values were calculated between the mean numbers of blast colonies in Cre expressing wildtype cells compared to Cre expressing Af10 floxed counterparts to account for Cre-mediated toxicity. *p<0.05. Error bars represent SEM. (I) Western blots showing H3K79me2 in Af10 floxed or deleted immortalized fibroblasts with or without retroviral AF10 overexpression. (J) Bar graph showing levels of H3K79me0, H3K79me1 and H3K79me2 in HOXA9-MEIS1 transformed single cell clones in the presence or absence of DOT1L. All values are shown as a percentage of total H3 peptides. On the right panel, Western blot in one of the single clones used for MS-MS is shown for DOT1L wi alpha-Tubulinth (TUBA1A) as a loading control. (K) Mass Spectrometric assessment of H3K79me0, H3K79me1 and H3K79me2 in HOXA9-MEIS1 transformed cells in the Af10fl/fl compared to Af10−/− backgrounds.
Figure S3 related to Figure 3: Effect of Af10 deletion on MLL-AF10 transformation
Colony numbers obtained 7 days after expression of the Cre recombinase in wildtype or Af10 floxed MLL-AF10 transformed cells are plotted. MLL-fusion transformed colonies are almost exclusively comprised of tight, round, hyper-cellular colonies or scattered, small colonies; n.s.= not significant. Figure represents mean from 3 independent experiments. p values were calculated between the mean numbers of blast colonies in Cre expressing wildtype cells compared to Cre expressing Af10fl/fl counterparts to account for Cre-mediated toxicity. Error bars represent SEM.
Figure S4 related to Figure 4: Abrogation of the DOT1L-AF10 interaction impairs MLL-AF9 leukemia (A) Survival curves for primary leukemia from MIT or Cre expressing MLL-AF9 transformed cells in the Af10fl/fl background. Af10 deletion significantly increases the latency of leukemia initiation. (B) Expression of Hoxa/Meis1 transcripts normalized to Actb in the Af10fl/fl compared to Af10−/− leukemias is shown. (C) Bar graph showing levels of H3K79me0, H3K79me1 and H3K79me2 in the Af10 floxed or deleted MLL-AF9 leukemias. All values are shown as a percentage of total H3 peptides. (D) Expression of Actb normalized fold change in Hoxa/Meis1 transcripts in the Af10fl/fl compared to Af10−/− leukemias in the presence of 10 μM of the EPZ004777 inhibitor are shown as a fold of DMSO vehicle control values. Error bars represent SD.
Figure S5 related to Figure 5: Epigenomic changes in MLL-AF9 leukemia upon Af10 deletion (A) A representative profile showing H3K79me3 levels upon Af10 deletion in MLL-AF9 leukemia. Y-axis scale represents read density per million. (B) Each dot in the scatter plot represents the change in average H3K79me3 levels of all genes in Af10 deficient compared to Af10 non-excised MLL-AF9 leukemias (x-axis) plotted against the absolute levels of H3K79me3 in wild-type MLL-AF9 leukemias (y-axis). Dots are coloured according to absolute H3K79me3 value in MLL-AF9 leukemia with blue being the lowest and red being the highest. (C) Changes in H3K79me1, me2 and me3 in Af10fl/fl (blue shades) compared to Af10−/− leukemias (red shades). The Hoxa7-10 genic region is marked with a dotted red rectangle. Y-axis scale represents read density per million. Values used to plot (B) and (C) are from reads per million per kb values averaged around the transcription start sites (−2 kb to +2 kb) of all genes. (D) Enrichment of H3K79me1, H3K79me2 and H3K79me3 at the promoter associated regions of Hox/Meis1 genes and controls by ChIP-qPCR. (E) A representative example of genes that gain H3K79 methylation in the Af10−/− leukemias. Y-axis scale represents read density per million. (F) GSEA showing enrichment of the DOT1L_down signature in Af10−/− compared to Af10fl/fl leukemias. (G) ChIP-qPCR for biotinylated MLL-AF9 in Af10fl/fl compared to Af10−/− deleted cells is shown at the Hoxa7-10 genes and Meis1. (n.s: not significant.) Data is presented as mean of 2 independent experiments. (H) ChIP-qPCR for bio-DOT1L at the Eya1 promoter-proximal region in Af10fl/fl compared to Af10−/− deleted cells is shown as a mean of 2 independent experiments. (p= 0.026). All p values were calculated using the Student’s T-Test. (I) Enrichment of H3K27me3 at the promoter associated regions of Hoxa genes and controls by ChIP-qPCR. (J) ChIP-seq data showing H3K27me3 deposition at select Hoxa genes in the absence of AF10. (K) Scatter plot showing changes in the values of promoter-proximal H3K79me2 (x-axis) or H3K27me3 (y-axis) in Af10fl/fl Vs Af10−/− leukemias. Each dot represents difference in averaged methylation around the promoter regions; positive and negative values reflect increases and decreases respectively in Af10−/− leukemias compared to Af10fl/fl controls. Select Hoxa cluster genes are colored and annotated. The values presented are representative of 3–5 independent sequenced samples. A similar plot with H3K79me3 changes (x-axis) plotted against H3K27me3 changes (y-axis) in Af10fl/fl Vs Af10−/− leukemias is presented on the right hand side. (L) Scatter plot showing absolute H3K79me1 levels (x-axis) plotted against corresponding absolute levels of H3K27me3 (y-axis) in Af10fl/fl compared to Af10−/− backgrounds. Error bars represent +/− SD.
Figure S6 related to Figure 6: Specificity of the H3K79me0/me1 and me2 identification by MS-MS (A–C) Unmodified (A), mono-methylated (B), and di-methylated (C) histone H3 (H33_XENLA) tryptic peptide that spans residues 74–84 are identified in the reconstituted nucleosomes incubated with DOT1L or DOT1L+ AF10 proteins. CAD mass spectrum of the (M+2H)2+ ions at m/z 659.34, 666.35, and 673.35 are shown in panels A, B and C, respectively. MS/MS spectra were collected in the Orbitrap at 30,000 resolution. Fragment ions of type b contain the amino terminus plus one or more additional residues. Fragment ions of type y contain the carboxy terminus plus one or more additional residues. b and y ions identified in the spectra are labelled.
Figure S7 related to Figure 7: Deletion of Af10 in NUP98-NSD1 transformed cells (A) Flow cytometry analysis of NUP98-NSD1 mediated transformation from BM derived LSKs cultured for 4 weeks is shown. Top panel shows empty-MSCV-puro vector transduced BM cells compared to MSCV-puro-NUP98-NSD1 derived BM cells at the bottom. NUP98-NSD1 transduced cells show an immature myeloid immunophenotype (cKitlowGr-1highMac-1high) whereas MSCV puro transduced cells show the outgrowth of contaminating mast cells (cKithighGr-1−Mac-1−). (B) Cytomorphology of Wright-Giemsa stained NUP98-NSD1 transformed cells is presented at different magnifications. (C) Western blot showing changes in global H3K79me1 and H3K79me2 5 days after Af10 deletion in NUP98-NSD1 transformed cells. Total H3 is shown as a loading control. (D) Hox gene expression in NUP98-NSD1 transformed murine BM cells in the presence or absence of the Af10 allele. Actb normalized fold expression change relative to Af10fl/fl cells is shown. * p<0.05. (E) Percentage Annexin V positive Sytox Blue negative cells (pro-apoptotic) in NUP98-NS 1 D cells with and without Af10 deletion are shown * p<0.05. Error bars represent +/− SD.
Figure S8 related to Figure 8: Sensitivity of NUP98-NSD1 cells to DOT1L inhibition (A) Global H3K79me2 levels in NUP98-NSD1 compared to control MLL-AF9 transformed hematopoietic progenitors following 10 days of treatment with 10 μM EPZ004777 as assessed by Western blotting with H3K79me2 specific antibody. Total H3 is shown as loading control. (B) Percentage Annexin V positive Sytox Blue negative cells (pro-apoptotic) in NUP98-NSD1 cells treated with 10 μM of EPZ004777 or the DMSO control are shown as mean values. * p<0.05. (C) Relative proliferation of NUP98-NSD1 cells or NUP98-NSD1 cells with retroviral HOXA9/MEIS1 overexpression in the presence of 10 μM EPZ004777 or DMSO vehicle control is depicted. Mean of 2–6 experiments is plotted in each case. Error bars represent +/− SD.
Table S1 related to Figure 1
Table S2 related to Figure 2
Table S3 related to Figure 5
SIGNIFICANCE.
A broad array of human malignancies shows aberrant expression of the clustered homeobox genes, particularly the genes of the HOXA gene cluster. Our study demonstrates that the DOT1L interacting protein AF10 regulates the HMTase activity and H3K79me1 to H3K79me2 conversion by DOT1L. AF10 inactivation causes widespread changes to the leukemia epigenome, suppressing the ability of multiple HOX-activating oncogenes to induce leukemic transformation. Our results provide evidence that transformation driven by MLL-fusions as well as the recurrent AML-associated NUP98-NSD1 fusion oncogene is critically dependent on the ability of AF10 to stimulate DOT1L activity. These findings have profound therapeutic implications as they provide a strong rationale for therapeutically targeting the DOT1L-AF10 complex in multiple types of refractory leukemias.
Acknowledgments
The authors acknowledge Ronald Hendrickson, Yuko Fujiwara and Stuart Orkin for valuable assistance with MS analysis and knockout mouse generation. This work was supported by the American Cancer Society, the Leukemia and Lymphoma Society (LLS), Gabrielle’s Angel Foundation and the NCI (CA105423, CA140575, CA176745) to S.A.A. and the NCI K99 Award to A.J.D.. S.A.A. is a consultant for Epizyme Inc.. The protein complex purification work and the MS studies were supported by the LLS SCOR grant 7132-08 to R.G.R. and an NCI Cancer Center support grant P30 CA08748 to MSKCC.
Footnotes
Supplemental data including detailed experimental procedures can be found in the accompanying supplemental section.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Bernt K, Zhu N, Sinha A, Vempati S, Faber J, Krivtsov A, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–144. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandimarte L, Pierini V, Di Giacomo D, Borga C, Nozza F, Gorello P, Giordan M, Cazzaniga G, Te Kronnie G, La Starza R, Mecucci C. New MLLT10 gene recombinations in pediatric T-acute lymphoblastic leukemia. Blood. 2013;121:5064–5067. doi: 10.1182/blood-2013-02-487256. [DOI] [PubMed] [Google Scholar]
- Cecere G, Hoersch S, Jensen MB, Dixit S, Grishok A. The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol Cell. 2013;50:894–907. doi: 10.1016/j.molcel.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Garcia R, Cervera M, Arredondo JJ. AF10 plays a key role in the survival of uncommitted hematopoietic cells. PLoS One. 2012;7:e51626. doi: 10.1371/journal.pone.0051626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, Hemenway CS, Zhang W. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Deshpande AJ, Banka D, Bernt KM, Dias S, Buske C, Olhava EJ, Daigle SR, Richon VM, Pollock RM, Armstrong SA. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 2013;27:813–822. doi: 10.1038/leu.2012.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AJ, Bradner J, Armstrong SA. Chromatin modifications as therapeutic targets in MLL-rearranged leukemia. Trends Immunol. 2012;33:563–570. doi: 10.1016/j.it.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AJ, Chen L, Fazio M, Sinha AU, Bernt KM, Banka D, Dias S, Chang J, Olhava EJ, Daigle SR, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013;121:2533–2541. doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13; q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasan A, Haferlach C, Alpermann T, Kern W, Haferlach T, Schnittger S. A rare but specific subset of adult AML patients can be defined by the cytogenetically cryptic NUP98-NSD1 fusion gene. Leukemia. 2013;27:245–248. doi: 10.1038/leu.2012.230. [DOI] [PubMed] [Google Scholar]
- Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, van Galen JF, Beverloo HB, Sonneveld E, Kaspers GJ, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- Linder B, Newman R, Jones LK, Debernardi S, Young BD, Freemont P, Verrijzer CP, Saha V. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J Mol Biol. 2000;299:369–378. doi: 10.1006/jmbi.2000.3766. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt LM, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012a;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012b;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G, Gong Z, Chen J, Kim JE. Characterization of the DOT1L network: implications of diverse roles for DOT1L. Protein J. 2010;29:213–223. doi: 10.1007/s10930-010-9242-8. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin L, Bloyer S, Ferraz C, Agrawal N, Sinha P, Dura JM. The leucine zipper motif of the Drosophila AF10 homologue can inhibit PRE-mediated repression: implications for leukemogenic activity of human MLL-AF10 fusions. Mol Cell Biol. 2003;23:119–130. doi: 10.1128/MCB.23.1.119-130.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler G, Kaltenbach S, Dobbelstein S, Broccardo C, Radford I, Mozziconacci MJ, Bernard OA, Penard-Lacronique V, Delabesse E, Romana SP. Identification of GSX2 and AF10 as NUP98 partner genes in myeloid malignancies. Blood Cancer J. 2013;3:e124. doi: 10.1038/bcj.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Kolking B, Hollink IH, Damm F, van den Heuvel-Eibrink MM, Michel Zwaan C, Bug G, Ottmann O, Wagner K, Morgan M, et al. Analysis of NUP98/NSD1 translocations in adult AML and MDS patients. Leukemia. 2013;27:750–754. doi: 10.1038/leu.2012.249. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 related to Figure 1: (A) ChIP-competition experiment to assess the cross-reactivity of H3K79me1, H3K79me2 and H3K79me3 antibodies by ChIP-qPCR at the Hoxa9 locus. Enrichment relative to input at the Hoxa9 promoter-proximal region after addition of different synthetic peptides described below are shown. Values are plotted as mean +/− SD. (B) Upper panel shows meta-analysis profiles of H3K79 mono, di and tri-methylation across +/−10 kb around the transcription start site (TSS) of LSK gene sets divided based on expression. Gene expression data from microarray experiments on LSKs was used to derive these four equally sized sets of genes. Bottom 25 % (non expressed) are shown by the blue line and the other three quartiles are divided into pink- low expressed genes, red- medium expressed genes, and top 25 % (dark red) highly expressed genes; Lower panel depicts heat maps of ChIP sequencing data across +/−10 kb around the transcription start site (TSS) of all genes sorted from highest methylation levels (top) to lowest (bottom); (C) Heat maps of ChIP sequencing data for H3K79me3 a the gene bodies of MLL-AF9 targets and control genes; (D) Meta- cross analysis of H3K79me2 (dark blue) and H3K79me1(light blue) across the gene body of MLL-AF9 target genes compared to controls. Control genes in both C and D are a size and expression matched set of non MLL-AF9 target genes.
Figure S2 related to Figure 2: (A) Normalized expression of Hoxa5-10, Actb (upper panel), and Dot1l, Af10 and Af17 (lower panel) from microarray profiling of LSK vs GMP cells in C57BL6 mice. Microarray data can be obtained from (Krivtsov et al., 2006) or from GEO accession ID GSE54500. The box plot shows normalized expression values from 6 independent replicates. Whiskers represent the upper and lower limits of the range. Boxes represent the first and third quartile, and the line represents the median. (B) Schematic representation of proteins identified in the human DOT1L complex purified from MLL-rearranged SEMK2 cells and non MLL-rearranged REH cells. VENN diagram shows the overlapping proteins DOT1L, AF10, AF17 and NPM1 found in both cell lines. NPM1, which was found at low representation, was also found in at least one empty vector control cell line. (C) Levels of AF10 or AF17 after knockdown with corresponding shRNAs relative to sh-Luc as a control are shown on the left and Western Blots for AF10 or AF17 protein after shRNA knockdown is shown on the right with LaminB1 (LMNB1) as a protein loading control. The bottom panel shows Western blots for H3K79me1 or H3K79me2 upon AF10 or AF17 knockdown in SEMK2 and MOLM13 cells with total H3 as a loading control. (D) Western blot showing H3K79me1 and H3K79me2 after DOT1L or AF10 knockdown in the MLL-rearranged MOLM13 cell line compared to cells expressing anti-Luciferase (Luc) of anti-GFP shRNAs. (E) Cell cycle changes in various cell lines upon AF10 knockdown. Error bars represent SD. (F) Mouse exons harboring the octapeptide-motif leucine zipper (OM-LZ) domain of mouse Af10 were flanked by loxp sites. Lower panel shows gel electrophoresis of representative PCR samples showing Af10 floxed and deleted amplicons. (G) Western blots for AF10, DOT1L, AF17 in nuclear lysates of HOXA9-MEIS1 transformed cells (left panel) or wh ole cell lysates of immortalized fibroblasts in the Af10fl/fl compared to the Af10−/− backgrounds. (right panel) Lamin B1 (LMNB1) or alpha-Tubulin TUBA1A are used as loading controls respectively. (H) Number of blast-like or differentiated colonies obtained from HOXA9-MEIS1 transformed cells 1 week after expression of the Cre recombinase or the empty vector in the Af10WT/WT or Af10fl/fl background are plotted; p values were calculated between the mean numbers of blast colonies in Cre expressing wildtype cells compared to Cre expressing Af10 floxed counterparts to account for Cre-mediated toxicity. *p<0.05. Error bars represent SEM. (I) Western blots showing H3K79me2 in Af10 floxed or deleted immortalized fibroblasts with or without retroviral AF10 overexpression. (J) Bar graph showing levels of H3K79me0, H3K79me1 and H3K79me2 in HOXA9-MEIS1 transformed single cell clones in the presence or absence of DOT1L. All values are shown as a percentage of total H3 peptides. On the right panel, Western blot in one of the single clones used for MS-MS is shown for DOT1L wi alpha-Tubulinth (TUBA1A) as a loading control. (K) Mass Spectrometric assessment of H3K79me0, H3K79me1 and H3K79me2 in HOXA9-MEIS1 transformed cells in the Af10fl/fl compared to Af10−/− backgrounds.
Figure S3 related to Figure 3: Effect of Af10 deletion on MLL-AF10 transformation
Colony numbers obtained 7 days after expression of the Cre recombinase in wildtype or Af10 floxed MLL-AF10 transformed cells are plotted. MLL-fusion transformed colonies are almost exclusively comprised of tight, round, hyper-cellular colonies or scattered, small colonies; n.s.= not significant. Figure represents mean from 3 independent experiments. p values were calculated between the mean numbers of blast colonies in Cre expressing wildtype cells compared to Cre expressing Af10fl/fl counterparts to account for Cre-mediated toxicity. Error bars represent SEM.
Figure S4 related to Figure 4: Abrogation of the DOT1L-AF10 interaction impairs MLL-AF9 leukemia (A) Survival curves for primary leukemia from MIT or Cre expressing MLL-AF9 transformed cells in the Af10fl/fl background. Af10 deletion significantly increases the latency of leukemia initiation. (B) Expression of Hoxa/Meis1 transcripts normalized to Actb in the Af10fl/fl compared to Af10−/− leukemias is shown. (C) Bar graph showing levels of H3K79me0, H3K79me1 and H3K79me2 in the Af10 floxed or deleted MLL-AF9 leukemias. All values are shown as a percentage of total H3 peptides. (D) Expression of Actb normalized fold change in Hoxa/Meis1 transcripts in the Af10fl/fl compared to Af10−/− leukemias in the presence of 10 μM of the EPZ004777 inhibitor are shown as a fold of DMSO vehicle control values. Error bars represent SD.
Figure S5 related to Figure 5: Epigenomic changes in MLL-AF9 leukemia upon Af10 deletion (A) A representative profile showing H3K79me3 levels upon Af10 deletion in MLL-AF9 leukemia. Y-axis scale represents read density per million. (B) Each dot in the scatter plot represents the change in average H3K79me3 levels of all genes in Af10 deficient compared to Af10 non-excised MLL-AF9 leukemias (x-axis) plotted against the absolute levels of H3K79me3 in wild-type MLL-AF9 leukemias (y-axis). Dots are coloured according to absolute H3K79me3 value in MLL-AF9 leukemia with blue being the lowest and red being the highest. (C) Changes in H3K79me1, me2 and me3 in Af10fl/fl (blue shades) compared to Af10−/− leukemias (red shades). The Hoxa7-10 genic region is marked with a dotted red rectangle. Y-axis scale represents read density per million. Values used to plot (B) and (C) are from reads per million per kb values averaged around the transcription start sites (−2 kb to +2 kb) of all genes. (D) Enrichment of H3K79me1, H3K79me2 and H3K79me3 at the promoter associated regions of Hox/Meis1 genes and controls by ChIP-qPCR. (E) A representative example of genes that gain H3K79 methylation in the Af10−/− leukemias. Y-axis scale represents read density per million. (F) GSEA showing enrichment of the DOT1L_down signature in Af10−/− compared to Af10fl/fl leukemias. (G) ChIP-qPCR for biotinylated MLL-AF9 in Af10fl/fl compared to Af10−/− deleted cells is shown at the Hoxa7-10 genes and Meis1. (n.s: not significant.) Data is presented as mean of 2 independent experiments. (H) ChIP-qPCR for bio-DOT1L at the Eya1 promoter-proximal region in Af10fl/fl compared to Af10−/− deleted cells is shown as a mean of 2 independent experiments. (p= 0.026). All p values were calculated using the Student’s T-Test. (I) Enrichment of H3K27me3 at the promoter associated regions of Hoxa genes and controls by ChIP-qPCR. (J) ChIP-seq data showing H3K27me3 deposition at select Hoxa genes in the absence of AF10. (K) Scatter plot showing changes in the values of promoter-proximal H3K79me2 (x-axis) or H3K27me3 (y-axis) in Af10fl/fl Vs Af10−/− leukemias. Each dot represents difference in averaged methylation around the promoter regions; positive and negative values reflect increases and decreases respectively in Af10−/− leukemias compared to Af10fl/fl controls. Select Hoxa cluster genes are colored and annotated. The values presented are representative of 3–5 independent sequenced samples. A similar plot with H3K79me3 changes (x-axis) plotted against H3K27me3 changes (y-axis) in Af10fl/fl Vs Af10−/− leukemias is presented on the right hand side. (L) Scatter plot showing absolute H3K79me1 levels (x-axis) plotted against corresponding absolute levels of H3K27me3 (y-axis) in Af10fl/fl compared to Af10−/− backgrounds. Error bars represent +/− SD.
Figure S6 related to Figure 6: Specificity of the H3K79me0/me1 and me2 identification by MS-MS (A–C) Unmodified (A), mono-methylated (B), and di-methylated (C) histone H3 (H33_XENLA) tryptic peptide that spans residues 74–84 are identified in the reconstituted nucleosomes incubated with DOT1L or DOT1L+ AF10 proteins. CAD mass spectrum of the (M+2H)2+ ions at m/z 659.34, 666.35, and 673.35 are shown in panels A, B and C, respectively. MS/MS spectra were collected in the Orbitrap at 30,000 resolution. Fragment ions of type b contain the amino terminus plus one or more additional residues. Fragment ions of type y contain the carboxy terminus plus one or more additional residues. b and y ions identified in the spectra are labelled.
Figure S7 related to Figure 7: Deletion of Af10 in NUP98-NSD1 transformed cells (A) Flow cytometry analysis of NUP98-NSD1 mediated transformation from BM derived LSKs cultured for 4 weeks is shown. Top panel shows empty-MSCV-puro vector transduced BM cells compared to MSCV-puro-NUP98-NSD1 derived BM cells at the bottom. NUP98-NSD1 transduced cells show an immature myeloid immunophenotype (cKitlowGr-1highMac-1high) whereas MSCV puro transduced cells show the outgrowth of contaminating mast cells (cKithighGr-1−Mac-1−). (B) Cytomorphology of Wright-Giemsa stained NUP98-NSD1 transformed cells is presented at different magnifications. (C) Western blot showing changes in global H3K79me1 and H3K79me2 5 days after Af10 deletion in NUP98-NSD1 transformed cells. Total H3 is shown as a loading control. (D) Hox gene expression in NUP98-NSD1 transformed murine BM cells in the presence or absence of the Af10 allele. Actb normalized fold expression change relative to Af10fl/fl cells is shown. * p<0.05. (E) Percentage Annexin V positive Sytox Blue negative cells (pro-apoptotic) in NUP98-NS 1 D cells with and without Af10 deletion are shown * p<0.05. Error bars represent +/− SD.
Figure S8 related to Figure 8: Sensitivity of NUP98-NSD1 cells to DOT1L inhibition (A) Global H3K79me2 levels in NUP98-NSD1 compared to control MLL-AF9 transformed hematopoietic progenitors following 10 days of treatment with 10 μM EPZ004777 as assessed by Western blotting with H3K79me2 specific antibody. Total H3 is shown as loading control. (B) Percentage Annexin V positive Sytox Blue negative cells (pro-apoptotic) in NUP98-NSD1 cells treated with 10 μM of EPZ004777 or the DMSO control are shown as mean values. * p<0.05. (C) Relative proliferation of NUP98-NSD1 cells or NUP98-NSD1 cells with retroviral HOXA9/MEIS1 overexpression in the presence of 10 μM EPZ004777 or DMSO vehicle control is depicted. Mean of 2–6 experiments is plotted in each case. Error bars represent +/− SD.
Table S1 related to Figure 1
Table S2 related to Figure 2
Table S3 related to Figure 5