Abstract
In the CNS, the transcription factor NF-κB is a key regulator of inflammation and secondary injury processes. Following trauma or disease, the expression of NF-κB-dependent genes is activated, leading to both protective and detrimental effects. In this study, we show that transgenic inactivation of astroglial NF-κB (glial fibrillary acidic protein-IκBα-dominant-negative mice) resulted in reduced disease severity and improved functional recovery following experimental autoimmune encephalomyelitis. At the chronic stage of the disease, transgenic mice exhibited an overall higher presence of leukocytes in spinal cord and brain, and a markedly higher percentage of CD8+CD122+ T regulatory cells compared with wild type, which correlated with the timing of clinical recovery. We also observed that expression of proinflammatory genes in both spinal cord and cerebellum was delayed and reduced, whereas the loss of neuronal-specific molecules essential for synaptic transmission was limited compared with wild-type mice. Furthermore, death of retinal ganglion cells in affected retinas was almost abolished, suggesting the activation of neuro-protective mechanisms. Our data indicate that inhibiting NF-κB in astrocytes results in neuroprotective effects following experimental autoimmune encephalomyelitis, directly implicating astrocytes in the pathophysiology of this disease.
Multiple sclerosis (MS)3 and its animal model experimental autoimmune encephalomyelitis (EAE) are believed to be initiated by T cell-mediated immune responses to myelin Ags. In recent years, however, a significant body of evidence has been compiled indicating the contribution of various cell populations within the CNS, namely astrocytes and microglia, to the development and progression of the disease. Nevertheless, the role of these cell types is far from being fully elucidated. In the so-called axonal hypothesis of MS, it has been postulated that chronic neurological deficits associated with MS and EAE are due to cumulative loss of axons (1). Because axonal damage is observed starting at the early stages of the disease, it is believed that accumulation of axonal degeneration is a major correlate of permanent disability (1–4). Additionally, chronic neuroinflammation, cell death, inhibition of regenerative sprouting, and remyelination may also contribute to disease progression. In all of these processes, in MS as well as in other neurodegenerative diseases, reactive astrocytes play an active role (5–9), leading us to hypothesize that this cell type may significantly contribute to disease progression and development of chronic neurological deficits in EAE and MS. Because many of the processes occurring in reactive astrocytes are regulated by the NF-κB family of transcription factors, we designed the present study to investigate the role of astroglial NF-κB in the pathophysiology of EAE. We used a transgenic model previously generated in our laboratory (glial fibrillary acidic protein (GFAP)-IκBα-dominant-negative (dn) mice) (10) in which NF-κB is inactivated in astrocytes through overexpression of a dn form of the NF-κB superrepressor (IκBα-dn) under the control of the GFAP promoter. In this study, we show that blocking astroglial NF-κB significantly reduces disease severity and improves functional recovery following EAE, providing a direct demonstration of the critical role of astrocytes in the pathophysiology of a disease long considered to be mediated solely by leukocytes.
Materials and Methods
Mice
Experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the University of Miami. GFAP-IκBα-dn mice were generated in the Transgenic Core Facility of the University of Miami, as previously described (10). All mice used were 2–4 mo old and obtained by breeding heterozygous GFAP-IκBα-dn males with wild-type (WT) females. WT littermates were used as controls. Animals were housed in a 12-h light/dark cycle in a virus/Ag-free facility with controlled temperature and humidity, and provided with water and food ad libitum.
Induction of EAE
Active EAE were induced with myelin oligodendrocyte glycoprotein (MOG)35–55 peptide, as previously described (11). Clinical signs of EAE were assessed daily using a standard scale of 0–6, as follows: 0, no clinical signs; 1, loss of tail tone; 2, flaccid tail; 3 complete hind limb paralysis; 4, complete fore limb paralysis; 5, moribund; and 6, death. Mice were considered to have onset of EAE when they reached a score of 2 or more for at least 2 consecutive days. For each animal, a cumulative disease index was calculated as sum of the daily clinical scores recorded between days 7 and 60.
T cell proliferation assay
Ag-specific T cell proliferation assays were performed, as previously described (11).
Isolation of leukocytes from spinal cords and spleens
Following perfusion with PBS, spinal cords and spleens were harvested and placed in cold HBSS. Samples were homogenized into single-cell suspensions by passing the tissues through a 70-μm cell strainer and washed in HBSS. Suspensions were spun at 400 × g for 5 min, and cells were collected, resuspended in 40% Percoll, and layered on 70% Percoll. Samples were centrifuged at 400 × g for 25 min at room temperature. Cells at the gradient interface were removed, washed in FACS buffer (eBio-science), and stained, as described below. Before staining, RBCs were lysed with 1× RBC lysis buffer (eBioscience), according to manufacturer's protocols. Spinal cords from eight animals were pooled to obtain a sufficient number of cells for flow cytometric analysis.
Staining and flow cytometry
Cells obtained from spinal cords and spleens were incubated with anti-CD16/32 (FcR block; eBioscience) to prevent nonspecific staining. Cells were then incubated for 30 min in the dark at 4°C with Alexa488-conjugated anti-CD3, Pacific Blue-conjugated or PE-conjugated anti-CD4, allophycocyanin-conjugated anti-CD8, PE-Cy7-conjugated anti-CD25, PE-conjugatedanti-CD122, and allophycocyanin-Alexa750-conjugatedanti-B220, all from eBioscience. Following staining of cell surface Ags, intra-cellular Foxp3 staining was performed with PE-conjugated anti-Foxp3 (FJK-16s; eBioscience), according to manufacturer's instructions. The cells were analyzed using an LSR II flow cytometer with FACSDiva 6.0 software (BD Biosciences).
Immunohistochemistry
Animals were perfused with 4% paraformaldehyde in 0.1 M PBS. Tissues were cryoprotected in 0.1 M PBS plus 25% sucrose, and cut with cryostat into 15-μm-thick sections. Sections were incubated overnight at 4°C with Abs against the following proteins: GFAP (rat, 1:1000, Zymed Laboratories; mouse, 1:1000, Chemicon International), CD11b (rat, 1:200; Serotec), CD45 (rat, 1:200; eBioscience), Aquaporin-4 (rabbit, 1:200; Alpha Diagnostics), TNF-α (rabbit, 1:200; Santa Cruz Biotechnology), CXCR3 (rabbit, 1:200; Novus Biologicals), CCR2 (rabbit, 1:200; Epitomics), followed by secondary fluorescent Abs (Alexa Fluor 488, 594, and 660, 1:750; Invitrogen) for 1 h at room temperature. Images were obtained with a Zeiss LSM 510 confocal microscope. For these experiments, four animals/group were used.
Microarray analysis and data processing
Microarray experiments were conducted at the University of Miami DNA and Microarray Core Facility (www.biomed.miami.edu/arrays) using Agilent Whole Mouse Genome Oligo microarrays (Agilent Technologies). Arrays were scanned at a 10-μm resolution using a GenePix 4000A scanner (Axon Instruments at Molecular Devices), and images were analyzed with the software GenePix Pro 5.1 (Axon Instruments at Molecular Devices). Extracted data were transferred to the software Acuity 4.0 (Axon Instruments at Molecular Devices) for normalization and statistical analysis. Each array was normalized for signal intensities across the whole array and locally using Lowess normalization. Features for further analysis were selected according to the following quality criteria: at least 90% of the pixels in the spot with intensity higher than background plus 2 SDs; less than 2% saturated pixels in the spot; signal to noise ratio (ratio of the background subtracted mean pixel intensity to SD of background) 3 or above for each channel; spot diameter between 80 and 110 μm; regression coefficient of ratios of pixel intensity 0.6 or above.
To identify significantly expressed genes, the R software LIMMA (12) was used. Within array normalization was conducted with Lowess normalization, and between arrays normalization with the quantile algorithm in the LIMMA package. Differential expression and false discovery rate (FDR) were assessed using a linear model and empirical Bayes moderated F statistics (13, 14). Genes with FDR below 1% were considered statistically significant. All primary microarray data were submitted to the public database at the Gene Expression Omnibus website (http://www.ncbi.nih.gov/geo; record number: GSE10235). Selected genes were classified according to Gene Ontology category biological process using Onto-Express (http://vortex.cs.wayne.edu/Projects.html). Pathway analysis was performed with Gene Spring 7.3 (Agilent Technologies). All experiments were performed in three replicates/group/time point.
Total RNA isolation and real-time RT-PCR
Total RNA was extracted with TRIzol (Invitrogen). Reverse transcription was performed with Superscript II (Invitrogen), according to manufacturer's instructions. cDNA equal to 10 –50 ng of total RNA was used as template in each PCR. Real-time PCR was performed in the Rotor-Gene 3000 Real Time Cycler (Corbett Research) with TAQurate Green Real-Time PCR MasterMix (Epicentre Biotechnologies). Relative expression was calculated by comparison with a standard curve after normalization to β-actin. Primers for gene amplification are listed in Supplementary Table III.4
Western blot
Proteins were resolved by SDS-PAGE on 6–15% gels, transferred to nitrocellulose, and blocked in 5% milk. Membranes were probed with Abs against the following proteins: GAP43 (rabbit, 1:1000; GeneTex), myelin basic protein (MBP; rat, 1:5000; Chemicon International), and CD45 (rat, 1:1000; eBioscience), followed by HRP-conjugated secondary Abs. Proteins were visualized with ECL (GE Healthcare/Amersham), and bands were quantified with the software Quantity One (Bio-Rad). Data were normalized against β-actin and expressed as percentage of WT.
Retinal stereology
Retinal ganglion cell (RGC) count was performed by confocal imaging in fixed (2% paraformaldehyde in PBS (pH 7.5), 2 h) flat-mounted retinas stained with Cy3-conjugated anti-NeuN Ab (Millipore). Retinas were sampled randomly to collect a total of 20 images 1–2 mm away from the optic disk in the four retinal quadrants using a ×20 objective. Neurons with the size range of 6–30 μm were counted semiautomatically using MetaMorph (Molecular Devices) after image thresholding and manual exclusion of artifacts.
Statistical analysis
Statistical analysis of EAE experiments was conducted with the Mann-Whitney U test. Real-time PCR and cell density data were analyzed with one-way ANOVA, followed by Tukey test for multiple comparisons. For single comparisons, Student's t test was applied. Values of p equal to or less than 0.05 were considered statistically significant.
Results
Inhibition of astroglial NF-κB resulted in reduced disease severity and improved functional recovery following EAE
To determine whether blocking NF-κB in astrocytes would alter the development and progression of EAE, we induced MOG35–55- mediated EAE in WT and GFAP-IκBα-dn mice, as previously described (11). The GFAP-IκBα-dn transgenic mice were generated in our laboratory by overexpressing in astrocytes a dn form of IκBα with the GFAP promoter. After extensive characterization, which has been previously published (10, 15), we determined that the GFAP-IκBα-dn mice do not have any phenotypic alterations and they are in every aspect indistinguishable from the WT littermates. The expression of the transgene is localized to astrocytes and GFAP-expressing nonmyelinating Schwann cells. No ectopic expression of the transgene was found outside of the nervous system. Furthermore, we analyzed by flow cytometry with a standard panel of cell surface markers (CD4, CD8, B220, NK1.1, and CD11b) the immune cell profile of the naive transgenic mice in spleen, thymus, bone marrow, and lymph nodes, and found no differences in the numbers and phenotypes between the GFAP-IκBα-dn mice and their WT counterpart (data not shown).
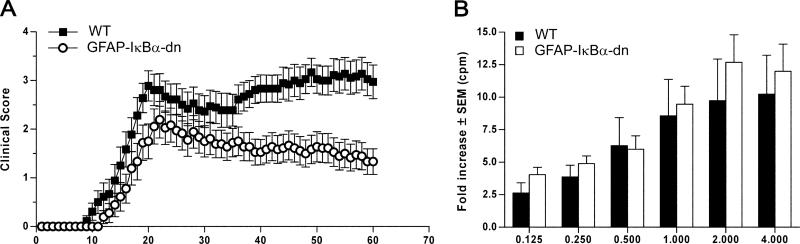
Following induction of EAE, we observed a significant reduction of disease severity in GFAP-IκBα-dn mice compared with WT (CDI: WT = 121.3 ± 8.5 vs GFAP-IκBα-dn = 61.3 ± 10.6, p < 0.001) (Table I), resulting in dramatic improvement in functional recovery from 37 to 60 days postinduction (dpi) (Fig. 1A). In a separate study in which animals were allowed to survive up to 80 dpi, GFAP-IκBα-dn mice steadily maintained the same improved functional recovery over WT mice as assessed at 60 dpi (data not shown). Aside from a lower incidence of the disease (Table I), GFAP-IκBα-dn mice developed EAE similarly to WT mice (onset: WT = 16.2 ± 0.8 vs GFAP-IκBα-dn = 18.2 ± 0.8) (Table I) with acute symptoms of reduced tail tone and impaired locomotion virtually indistinguishable from those shown by WT mice. Chronically, however, GFAP-IκBα-dn mice displayed a steady functional improvement, whereas WT mice showed no improvement (final scores at 60 dpi: WT = 2.97 ± 0.35 vs GFAPIκBα-dn = 1.33 ± 0.26, p < 0.001).
Table I.
Clinical parameters of EAEa
| Genotype | Incidence | Onset (day) | CDI |
|---|---|---|---|
| WT | 100% (18 of 18) | 16.2 ± 0.8 | 121.3 ± 8.5 |
| GFAP-IκBa-dn | 72% (13 of 18) | 18.2 ± 0.8 | 61.3 ± 10.6* |
Results represent the mean ± SEM of 18 animals/group from two independent experiments. Onset is considered when animals reach a clinical score of 2 for 2 consecutive days. CDI is calculated as the sum of the clinical scores of each animal between days 7 and 60 and is a measure of disease severity.
p < 0.001 with respect to WT; Student's t test.
FIGURE 1.
Inhibition of astroglial NF-κB improves functional outcome following EAE. A, Clinical course of MOG35–55-induced EAE in WT and GFAP-IκBα-dn mice. EAE symptoms were scored daily for 60 days, as described in Materials and Methods. Results are expressed as the daily mean clinical score ± SEM of 18 animals/group from two independent experiments. Curves are significantly different (p < 0.0001, Mann-Whitney U test). B, Cell proliferation assay on T cells isolated from WT and GFAP-IκBα-dn mice undergoing active EAE 2 wk after immunization. Following exposure to increasing MOG concentrations, cells were pulsed with [3H]thymidine, and proliferation was assessed in cpm of incorporated tritiated thymidine. Results are expressed as mean fold increase incorporation relative to background ± SEM of four samples/group.
Because previous characterization of the GFAP-IκBα-dn transgenic model revealed expression of the transgene in the peripheral nervous system (Schwann cells) (10), to rule out the possibility that interfering with NF-κB in Schwann cells could impair the T cell response due to physiologic cross-talk with the immune system, thereby facilitating functional improvements in GFAPIκBα-dn mice, we performed cell proliferation assays on T cells obtained from both WT and GFAP-IκBα-dn mice (Fig. 1B). No differences were detected between the two genotypes at any concentrations of MOG peptide used.
GFAP-IκBα-dn mice show higher presence of leukocytes than WT mice at chronic disease
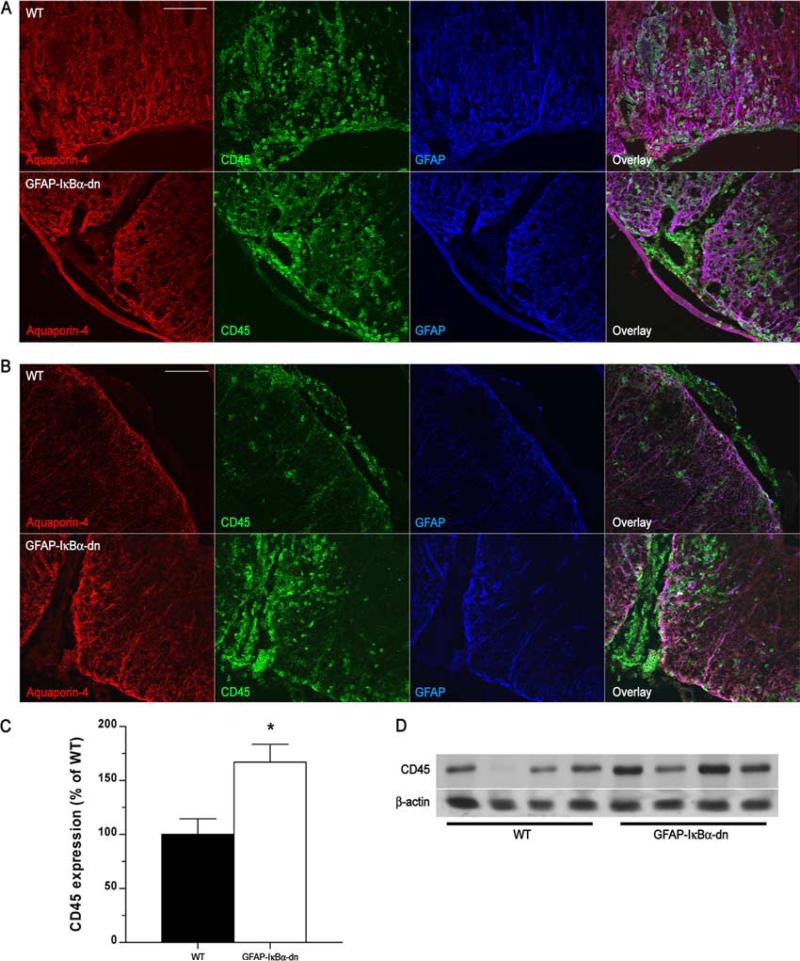
It is generally accepted that the development of EAE is associated with infiltration of leukocytes (particularly encephalitogenic CD4+ and CD8+ T cells, macrophages) into the CNS, which initiate an inflammatory response resulting in demyelination, oligodendrocyte loss, and neuronal degeneration (16). To assess the extent of leukocyte infiltration, we performed immunohistochemical studies on spinal cords and brains from naive and diseased mice at 20 and 40 dpi. The leukocyte population was labeled with an anti-CD45 Ab, and astrocytes and astrocyte end-feet were identified with anti-GFAP and anti-Aquaporin-4, respectively (Fig. 2, A and B). Strong astrocytic activation was observed in both geno-types, as indicated by intense GFAP and Aquaporin-4 immuore-activity. In naive tissue, CD45+ leukocytes were not detected (data not shown). In the spinal cord of both genotypes, massive leukocyte infiltration was evident at 20 dpi, coinciding with the highest degree of motor impairment (Fig. 2A). Both localized perivascular and diffuse parenchymal infiltrates were observed. At 40 dpi, contrary to our expectations, a higher presence of infiltrates was detected in transgenic mice compared with WT (Fig. 2B). Indeed, few leukocytes were found in the spinal cord parenchyma of WT mice, whereas large clusters were concentrated in the perivascular cuffs and parenchyma of transgenic mice (Fig. 2B). A similar pattern was observed in the cerebellum (Supplementary Fig. 1).4 In support of the immunohistochemical assessment, a significantly higher expression of CD45 was measured by Western blot in GFAP-IκBα-dn mice compared with WT at 40 dpi (Fig. 2, C and D). These data indicate an inverse correlation between disease severity and leukocyte presence in the CNS. Indeed, an increased leukocyte presence parallels a reduced pathology in GFAPIκBα-dn mice at chronic stages of EAE, raising the possibility that populations of protective anti-inflammatory leukocytes (e.g., T regulatory cells (Tregs)) may represent a significant subset of these cells in the GFAP-IκBα-dn mice.
FIGURE 2.
Leukocyte infiltration in WT and GFAP-IκBα-dn spinal cords following EAE. A, Immunofluorescent labeling of WT and GFAP-IκBα-dn tissue sections at 20 dpi (acute disease). B, Immunofluorescent labeling of WT and GFAP-IκBα-dn tissue sections at 40 dpi (chronic disease). Red: Aquaporin-4 (localized to astrocyte end-feet in contact with the vascular endothelium). Green: pan-leukocyte marker CD45. Blue: astrocyte-specific marker GFAP. Scale bar: 100 μm. C, Quantification of CD45 protein expression in spinal cord tissue of WT and GFAP-IκBα-dn mice at 40 dpi. Data are normalized to β-actin. Results represent the mean ± SEM of six animals/group and are expressed as percentage of WT. *, p < 0.05; Student's t test. D, Representative Western blot probed for CD45 and β-actin as loading control.
GFAP-IκBα-dn mice show increased CD8+ CD122+ Tregs compared with WT mice at chronic disease
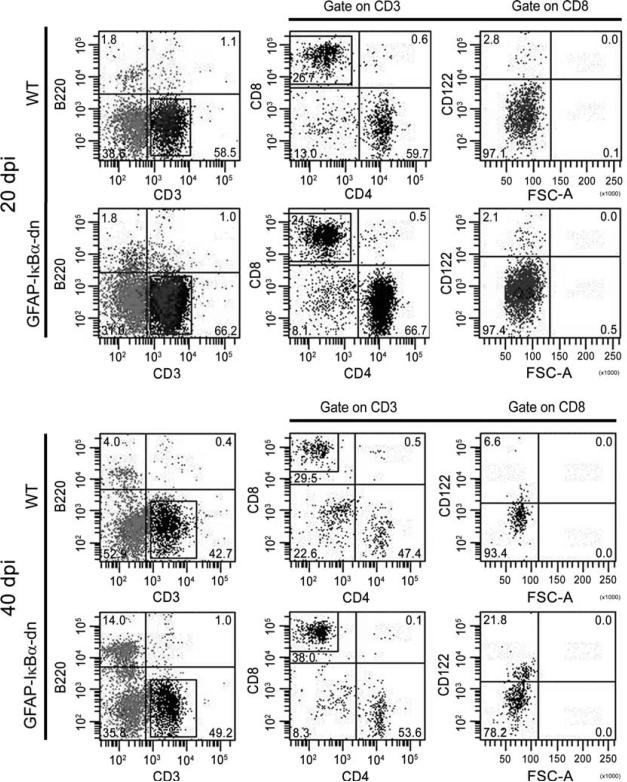
To assess the profile of T cell infiltration, we performed flow cytometric analysis of cell infiltrates into the cord of WT and transgenic mice at acute (20 dpi) and chronic (40 dpi) disease. Due to the very low number of infiltrating cells, eight animals/group/time point were pooled to obtain samples suitable for analysis. Acutely (20 dpi), almost no differences were found in the leukocyte profiles of WT and transgenic mice (Table II). Indeed, equal percentages of B cells (B220+) and T cells (CD3+) as well as CD4+ and CD8+ T cell subsets were found. The Treg populations of CD4+CD25+Foxp3+ and CD8+CD122+ cells were also equal (Table II; Fig. 3). These data correlate with the observation that WT and transgenic mice develop a similar pattern of EAE up to the acute phase of the disease (Fig. 1A). In contrast, at the chronic stage of the disease (40 dpi), the distribution of various leukocyte populations resulted to be different between WT and GFAPIκBα-dn mice (Table II; Fig. 3). Specifically, a higher percentage of B cells (WT 4.0% vs GFAP-IκBα-dn 14.0%) and a lower percentage of B220−CD3− cells (WT 51.8% vs GFAP-IκBα-dn 36.7%), which are mainly a combination of NKs and macrophages, were found in transgenic mice. Furthermore, although we did not detect differences in CD4+ and CD8+ subsets nor in CD4+CD25+Foxp3+ Tregs, we observed in the transgenic mice a marked increase in CD8+CD122+ Tregs with respect to WT. These data indicate that whereas the CD4+CD25+Foxp3+ Treg population, normally involved in protection against autoimmunity, does not appear to be associated with the improved clinical outcome of GFAP-IκBα-dn mice, it is the subset of naturally occurring CD8+CD122+ Tregs that could be directly associated to this effect.
Table II.
Flow cytometric analysis of leukocyte infiltrates into the spinal cord of EAE-induced mice at 20 and 40 dpia
| CD3 Gate |
CD4 Gate |
CD8 Gate |
|||||
|---|---|---|---|---|---|---|---|
| Time & Experimental Group | B Cells (B220+) | T Cells (CD3+) | B220–CD3– Cells | CD4+ | CD8+ | CD25+Foxp3+ | CD122+ |
| 20 dpi | |||||||
| WT | 1.9 | 59.3 | 39.8 | 57.9 | 25.9 | 17.4 | 2.6 |
| GFAP-Iκ Ba-dn | 1.7 | 65.4 | 28.8 | 60.3 | 29.4 | 17.7 | 2.1 |
| 40 dpi | |||||||
| WT | 3.8 | 45.6 | 51.8 | 45.5 | 32.9 | 1.2 | 6.4 |
| GFAP-Iκ Ba-dn | 13.5 | 51.3 | 36.7 | 47.8 | 38.0 | 0.8 | 20.5 |
Results represent the mean of two independent experiments each conducted by pooling eight animals/group.
FIGURE 3.
Flow cytometric analysis of leukocyte infiltrates into the spinal cord of EAE-induced mice at 20 and 40 dpi. A representative experiment conducted by pooling eight animals/group is shown. The numbers in the quadrants represent the relative percentages of the various cell populations.
Gene expression analysis with Whole Mouse Genome microarrays following EAE
To elucidate the molecular mechanisms responsible for the functional recovery in GFAP-IκBα-dn mice, we investigated changes in gene expression using Whole Mouse Genome microarrays, which included 41,000 genes and transcripts. The experiment was conducted on naive animals, as well as animals at pre- (10 dpi), acute (17 dpi), and chronic (80 dpi) disease stages. Behaviorally, the animals exhibited similar scores, as reported in Fig. 1A, with a significant functional improvement of transgenic mice over WT at chronic (40–80 dpi) time points (data not shown). Three animals per group were compared. Analysis of microarrays demonstrated that there were no significant differences in gene expression between noninduced naive WT and transgenic mice (only 13 genes were changed, and all below the threshold of 2.0; Supplementary Table I).4 However, significant changes were present at pre-, acute, and chronic disease time points in genes involved in inflammation, immunity, cell death, signal transduction, axonal growth, synaptogenesis, metabolism, etc., all processes that may influence functional recovery (Table III; National Center for Biotechnology Information Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/, record number: GSE10235). We concentrated on genes with a fold-change greater than 2.0 and a FDR <0.1%. The highest number of differentially expressed genes was observed at 17 dpi, which corresponds to the acute phase of the disease (Supplementary Table I).4 Remarkably, at this time point, over 88% of the differentially expressed genes were down-regulated in GFAPIκBα-dn mice compared with WT (chemokines, cytokines, and TLRs were the families of molecules most significantly affected). Furthermore, many of those genes are expressed by astrocytes and directly regulated by NF-κB, such as, for example, the stress-associated gene nuclear protein 1 (better known as p8), which was found dramatically down-regulated in transgenic mice compared with WT at 17 dpi (Table III) and 80 dpi (see data at National Center for Biotechnology Information Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/). This is an intriguing finding because increased expression of nuclear protein 1 in the brain has been associated with inflammation and demyelination (17). Interestingly, the small set of genes expressed at higher levels in transgenic mice at both 17 and 80 dpi represented molecules involved in metabolic processes or axonal growth and synaptogenesis, which correlate with the improved functional recovery seen in GFAP-IκBα-dn mice (Table III; National Center for Biotechnology Information Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/). Additionally, we performed a pathway analysis with GeneSpring that allowed for the identification of signaling pathways showing, in their entirety, a significant change between the two genotypes. No significantly different pathways were found in naive mice and at 10 dpi, whereas several pathways related to immune/inflammatory processes were changed at 17 and 80 dpi (Supplementary Table II).4 Interestingly, the chemokine-chemokine receptor interaction pathway showed the highest number of differentially expressed genes (41 genes at 17 dpi), and was relatively down-regulated in GFAP-IκBα-dn mice compared with WT at both 17 and 80 dpi (Supplementary Fig. 2).4 This implies that inhibition of NF-κB in astrocytes leads to dramatic reduction in the expression of molecules (such as chemokines) necessary for T cell recruitment and activation in the injured CNS, where they propagate the detrimental inflammatory response.
Table III.
Microarray analysis of GFAP-Iκ Ba-dn mice vs WT at 17 dpi of EAEa
| Gene Bank ID | Symbol | Description | Fold Changeb | FDR (%)c |
|---|---|---|---|---|
| Immune Related | ||||
| NM_007535 | Bcl2a1c | B cell leukemia/lymphoma 2-related protein A1c | –5.15 | 4.5e-7 |
| NM_178611 | Lair1 | Leukocyte-associated Ig-like receptor 1 | –2.97 | 2.6e-6 |
| NM_010382 | H2-Eb1 | Histocompatibility 2, class II Ag Eβ | –3.29 | 4.1e-6 |
| NM_134158 | Cd300d | Cd300D Ag | –3.15 | 6.2e-6 |
| NM_013489 | Cd84 | CD84 Ag | –4.21 | 1.1e-5 |
| NM_011095 | Pirb | Paired Ig-like receptor B | –8.47 | 3.1e-5 |
| NM_007639 | Cd1d1 | CD1d1 Ag | –3.41 | 3.1e-5 |
| NM_010511 | IfngR1 | IFN-γ receptor 1 | –3.22 | 4.6e-5 |
| NM_008329 | Ifi204 | IFN-activated gene 204 | –3.83 | 4.8e-5 |
| NM_008620 | Mpa2 | Macrophage activation 2 | –5.44 | 7.3e-5 |
| NM_007651 | Cd53 | CD53 Ag | –3.14 | 8.2e-5 |
| NM_008326 | Ifi1 | IFN-inducible protein 1 | –3.62 | 9.7e-5 |
| NM_010378 | H2-Aa | Histocompatibility 2, class II Ag A, α | –6.75 | 2.0e-4 |
| NM_009779 | C3ar1 | Complement component 3a receptor 1 | –3.20 | 1.9e-4 |
| NM_021325 | Cd200r1 | CD200 receptor 1 | –3.12 | 2.7e-4 |
| NM_010379 | H2-Ab1 | Histocompatibility 2, class II Ag A, β1 | –6.60 | 9.5e-4 |
| NM_023143 | C1r | Complement component 1, r subcomponent | –5.29 | 1.4e-3 |
| NM_009778 | C3 | Complement component 3 | –3.15 | 2.2e-2 |
| TLRs | ||||
| NM_133211 | Tlr7 | TLR7 | –3.60 | 1.2e-5 |
| NM_030682 | Tlr1 | TLR1 | –2.95 | 6.2e-5 |
| NM_011905 | Tlr2 | TLR2 | –3.80 | 2.5e-4 |
| NM_011604 | Tlr6 | TLR6 | –2.48 | 5.1e-4 |
| NM_126166 | Tlr3 | TLR3 | –2.09 | 5.2e-1 |
| NM_021297 | Tlr4 | TLR4 | –1.80 | 5.7e-1 |
| Chemokines | ||||
| BC033508 | CCL5 | Chemokine (C-C motif) ligand 5 | –8.65 | 2.1e-5 |
| NM_023158 | CXCL16 | Chemokine (C-X-C motif) ligand 16 | –4.88 | 6.2e-5 |
| NM_009915 | CCR2 | Chemokine (C-C motif) receptor 2 | –5.79 | 1.4e-4 |
| NM_011888 | CCL19 | Chemokine (C-C motif) ligand 19 | –2.08 | 1.4e-4 |
| NM_011329 | CCL1 | Chemokine (C-C motif) ligand 1 | –1.93 | 1.8e-4 |
| NM_011338 | CCL9 | Chemokine (C-C motif) ligand 9 | –1.97 | 4.0e-4 |
| NM_008599 | CXCL9 | Chemokine (C-X-C motif) ligand | –7.11 | 8.8e-4 |
| NM_009917 | CCR5 | Chemokine (C-C motif) receptor 5 | –2.33 | 1.1e-3 |
| NM_009911 | CXCR4 | Chemokine (C-X-C motif) receptor 4 | –2.61 | 1.9e-3 |
| NM_030712 | CXCR6 | Chemokine (C-X-C motif) receptor 6 | –4.85 | 3.2e-3 |
| NM_017466 | CCRl2 | Chemokine (C-C motif) receptor-like 2 | –2.07 | 1.2e-2 |
| NM_021704 | CXCL12 | Chemokine (C-X-C motif) ligand 12 | –2.58 | 2.3e-2 |
| NM_011332 | CCL17 | Chemokine (C-C motif) ligand 17 | –1.99 | 3.2e-2 |
| NM_011330 | CCL11 | Small chemokine (C-C motif) ligand 11 | –1.94 | 9.3e-2 |
| NM_009139 | CCL6 | Chemokine (C-C motif) ligand 6 | –4.57 | 1.2e-1 |
| NM_019577 | CCL24 | Chemokine (C-C motif) ligand 24 | –1.76 | 7.1e-1 |
| NM_013654 | CCL7 | Chemokine (C-C motif) ligand 7 | –2.29 | 7.5e-1 |
| Cytokines | ||||
| NM_008353 | IL12rb1 | IL-12R, β1 | –5.25 | 3.4e-6 |
| NM_008349 | IL10rb | IL-10R, β | –2.62 | 4.2e-6 |
| NM_008361 | IL1b | IL-1β | –3.13 | 2.4e-4 |
| NM_010555 | IL1r2 | IL-1R, type II | –4.30 | 5.8e-4 |
| NM_008372 | IL7R | IL-7R | –3.48 | 6.1e-4 |
| NM_009371 | TgfbR2 | TGF, β receptor II | –2.35 | 1.3e-3 |
| NM_172161 | Irak2 | IL-1R-associated kinase 2 | –1.57 | 5.2e-3 |
| NM_009369 | Tgfbi | TGF, β induced | –7.82 | 5.6e-3 |
| NM_010531 | IL18bp | IL-18-binding protein | –1.55 | 7.8e-3 |
| NM_013584 | LIFr | Leukemia-inhibitory factor receptor | 1.67 | 1.2e-2 |
| NM_013563 | IL2rg | IL-2R, γ-chain | –2.94 | 1.3e-2 |
| Stress/Apoptosis | ||||
| NM_019738 | Nupr1 (p8) | Nuclear protein 1 (also p8, com1) | –5.07 | 9.8e-6 |
| NM_207653 | Cflar | CASP8 and Fas-associated death domain protein-like apoptosis regulator | –1.93 | 4.5e-5 |
| NM_007609 | Casp11 | Caspase 11, apoptosis-related cysteine protease | –3.93 | 1.1e-3 |
| NM_009807 | Casp1 | Caspase 1 | –2.63 | 2.9e-3 |
| NM_007611 | Casp7 | Caspase 7 | –1.94 | 4.6e-3 |
| Signaling | ||||
| NM_008689 | NFκ B1 | NF-κB1, p105 | –1.99 | 2.4e-5 |
| NM_145133 | MGI:2182965 | TNFR-associated factor 2-binding protein | –2.30 | 4.9e-5 |
| NM_009283 | Stat1 | STAT1 | –2.59 | 1.2e-4 |
| BC029318 | Stat6 | STAT6 | –3.49 | 4.2e-4 |
| NM_007486 | Arhgdib | Rho, GDP dissociation inhibitor (GDI) β | –3.25 | 1.2e-3 |
| NM_019401 | Nmi | N-myc (and STAT) interactor | –1.88 | 3.6e-3 |
| NM_013498 | Crem | cAMP-responsive element modulator | –1.75 | 4.3e-3 |
| Miscellaneous | ||||
| NM_008614 | Mobp | Myelin-associated oligodendrocyte basic protein | 1.77 | 2.1e-3 |
| NM_008747 | Ntsr2 | Neurotensin receptor 2 | 1.64 | 2.1e-3 |
| NM_009898 | Coro1a | Coronin, actin-binding protein 1A | –1.72 | 3.1e-3 |
| NM_009115 | S100b | S100 protein, β polypeptide, neural | 1.68 | 8.8e-3 |
| NM_011428 | Snap25 | Synaptosomal-associated protein 25 | 1.56 | 8.8e-3 |
| NM_008220 | Hbb-b1 | Hemoglobin, β adult major chain | 2.21 | 1.0e-2 |
List of representative differentially expressed genes.
Values in regular font: GFAP-Iκ Ba-dn down-regulated vs WT; values in boldface: GFAP-Iκ Ba-dn up-regulated vs WT.
Measure of statistical significance: Genes with FDR <1% were considered statistically significant.
Inhibition of astroglial NF-κB resulted in reduced expression of cytokines and chemokines following EAE
To validate the microarray results and investigate differences in expression of gene families specifically involved in the disease process, we evaluated by real-time PCR changes in the expression of chemokines, cytokines, integrins (Itg), and cell adhesion molecules, all of which play important roles in the initiation and maintenance of inflammatory responses in EAE.
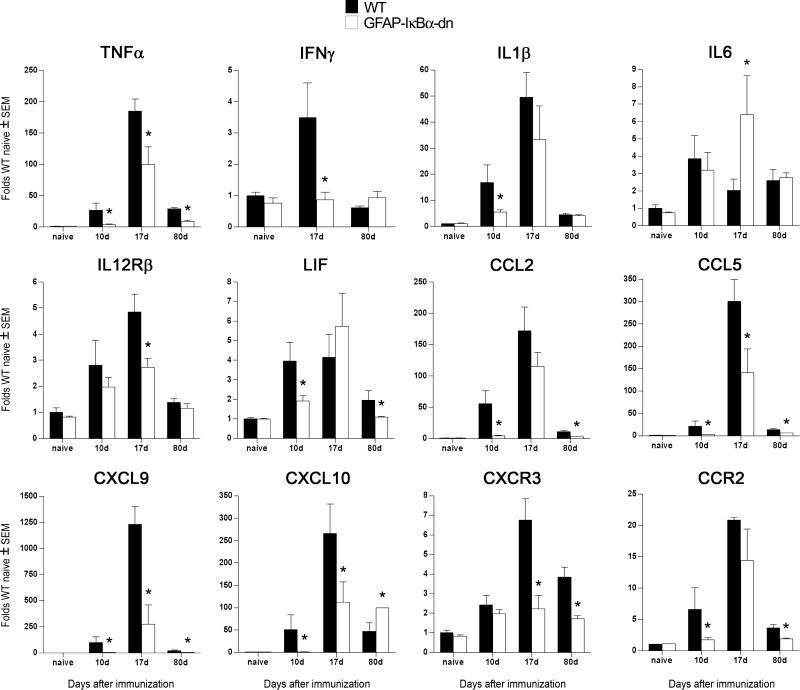
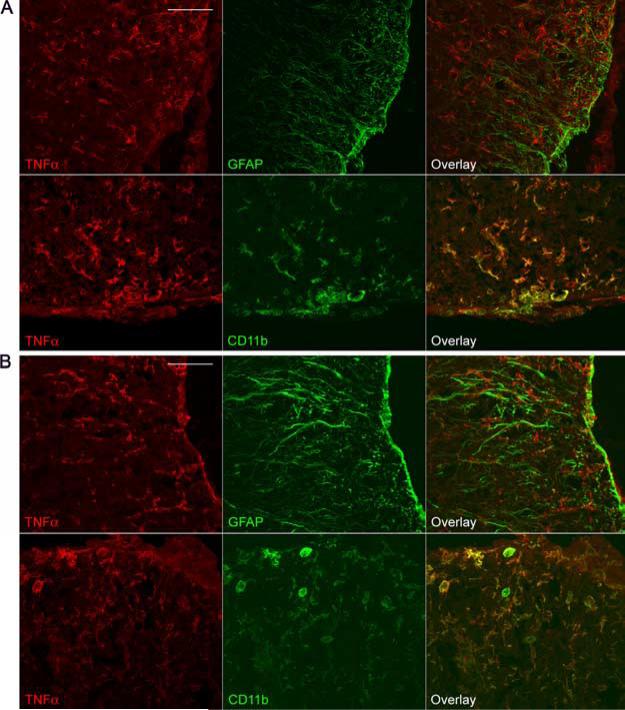
Cytokines like IL-1β and TNF-α have been demonstrated to act as proinflammatory mediators. They regulate the expression of cytokines, chemokines, cell adhesion molecules, and other immuno-regulatory molecules through NF-κB-dependent and independent pathways. TNF-α expression was elevated in WT spinal cords with EAE, but was significantly reduced in GFAP-IκBα-dn mice compared with WT at all time points (Fig. 4). To identify the cellular source of TNF-α, we conducted coimmunolabeling experiments with anti-TNF-α in combination with either the astrocyte-specific marker GFAP or the microglia/macrophage marker CD11b. In both genotypes, at 20 dpi, TNF-α was localized to microglia and macrophages, and was absent in astrocytes (Fig. 5). At 40 dpi, TNF-α was virtually undetectable in both WT and GFAP-IκBα-dn mice (data not shown). IFN-γ, which is known to be involved in the pathogenesis of EAE through its role in promoting the differentiation of T cells toward a Th1 proinflammatory phenotype (18), was significantly lower in GFAP-IκBα-dn mice compared with WT at 17 dpi (Fig. 4). Remarkably, no increase over naive levels was observed in GFAP-IκBα-dn mice at all times tested. As for IL-6, which can promote Th1 differentiation similarly to IFN-γ, a significant up-regulatation was detected in GFAPIκBα-dn mice compared with WT at 17 dpi. IL-12Rβ1, the receptor subunit that interacts with IL-12 and IL-23, two cytokines that have been recently linked to the etiology of EAE (19–22), was significantly reduced in GFAP-IκBα-dn mice at 17 dpi, in agreement with the microarray findings (Table II; National Center for Biotechnology Information Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/).
FIGURE 4.
Differential expression of cytokines and chemokines in WT and GFAP-IκBα-dn spinal cord tissue following EAE. Gene expression was assessed in naive animals and at 10, 17, and 80 dpi. For each gene, results are expressed as folds of corresponding WT naive ± SEM after normalization to β-actin. Four animals/group/time were analyzed. *, p < 0.05 with respect to corresponding WT; one-way ANOVA, Tukey test.
FIGURE 5.
TNF-α protein localization in WT and GFAP-IκBα-dn spinal cords following EAE. Immunofluorescent labeling of WT (A) and GFAP-IκBα-dn (B) tissue sections at 20 dpi (acute disease). Red: TNF-α. Green: GFAP or CD11b. Scale bar: 100 μm.
Chemokines are essential for trafficking of leukocytes into the CNS under physiological and pathological conditions (23). In line with previous reports, we found dramatic up-regulation of chemokine expression following EAE (Fig. 4). This up-regulation, however, was significantly limited in GFAP-IκBα-dn mice compared with WT. For example, CXCL9, CXCL10, and their receptor CXCR3 were down-regulated at all disease time points, whereas CCL2 and its receptor CCR2 were reduced in GFAP-IκBα-dn mice at pre- and chronic disease stages. Both chemokine receptors, CXCR3 and CCR2, were found to be localized in microglia and macrophages, but not in astrocytes, as indicated by coimmunolabeling experiments (Supplementary Figs. 3 and 4, respectively).4 Finally, CCL5 was significantly down-regulated in GFAPIκBα-dn mice at all time points. These data strongly indicate that improvements in functional recovery could be due, at least in part, to attenuated chemokine expression, which might reduce the infiltration of activated leukocytes into the CNS, thereby limiting disease severity. In addition, our results provide additional proof that CXCL10 and CCL2 in astrocytes are regulated by NF-κB, in agreement with previous studies (24).
Inhibition of astroglial NF-κB resulted in reduced expression of adhesion molecules and Itg following EAE
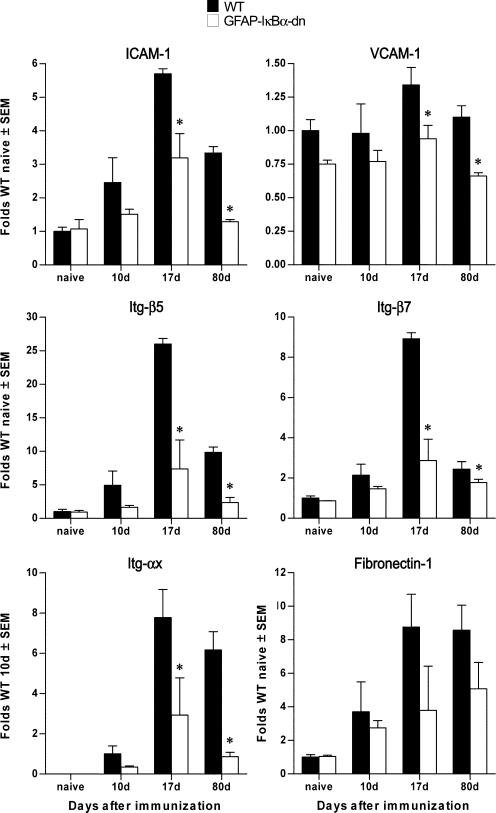
Cell adhesion molecules and receptors are critical for immune cell activation and trafficking of peripheral immune cells across the blood-brain barrier (BBB) into the CNS parenchyma (25–28). ICAM-1, VCAM-1, fibronectin-1, Itg-β5, Itg-β7, and Itg-αx were found to be differentially expressed in our microarray experiments. Our real-time PCR data confirmed a significant reduction in transcripts encoding for ICAM-1, VCAM-1, Itg-β5, and Itg-β7, which were significantly reduced in GFAP-IκBα-dn mice compared with WT at pre-, acute, and chronic disease (Fig. 6). These results demonstrated that astrocytes directly control the expression of some of the molecules required for T cell activation (e.g., ICAM-1) and maintenance of BBB integrity (e.g., ICAM-1, VCAM, and Itg). This opens the possibility that one of the mechanisms responsible for the reduced disease severity and improved functional recovery in GFAP-IκBα-dn mice could be a reduction in immune cell activation following entry into the CNS.
FIGURE 6.
Differential expression of adhesion molecules and Itg in WT and GFAP-IκBα-dn mice following EAE. Gene expression was assessed in naive animals and at 10, 17, and 80 dpi. For each gene, results are expressed as folds of corresponding WT naive ± SEM after normalization to β-actin. Four animals/group/time were analyzed. *, p < 0.05 with respect to corresponding WT; one-way ANOVA, Tukey test.
Inhibition of astroglial NF-κB resulted in neuroprotective effects following EAE
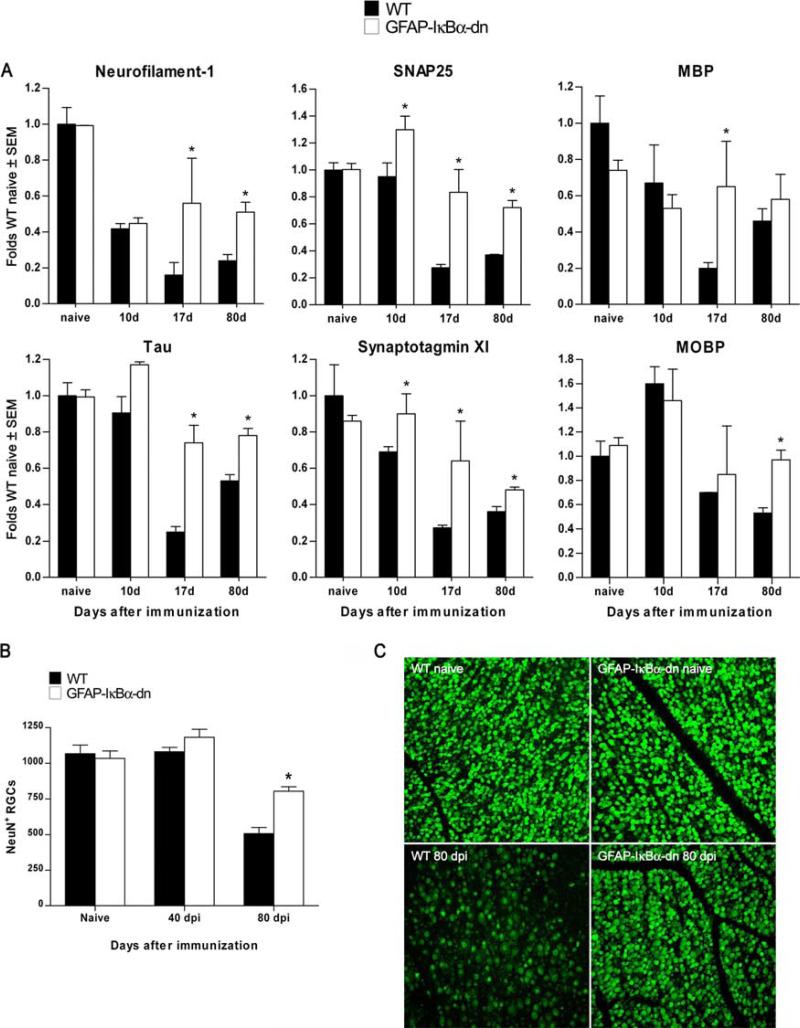
One of the most striking observations from the microarray experiments was that genes associated with synapse formation/function and neuronal growth were not as markedly reduced in GFAPIκBα-dn mice as they were in WT mice at acute and chronic stages of EAE (Fig. 7). In contrast to WT mice, in which Neurofilament-1, SNAP-25, Tau, and Synaptotagmin were reduced at all time points compared with naive animals, GFAP-IκBα-dn mice maintained significantly higher levels of expression compared with WT animals (Fig. 7A). This is most likely due to neuroprotective mechanisms leading to preservation of axons and synapses, which correlate with the improved functional recovery of GFAP-IκBα-dn mice. To test this hypothesis, we evaluated neuronal cell death by counting the number of RGCs in the retinas of WT and GFAPIκBα-dn mice at 40 and 80 dpi. We chose to evaluate cell death in the retina because it is well documented that acute optic neuritis is one of the first clinical symptoms and diagnostic indication of MS (29). Although the numbers of RGCs were not different between WT and transgenic animals at 40 dpi, and very similar to what was assessed in age-matched naive animals, retinas isolated from WT mice had a significantly lower number of RGCs compared with that of GFAP-IκBα-dn retinas at 80 dpi (Fig. 7, B and C). This result suggests that GFAP-IκBα-dn mice have in place neuroprotective mechanisms, which can contribute to the overall recovery of function observed in the chronic phase of EAE.
FIGURE 7.
Inhibition of astroglial NF-κB results in neuroprotective effects following EAE. A, Gene expression was assessed in naive animals and at 10, 17, and 80 dpi. For each gene, results are expressed as folds of corresponding WT naive ± SEM after normalization to β-actin. Four animals/group/time were analyzed. *, p < 0.05 with respect to corresponding WT; one-way ANOVA, Tukey test. B, Evaluation of RGC number on flat-mounted retinas labeled with FITC-conjugated NeuN Ab. C, Representative photomicrographs of flat-mounted retinas from WT and transgenic mice in naive and diseased (80 dpi) conditions. Nine animals per group were counted. *, p < 0.001 vs WT 80 days; one-way ANOVA, Tukey test. Scale bar = 100 μm.
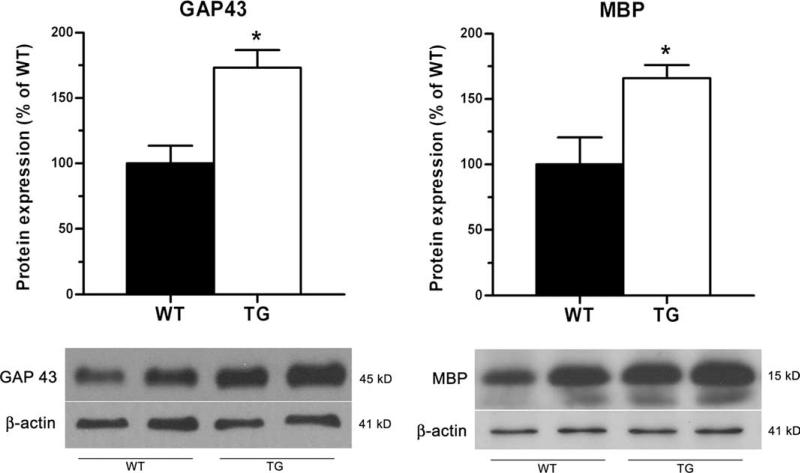
To further confirm this assessment, the expression of the neuronal-specific protein GAP43, which is normally expressed at high levels by developing neurons and re-expressed by regenerating neurons following injury to the CNS, was significantly up-regulated in the spinal cord of GFAP-IκBα-dn mice at 40 dpi (Fig. 8). Similarly, MBP, one of the essential components of central myelin, was also elevated in transgenic mice compared with WT at 40 dpi (Fig. 8). These data are indicative of the presence of a higher number of functional myelinated axons or, possibly, of an increased presence of regenerating healing axons in GFAP-IκBα-dn mice.
FIGURE 8.
Western blot quantification of CNS-specific proteins in WT and GFAP-IκBα-dn mice at 40 dpi. Quantification of GAP43 and MBP in spinal cord tissue of WT and GFAP-IκBα-dn mice at 40 dpi. Data are normalized to β-actin. Results represent the mean ± SEM of six animals/group and are expressed as percentage of WT. *, p < 0.05; Student's t test. Representative Western blots for each protein are shown.
Discussion
Our results provide a direct demonstration of the critical function of astrocytes and astrocytic NF-κB in the development of EAE. The role of astrocytes in the pathophysiology of many neurode-generative diseases has been largely demonstrated by work of our and other laboratories (10, 30, 31). Using the same model used in this study, we previously showed that NF-κB-dependent mechanisms initiated in astrocytes establish an unfavorable environment that prevents repair and recovery of function following spinal cord injury (10). Based on those data and the observation that EAE, although initiated by a Th-mediated response, develops into a widespread demyelinating condition with glial activation and neuroinflammation, we hypothesized that astrocytes, and specifically astrocytic NF-κB, played a prominent role in the pathophysiology of this disease. We induced MOG35–55-mediated EAE in GFAP IκBα-dn mice and WT littermates and scored the clinical signs of the pathology up to 60 dpi. Even though transgenic mice developed EAE similarly to WT, over time they exhibited striking recovery of function compared with WT mice. At acute disease, histopathological signs of demyelination and leukocyte infiltration were observed in both genotypes. However, at chronic disease, leukocyte infiltration was significantly higher in GFAP-IκBα-dn mice compared with their WT counterpart. In EAE, similarly to MS and other immune-inflammatory diseases, the presence of leukocytes in CNS tissue is typically associated with progression of the pathology, due to their ability to release proinflammatory mediators that ultimately cause demyelination and neuronal degeneration (16, 32). In our model, however, the higher abundance of leukocytes coincides with improvement of function in GFAPIκBα-dn mice, leading us to hypothesize that those CD45+ cells, or at least a select population, might be exerting anti-inflammatory, neuroprotective effects. In recent years, many have reported the presence of Tregs in the CNS of EAE mice, typically CD4+CD25+Foxp3+, which play a key role in inhibiting autoimmunity (33). Our flow cytometry data, however, show an equal distribution of CD4+CD25+Foxp3 Tregs in WT and transgenic animals both at acute and chronic stages of the disease, suggesting that the improved functional outcome of the GFAP-IκBα-dn mice is unlikely to be attributed to the protective role of this specific T cell population. In contrast, the flow cytometry studies have revealed a marked increase in CD8+CD122+ T cells, which have regulatory properties (34, 35) and have been shown to promote recovery from EAE (36). Indeed, depletion of CD122+ T cells prolongs the duration of EAE symptoms, and transfer of CD8+CD122+ cells into EAE-induced mice significantly reduces EAE symptoms. Based on this evidence, we can speculate that the protective regulatory role of CD8+CD122+ T cells, found in higher percentage in GFAP-IκBα-dn mice at chronic disease, could represent one of the mechanisms at the basis of the improved clinical outcome of transgenic mice. Additionally, a higher percentage of B220+ B cells was found in GFAP-IκBα-dn mice at 40 dpi. Although the contribution of B cells to the pathophysiology of EAE is still unclear, several reports have indicated a positive role for certain types of B cells to the recovery from EAE. B cells can restrain autoaggressive T cell responses. Specifically, IL-10-producing B cells stimulate resolution of EAE (37), and this appears to be linked to TLR activation (38). On this basis, we cannot exclude the possibility that B cells could also participate in the clinical recovery of transgenic mice. Further studies are necessary to fully investigate these hypotheses; nevertheless, it is intriguing to observe that by inhibiting NF-κB in a CNS cell population, the astrocytes, we can interfere with and manipulate, at least partially, the function of the immune system, demonstrating the tight crosstalk and cross-regulation of these two compartments.
To dissect the molecular mechanisms for the functional recovery observed in the transgenic animals, we took a comprehensive approach of comparing gene expression profiles of WT and GFAPIκBα-dn mice at various times following EAE. No differences were detected between naive WT and GFAP-IκBα-dn mice, confirming that NF-κB inactivation in astrocytes did not alter the normal phenotype, as previously shown (10). The highest number of differentially expressed genes was found at acute disease (17 dpi). Most of those genes, primarily related to immune/inflammatory processes, were down-regulated in transgenic mice compared with WT, particularly chemokines, cytokines, and TLRs. TNF-α, which has been detected in MS lesions and linked to the development of EAE based on studies using TNF-α- and TNFR1-deficient mice (39, 40), was significantly reduced in GFAP-IκBα-dn mice at all time points. TNF-α protein expression was localized in microglial cells and macrophages, but not in astrocytes. This demonstrates that inhibition of NF-κB in astrocytes results in indirect effects on neighboring cells. We speculate that activated astrocytes might release NF-κB-regulated ligands that trigger an increased production of TNF-α and other proinflammatory molecules in neighboring microglia.
Similarly to our previous study in spinal cord injury (10), IL-6 production was increased in transgenic mice at the acute disease. This finding is in apparent contradiction with the notion that IL-6 is critical in the induction phase of EAE, as indicated by studies with IL-6−/− mice (41, 42). In fact, although GFAP-IκBα-dn mice exhibited higher IL-6 levels than WT animals, they showed a significant functional improvement. This may indicate that IL-6 has a dual effect: proinflammatory in the initial phase and antiinflammatory/neuroprotective at later stages of the disease. This hypothesis is in line with previous reports linking IL-6 with antiinflammatory/neuroprotective effects following CNS damage (43, 44).
In addition to reduced cytokine expression, even more striking was the reduced chemokine expression in GFAP-IκBα-dn mice compared with WT. CCL2, which in the CNS is highly expressed by astrocytes (5, 10, 45, 46), its receptor CCR2, as well as CCL5 were significantly down-regulated in GFAP-IκBα-dn mice following EAE. This is an important result because both CCL2 and CCL5 are essential for leukocyte adhesion to brain microvessels and migration into the CNS (47).
A key step in T cell migration into the CNS involves interaction with adhesion molecules on endothelium and astrocyte end-feet at the BBB level. We detected in GFAP-IκBα-dn mice a significant reduction of ICAM-1, VCAM-1, and Itg. Together these molecules combine to form adhesion complexes necessary for the transmigration process. Itg-β7, for example, is a key contributor to CNS demyelinating disease, because Abs against Itg-β7 attenuate EAE and Itg-β −/−7 mice develop a milder form of the disease (48). Furthermore, a recent report by Bullard et al. (49) described that ICAM-1null mice had significantly attenuated EAE with reduction in T cell infiltration and IFN-γ production. The reduced expression of these molecules in GFAP-IκBα-dn mice could suggest a reduced entry of peripheral leukocytes into the CNS, which apparently contradicts the observation that transgenic mice show a higher presence of CD45+ cells at chronic disease. We can speculate that this is dependent not on the entry of leukocytes into the CNS, but rather on the differential survival and differentiation of such cells once they are inside the spinal cord. Indeed, the environment set in place by the inactivation of NF-κB in astrocyte could favor the selective differentiation of that population of CD8+CD122+ Tregs that ultimately participate in the recovery process shown by transgenic mice over WT.
Collectively, these data demonstrate that astrocytes, due to their ability to secrete inflammatory mediators, trophic factors, and other immune modulators, control the composition of the extra-cellular milieu and set the stage for sustaining the chronic phase of the disease. By blocking these processes through inactivation of NF-κB in astrocytes, we could prevent the establishment of the chronic proinflammatory neurodestructive environment necessary for the long-term progression of EAE. Our transgenic approach functions as a multitarget neuroprotective strategy, based on the maintenance of neuronal-specific and myelin-associated gene expression, as well as reduced neuronal loss, as indicated by the RGC survival data. This latter result is especially significant because a decrease in RGC density has been correlated with the severity of demyelination and inflammation in MOG-induced EAE in rats (50). As a further indication of neuroprotection, the expression of neuronal-specific molecules (SNAP-25, Neurofilament-1, Tau, Synaptotagmin) was significantly higher in transgenic mice. In addition, GAP43, which has been linked to axonal regeneration and sprouting (51–53), and MBP protein levels were also elevated in GFAP-IκBα-dn mice at chronic disease stages, indicating that regenerative sprouting and remyelination may be occurring.
In summary, our studies directly demonstrate that by targeting two essential components of CNS inflammation, astrocytes and the NF-κB pathway, positive effects on tissue damage and functional recovery can be achieved following injury. Our findings are in agreement with a recent report by van Loo et al. (54), showing that CNS-restricted ablation of NEMO (NF-κB essential modulator) and IKK2 (IκB kinase 2) ameliorates the course of EAE through reduced expression of chemokines and adhesion molecules. Although NF-κB inhibition is extended to neurons, astrocytes, and oligodendrocytes, the results indirectly indicate that major contributors to the pathology of EAE are astrocytes.
Furthermore, our studies clearly underscore the importance of the cross-talk between the CNS and the immune system, and how by manipulating a molecule central to the response of astrocytes to injury and disease, we can alter the course of a disease like EAE that has long been considered to be purely driven by the activation of an autoimmune response.
In conclusion, our data suggest that strategies aimed at inhibiting NF-κB signaling in the CNS may represent an alternative therapeutic approach for the treatment of MS. Provided that NF-κB is a ubiquitous factor essential for a number of physiologic functions, including development and function of the immune system (55), we need to take into account that global NF-κB inactivation could result in deleterious side effects. Nevertheless, the feasibility and efficacy of treatments based on NF-κB inhibition (e.i., proteasome inhibitors) have been documented in various animal models, and clinical trials for cancer and chronic inflammatory pathologies (e.g., rheumatoid arthritis and psoriasis) are underway (56–58). In this perspective, our studies provide the preclinical evidence that such molecules could potentially be used in the treatment of MS.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grants NS051709 (to J.R.B.), NS46032 (to S.R.B.), and EY017991 (to V.I.S.), and The Miami Project To Cure Paralysis.
Abbreviations used in this paper: >MS, multiple sclerosis; BBB, blood-brain barrier; dn, dominant negative; dpi, days postinduction; EAE, experimental autoimmune encephalomyelitis; FDR, false discovery rate; GFAP, glial fibrillary acidic protein; Itg, integrin; MBP, myelin basic protein; MOG, myelin oligodendrocyte glycoprotein; RGC, retinal ganglion cell; Treg, T regulatory cell; WT, wild type; CDI, cumulative disease index.
The online version of this article contains supplementary material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kornek B, Storch MK, Weissert R, Wallstroem E, Stefferl A, Olsson T, Linington C, Schmidbauer M, Lassmann H. Multiple sclerosis and chronic autoimmune encephalomyelitis: a comparative quantitative study of axonal injury in active, inactive, and remyelinated lesions. Am. J. Pathol. 2000;157:267–276. doi: 10.1016/S0002-9440(10)64537-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayers MM, Hazelwood LJ, Catmull DV, Wang D, McKormack Q, Bernard CC, Orian JM. Early glial responses in murine models of multiple sclerosis. Neurochem. Int. 2004;45:409–419. doi: 10.1016/j.neuint.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Bjartmar C, Trapp BD. Axonal and neuronal degeneration in multiple sclerosis: mechanisms and functional consequences. Curr. Opin. Neurol. 2001;14:271–278. doi: 10.1097/00019052-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Wujek JR, Bjartmar C, Richer E, Ransohoff RM, Yu M, Tuohy VK, Trapp BD. Axon loss in the spinal cord determines permanent neurological disability in an animal model of multiple sclerosis. J. Neuropathol. Exp. Neurol. 2002;61:23–32. doi: 10.1093/jnen/61.1.23. [DOI] [PubMed] [Google Scholar]
- 5.Babcock AA, Kuziel WA, Rivest S, Owens T. Chemokine expression by glial cells directs leukocytes to sites of axonal injury in the CNS. J. Neurosci. 2003;23:7922–7930. doi: 10.1523/JNEUROSCI.23-21-07922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagele RG, Wegiel J, Venkataraman V, Imaki H, Wang KC. Contribution of glial cells to the development of amyloid plaques in Alzheimer's disease. Neurobiol. Aging. 2004;25:663–674. doi: 10.1016/j.neurobiolaging.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Mrak RE, Griffin WS. Glia and their cytokines in progression of neurodegeneration. Neurobiol. Aging. 2005;26:349–354. doi: 10.1016/j.neurobiolaging.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Norris CM, Kadish I, Blalock EM, Chen KC, Thibault V, Porter NM, Landfield PW, Kraner SD. Calcineurin triggers reactive/inflammatory processes in astrocytes and is up-regulated in aging and Alzheimer's models. J. Neurosci. 2005;25:4649–4658. doi: 10.1523/JNEUROSCI.0365-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Ayers MM, Catmull DV, Hazelwood LJ, Bernard CC, Orian JM. Astrocyte-associated axonal damage in pre-onset stages of experimental autoimmune encephalomyelitis. Glia. 2005;51:235–240. doi: 10.1002/glia.20199. [DOI] [PubMed] [Google Scholar]
- 10.Brambilla R, Bracchi-Ricard V, Hu WH, Frydel B, Bramwell A, Karmally S, Green EJ, Bethea JR. Inhibition of astroglial nuclear factor κB reduces inflammation and improves functional recovery after spinal cord injury. J. Exp. Med. 2005;202:145–156. doi: 10.1084/jem.20041918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szalai AJ, Nataf S, Hu XZ, Barnum SR. Experimental allergic encephalomyelitis is inhibited in transgenic mice expressing human C-reactive protein. J. Immunol. 2002;168:5792–5797. doi: 10.4049/jimmunol.168.11.5792. [DOI] [PubMed] [Google Scholar]
- 12.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 13.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 14.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 15.Bracchi-Ricard V, Brambilla R, Levenson J, Hu WH, Bramwell A, Sweatt JD, Green EJ, Bethea JR. Astroglial nuclear factor-κB regulates learning and memory and synaptic plasticity in female mice. J. Neurochem. 2008;104:611–623. doi: 10.1111/j.1471-4159.2007.04993.x. [DOI] [PubMed] [Google Scholar]
- 16.Gold R, Linington C, Lassmann H. Understanding pathogenesis and therapy of multiple sclerosis via animal models: 70 years of merits and culprits in experimental autoimmune encephalomyelitis research. Brain. 2006;129:1953–1971. doi: 10.1093/brain/awl075. [DOI] [PubMed] [Google Scholar]
- 17.Plant SR, Wang Y, Vasseur S, Thrash JC, McMahon EJ, Bergstralh DT, Arnett HA, Miller SD, Carson MJ, Iovanna JL, Ting JP. Up-regulation of the stress-associated gene p8 in mouse models of demyelination and in multiple sclerosis tissues. Glia. 2006;53:529–537. doi: 10.1002/glia.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renno T, Taupin V, Bourbonniere L, Verge G, Tran E, De Simone R, Krakowski M, Rodriguez M, Peterson A, Owens T. Interferon-γ in progression to chronic demyelination and neurological deficit following acute EAE. Mol. Cell. Neurosci. 1998;12:376–389. doi: 10.1006/mcne.1998.0725. [DOI] [PubMed] [Google Scholar]
- 19.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Langrish CL, McKenzie B, Joyce-Shaikh B, Stumhofer JS, McClanahan T, Blumenschein W, Churakovsa T, Low J, Presta L, et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J. Clin. Invest. 2006;116:1317–1326. doi: 10.1172/JCI25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshpande P, King IL, Segal BM. IL-12 driven up-regulation of P-selectin ligand on myelin-specific T cells is a critical step in an animal model of autoimmune demyelination. J. Neuroimmunol. 2006;173:35–44. doi: 10.1016/j.jneuroim.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Ubogu EE, Cossoy MB, Ransohoff RM. The expression and function of chemokines involved in CNS inflammation. Trends Pharmacol. Sci. 2006;27:48–55. doi: 10.1016/j.tips.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Huang D, Han Y, Rani MR, Glabinski A, Trebst C, Sorensen T, Tani M, Wang J, Chien P, O'Bryan S, et al. Chemokines and chemokine receptors in inflammation of the nervous system: manifold roles and exquisite regulation. Immunol. Rev. 2000;177:52–67. doi: 10.1034/j.1600-065x.2000.17709.x. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhary P, Marracci GH, Bourdette DN. Lipoic acid inhibits expression of ICAM-1 and VCAM-1 by CNS endothelial cells and T cell migration into the spinal cord in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2006;175:87–96. doi: 10.1016/j.jneuroim.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann N, Lachnit N, Streppel M, Witter B, Neiss WF, Guntinas-Lichius O, Angelov DN. Increased expression of ICAM-1, VCAM-1, MCP-1, and MIP-1α by spinal perivascular macrophages during experimental allergic encephalomyelitis in rats. BMC Immunol. 2002;3:11. doi: 10.1186/1471-2172-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimenez MA, Sim JE, Russell JH. TNFR1-dependent VCAM-1 expression by astrocytes exposes the CNS to destructive inflammation. J. Neuroimmunol. 2004;151:116–125. doi: 10.1016/j.jneuroim.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Del Zoppo GJ, Milner R. Integrin-matrix interactions in the cerebral microvasculature. Arterioscler. Thromb. Vasc. Biol. 2006;26:1966–1975. doi: 10.1161/01.ATV.0000232525.65682.a2. [DOI] [PubMed] [Google Scholar]
- 29.Frohman EM, Frohman TC, Zee DS, McColl R, Galetta S. The neuro-ophthalmology of multiple sclerosis. Lancet Neurol. 2005;4:111–121. doi: 10.1016/S1474-4422(05)00992-0. [DOI] [PubMed] [Google Scholar]
- 30.Calderon TM, Eugenin EA, Lopez L, Kumar SS, Hesselgesser J, Raine CS, Berman JW. A role for CXCL12 (SDF-1α) in the pathogenesis of multiple sclerosis: regulation of CXCL12 expression in astrocytes by soluble myelin basic protein. J. Neuroimmunol. 2006;177:27–39. doi: 10.1016/j.jneuroim.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Maragakis NJ, Rothstein JD. Mechanisms of disease: astrocytes in neurodegenerative disease. Nat. Clin. Pract. Neurol. 2006;2:679–689. doi: 10.1038/ncpneuro0355. [DOI] [PubMed] [Google Scholar]
- 32.Ercolini AM, Miller SD. Mechanisms of immunopathology in murine models of central nervous system demyelinating disease. J. Immunol. 2006;176:3293–3298. doi: 10.4049/jimmunol.176.6.3293. [DOI] [PubMed] [Google Scholar]
- 33.Kohm AP, Carpentier PA, Anger HA, Miller SD. Cutting edge: CD4+CD25+ regulatory T cells suppress antigen-specific autoreactive immune responses and central nervous system inflammation during active experimental autoimmune encephalomyelitis. J. Immunol. 2002;169:4712–4716. doi: 10.4049/jimmunol.169.9.4712. [DOI] [PubMed] [Google Scholar]
- 34.Rifa'i M, Kawamoto Y, Nakashima I, Suzuki H. Essential roles of CD8+CD122+ regulatory T cells in the maintenance of T cell homeostasis. J. Exp. Med. 2004;200:1123–1134. doi: 10.1084/jem.20040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Endharti AT, Rifa'i M, Shi Z, Fukuoka Y, Nakahara Y, Kawamoto Y, Takeda K, Isobe K, Suzuki H. Cutting edge: CD8+CD122+ regulatory T cells produce IL-10 to suppress IFN-γ production and proliferation of CD8+ T cells. J. Immunol. 2005;175:7093–7097. doi: 10.4049/jimmunol.175.11.7093. [DOI] [PubMed] [Google Scholar]
- 36.Lee YH, Ishida Y, Rifa'i M, Shi Z, Isobe K, Suzuki H. Essential role of CD8+CD122+ regulatory T cells in the recovery from experimental autoimmune encephalomyelitis. J. Immunol. 2008;180:825–832. doi: 10.4049/jimmunol.180.2.825. [DOI] [PubMed] [Google Scholar]
- 37.Fillatreau S, Sweenie CH, McGeachy MJ, Gray D, Anderton SM. B cells regulate autoimmunity by provision of IL-10. Nat. Immunol. 2002;3:944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- 38.Lampropoulou V, Hoehlig K, Roch T, Neves P, Calderon Gomez E, Sweenie CH, Hao Y, Freitas AA, Steinhoff U, Anderton SM, Fillatreau S. TLR-activated B cells suppress T cell-mediated autoimmunity. J. Immunol. 2008;180:4763–4773. doi: 10.4049/jimmunol.180.7.4763. [DOI] [PubMed] [Google Scholar]
- 39.Sean Riminton D, Korner H, Strickland DH, Lemckert FA, Pollard JD, Sedgwick JD. Challenging cytokine redundancy: inflammatory cell movement and clinical course of experimental autoimmune encephalomyelitis are normal in lymphotoxin-deficient, but not tumor necrosis factor-deficient, mice. J. Exp. Med. 1998;187:1517–1528. doi: 10.1084/jem.187.9.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eugster HP, Frei K, Bachmann R, Bluethmann H, Lassmann H, Fontana A. Severity of symptoms and demyelination in MOG-induced EAE depends on TNFR1. Eur. J. Immunol. 1999;29:626–632. doi: 10.1002/(SICI)1521-4141(199902)29:02<626::AID-IMMU626>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 41.Eugster HP, Frei K, Kopf M, Lassmann H, Fontana A. IL-6-deficient mice resist myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. Eur. J. Immunol. 1998;28:2178–2187. doi: 10.1002/(SICI)1521-4141(199807)28:07<2178::AID-IMMU2178>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 42.Okuda Y, Sakoda S, Fujimura H, Saeki Y, Kishimoto T, Yanagihara T. IL-6 plays a crucial role in the induction phase of myelin oligodendrocyte glucoprotein 35–55 induced experimental autoimmune encephalomyelitis. J. Neuroimmunol. 1999;101:188–196. doi: 10.1016/s0165-5728(99)00139-3. [DOI] [PubMed] [Google Scholar]
- 43.Herrmann O, Tarabin V, Suzuki S, Attigah N, Coserea I, Schneider A, Vogel J, Prinz S, Schwab S, Monyer H, et al. Regulation of body temperature and neuroprotection by endogenous interleukin-6 in cerebral ischemia. J. Cereb. Blood Flow Metab. 2003;23:406–415. doi: 10.1097/01.WCB.0000055177.50448.FA. [DOI] [PubMed] [Google Scholar]
- 44.Penkowa M, Moos T, Carrasco J, Hadberg H, Molinero A, Bluethmann H, Hidalgo J. Strongly compromised inflammatory response to brain injury in interleukin-6-deficient mice. Glia. 1999;25:343–357. [PubMed] [Google Scholar]
- 45.Ambrosini E, Remoli ME, Giacomini E, Rosicarelli B, Serafini B, Lande R, Aloisi F, Coccia EM. Astrocytes produce dendritic cell-attracting chemokines in vitro and in multiple sclerosis lesions. J. Neuropathol. Exp. Neurol. 2005;64:706–715. doi: 10.1097/01.jnen.0000173893.01929.fc. [DOI] [PubMed] [Google Scholar]
- 46.Che X, Ye W, Panga L, Wu DC, Yang GY. Monocyte chemoattractant protein-1 expressed in neurons and astrocytes during focal ischemia in mice. Brain Res. 2001;902:171–177. doi: 10.1016/s0006-8993(01)02328-9. [DOI] [PubMed] [Google Scholar]
- 47.Dos Santos AC, Barsante MM, Arantes RM, Bernard CC, Teixeira MM, Carvalho-Tavares J. CCL2 and CCL5 mediate leukocyte adhesion in experimental autoimmune encephalomyelitis: an intravital microscopy study. J. Neuroimmunol. 2005;162:122–129. doi: 10.1016/j.jneuroim.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 48.Kanwar JR, Harrison JE, Wang D, Leung E, Mueller W, Wagner N, Krissansen GW. β7 integrins contribute to demyelinating disease of the central nervous system. J. Neuroimmunol. 2000;103:146–152. doi: 10.1016/s0165-5728(99)00245-3. [DOI] [PubMed] [Google Scholar]
- 49.Bullard DC, Hu X, Schoeb TR, Collins RG, Beaudet AL, Barnum SR. Intercellular adhesion molecule-1 expression is required on multiple cell types for the development of experimental autoimmune encephalomyelitis. J. Immunol. 2007;178:851–857. doi: 10.4049/jimmunol.178.2.851. [DOI] [PubMed] [Google Scholar]
- 50.Meyer R, Weissert R, Diem R, Storch MK, de Graaf KL, Kramer B, Bahr M. Acute neuronal apoptosis in a rat model of multiple sclerosis. J. Neurosci. 2001;21:6214–6220. doi: 10.1523/JNEUROSCI.21-16-06214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carmichael ST, Archibeque I, Luke L, Nolan T, Momiy J, Li S. Growth-associated gene expression after stroke: evidence for a growth-promoting region in peri-infarct cortex. Exp. Neurol. 2005;193:291–311. doi: 10.1016/j.expneurol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Tobias CA, Shumsky JS, Shibata M, Tuszynski MH, Fischer I, Tessler A, Murray M. Delayed grafting of BDNF and NT-3 producing fibroblasts into the injured spinal cord stimulates sprouting, partially rescues axotomized red nucleus neurons from loss and atrophy, and provides limited regeneration. Exp. Neurol. 2003;184:97–113. doi: 10.1016/s0014-4886(03)00394-7. [DOI] [PubMed] [Google Scholar]
- 53.Ghirnikar RS, Lee YL, Eng LF. Chemokine antagonist infusion promotes axonal sparing after spinal cord contusion injury in rat. J. Neurosci. Res. 2001;64:582–589. doi: 10.1002/jnr.1110. [DOI] [PubMed] [Google Scholar]
- 54.Van Loo G, De Lorenzi R, Schmidt H, Huth M, Mildner A, Schmidt-Supprian M, Lassmann H, Prinz MR, Pasparakis M. Inhibition of transcription factor NF-κB in the central nervous system ameliorates autoimmune encephalomyelitis in mice. Nat. Immunol. 2006;7:954–961. doi: 10.1038/ni1372. [DOI] [PubMed] [Google Scholar]
- 55.Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 56.Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J. Clin. Oncol. 2005;23:630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 57.Kropff M, Bisping G, Wenning D, Berdel WE, Kienast J. Proteasome inhibition in multiple myeloma. Eur. J. Cancer. 2006;42:1623–1639. doi: 10.1016/j.ejca.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 58.Elliott PJ, Zollner TM, Boehncke WH. Proteasome inhibition: a new anti-inflammatory strategy. J. Mol. Med. 2003;81:235–245. doi: 10.1007/s00109-003-0422-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.