SUMMARY
The cyanobacterial pentameric ligand-gated ion channel GLIC, a homolog of the Cys-loop receptor superfamily, has provided useful structural and functional information about its eukaryotic counterparts. X-ray diffraction data and site-directed mutagenesis have previously implicated a transmembrane histi-dine residue (His234) as essential for channel function. Here, we investigated the role of His234 via synthesis and incorporation of histidine analogs and α-hydroxy acids using in vivo nonsense suppression. Receptors were expressed heterologously in Xenopus laevis oocytes, and whole-cell voltage-clamp electrophysiology was used to monitor channel activity. We show that an interhelix hydrogen bond involving His234 is important for stabilization of the open state, and that the shape and basicity of its side chain are highly sensitive to perturbations. In contrast, our data show that two other His residues are not involved in the acid-sensing mechanism.
INTRODUCTION
Cys-loop receptors are a class of pentameric ligand-gated ion channels (pLGICs) that mediate synaptic transmission among neurons in the central and peripheral nervous systems of eukaryotes (daCosta and Baenziger, 2013; Thompson et al., 2010). The family has been extensively characterized using electrophysio-logical and biochemical techniques, and includes the nicotinic acetylcholine (nACh), glycine (Gly), serotonin (5-HT3), and γ-aminobutyric acid (GABAA) receptors (Rs). These receptors are essential for normal neuronal function, and an appreciation of the mechanisms by which they recognize ligands and undergo structural changes associated with channel gating is crucial for a full understanding of intercellular signaling in the nervous system. Moreover, many Cys-loop receptors have been identified as playing a role in various neurodegenerative diseases, establishing them as targets for drug development (Jensen et al., 2005; Lemoine et al., 2012). Structure-function studies of the Cys-loop family have been guided by electron microscopy images of nAChRs (Unwin, 2005), and by crystal structures of a family of soluble proteins, the acetylcholine-binding proteins, which display high sequence homology to the extracellular domain of nAChR subunits (Brejc et al., 2001; Sixma and Smit, 2003).
Crystallization of full-length Cys-loop receptors has proven to be challenging, and the only members of the family for which a high-resolution X-ray crystallography structure has been reported are the invertebrate, glutamate-gated chloride channel (GluCl) (Hibbs and Gouaux, 2011; Althoff et al., 2014), the human GABAA receptor (b3 subunit homopentamer) (Miller and Aricescu, 2014), and recently the mouse 5-HT3 receptor (A subunit homopentamer) (Hassaine et al., 2014). There are also two prokaryotic pentameric channels whose structures have been solved; they were identified in the cyanobacterium Gloeobacter violaceus (GLIC) and the bacterium Erwinia chrysanthemi (ELIC) (Bocquet et al., 2007; Hilf and Dutzler, 2008, 2009; Tasneem et al., 2005). GLIC is of particular interest for the study of gating transitions, in that its structure has been elucidated in multiple conformational states under a number of conditions (Bocquet et al., 2009; Gonzalez-Gutierrez et al., 2013; Hilf and Dutzler, 2009; Nury et al., 2011; Prevost et al., 2012; Sauguet et al., 2014). Although molecular details for these homologs likely differ from those of eukaryotic Cys-loop receptors (including the fact that the Cys-loop is absent), the global conformational changes that occur upon channel activation should be informative for the entire family. As such, the prokaryotic proteins provide an opportunity to study these energetic landscapes in a context where structural data are already available.
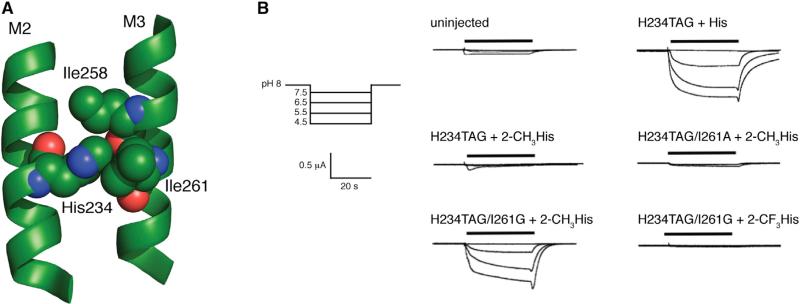
Unlike most Cys-loop receptors, activation of GLIC is mediated by titration of ionizable residues at low pH, rather than by binding of a small molecule. Direct observation of the ligand-binding site(s) via crystallography is therefore not possible, leaving mutagenesis and biochemical probes of the receptor as the best tools to discern which residues are responsible for detection of a pH change and how resulting conformational changes are propagated to give an ion-conducting state. Recent studies have implicated a transmembrane histidine residue (His234 or His11’ using the M2 prime-labeling system) as playing an essential role in the transition to the ion-conducting state (Prevost et al., 2012; Wang et al., 2012). Specifically, the histidine side chain was proposed to form an interhelix hydrogen bond with the backbone carbonyl of Ile258, stabilizing the closer association of the M2 and M3 helices that is observed in the open state of the channel, compared with locally closed mutants and structures obtained at neutral pH (Figure 1). Conventional mutagenesis at this histidine has revealed high sensitivity to perturbation, both for channel function and surface expression of the receptor. No receptors with point mutations at His234 were shown to access an ion-conducting state (Wang et al., 2012). Additionally, molecular dynamics simulations have suggested that protonation of His234 occurs concomitantly with a transition to the open state (Wang et al., 2012). In spite of these insights, a functional, proton-gated fusion consisting of the extracellular domain of GLIC and the transmembrane domain from the human α1 GlyR (which does not contain an analogous His residue) also has been reported (Duret et al., 2011). While functional, this chimera produced a shifted pH-response curve relative to full-length GLIC, a more dramatic shift than is typically seen for comparable chimeras of mammalian Cys-loop receptors where extracellular domain activation reasonably mimics that of the parent receptor (Eiselé et al., 1993; Grutter et al., 2005). Thus, while some aspects of the pH sensitivity of GLIC may be conferred by an as-yet-unidentified proton-binding region in the extracellular domain, such a region may play a major role only in the chimera. This is reminiscent of proton activation of KcsA, where there are two proton-sensing regions: (1) H25 in M1 (likely the major sensor), and (2) a network of ionizable residues at the transmembrane/cytoplasmic interface (Posson et al., 2013; Cuello et al., 2010; Thompson et al., 2008; Takeuchi et al., 2007).
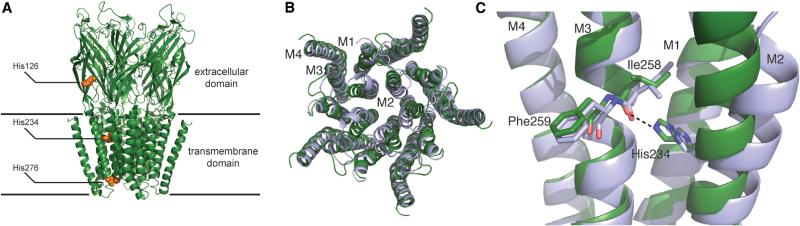
Figure 1. Several Views of the GLIC Channel.
(A) Crystal structure of GLIC in a presumed open state (PDB ID 3EHZ). Histidines studied here are highlighted.
(B) Top-down overlay of a locally closed GLIC mutant structure (blue, PDB ID 3TLU) with the open channel structure (green), omitting the extracellular domain. Note the movement of the M2 helix associated with channel opening.
(C) Side view overlay of locally closed (blue) and open (green) GLIC structures. The orientation of the depicted residues suggests the formation of a hydrogen bond (dashed line) between His234 and the backbone carbonyl of I258 upon channel activation.
Since conventional mutagenesis at His234 is disruptive to GLIC function, we have implemented the subtler probe of noncanonical amino acid mutagenesis in the transmembrane domain. None of the canonical amino acids closely resemble the properties of histidine, and conventional mutagenesis does not allow for modification of the protein backbone. The small systematic perturbations to steric and electrostatic interactions enabled by noncanonical amino acid mutagenesis could allow for a deeper understanding of the origins of its sensitivity to substitution. In this work, we describe the synthesis and incorporation of two noncanonical histidine analogs into GLIC, residues designed specifically to probe the protonation of His234. In addition, we used backbone mutagenesis to probe formation of the M2-M3 interhelix hydrogen bond. Our data suggest that both are essential for channel function.
RESULTS AND DISCUSSION
His234 Is the Only His Residue Critical for GLIC Function
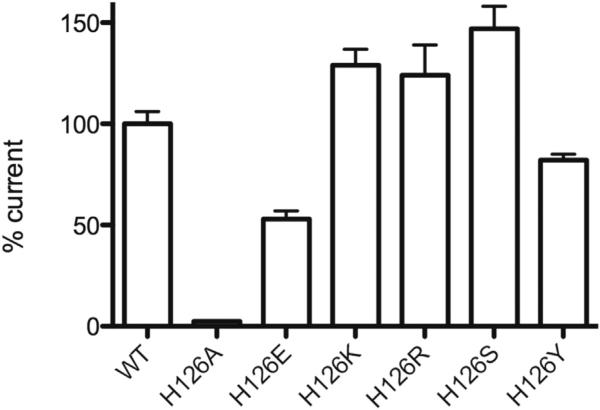
GLIC is activated when extracellular pH is lowered, and the half-maximal pH for activation (pH50) is 5.5, implicating a histidine (pKa ~6) as a critical ionizable residue. There are three His residues in GLIC (Figure 1A), and previous data have shown that mutation of either His126 or His234 ablates function, while alteration of His276 does not (Wang et al., 2012). Thus, His126 and His234 were initially proposed as candidates for a proton-binding site. However, subsequent expression studies in HEK cells showed that lack of function with His126 mutants was due to the role of this His in protein folding, subunit oligomerization, and/or transport to the cell surface (Wang et al., 2012). To further probe the importance of this site, we created a range of GLIC His126 mutants and evaluated them in Xenopus oocytes, which are often able to express proteins that are not amenable to expression in mammalian cells. Our data revealed that these mutant receptors are functional, with pH50 and nH values similar to those of wild-type, for nearly all the His126 mutants we tested (Table 1). The data therefore indicate that His126 is not important for proton activation. Our results also support the earlier study that suggested this residue has a role in expression, as we observed very different maximal currents, indicating different expression levels, for the mutant receptors (Figure 2).
Table 1.
Summary of Mutagenesis Experiments
| Mutant | pH50 | n H | n |
|---|---|---|---|
| WT GLIC | 5.50 ± 0.02 | 1.4 ± 0.1 | 16 |
| H126A | 5.24 ±0.10 | 2.3 ± 0.3 | 3 |
| H126E | 5.38 ± 0.07 | 2.2 ± 0.2 | 3 |
| H126K | 5.34 ± 0.06 | 2.0 ± 0.2 | 3 |
| H126L | NR | 3 | |
| H126R | 5.40 ± 0.14 | 2.0 ± 0.4 | 3 |
| H126S | 5.52 ± 0.05 | 2.0 ± 0.2 | 3 |
| H126Y | 5.35 ± 0.04 | 2.6 ± 0.3 | 3 |
| F259TAG + Phe | 5.54 ± 0.07 | 1.1 ± 0.1 | 14 |
| F259TAG + phenyllactic acid | 4.47 ± 0.07 | 2.4 ± 0.3 | 10 |
| A256TAG + Ala | 5.43 ± 0.03 | 1.6 ± 0.6 | 8 |
| A256TAG + Aah | NR | 8 | |
| H234F | NR | 17 | |
| H234L | NR | 12 | |
| H234TAG + His | 5.37 ± 0.04 | 1.5 ± 0.1 | 23 |
| H234TAG + 2-CH3His | NR | 20 | |
| H126TAG + His | 5.56 ± 0.03 | 1.5 ± 0.1 | 13 |
| H126TAG + 2-CH3His | 5.61 ± 0.02 | 1.6 ± 0.1 | 21 |
| H126TAG + 2-CF3His | 5.72 ±0.01 | 1.6 ± 0.1 | 18 |
| H276TAG + His | 5.41 ± 0.03 | 1.7 ± 0.1 | 12 |
| H276TAG + 2-CH3His | 4.52 ± 0.03 | 2.2 ±0.1 | 15 |
| H276TAG + 2-CF3His | 4.52 ± 0.08 | 2.7 ± 0.4 | 16 |
| I261A | 5.05 ± 0.03 | 1.6 ± 0.1 | 11 |
| I261A/H234TAG + His | 4.96 ± 0.02 | 1.3 ± 0.1 | 15 |
| I261A/H234TAG + 2-CH3His | NR | 19 | |
| I261Ga | ~5.0 | ~0.9 | 25 |
| I261G/H234F | NR | 17 | |
| I261G/H234L | NR | 10 | |
| I261G/H234TAG + Hisa | ≤4.5 | ~0.8 | 9 |
| I261G/H234TAG + 2-CH3Hisa | ≤4.5 | ~0.8 | 14 |
| I261G/H234TAG + 2-CF3His | NR | 23 |
The pH50 and nH data are shown as mean ± SEM. NR, nonresponsive. See also Figure S1.
Oocytes did not tolerate saturating proton concentrations; pH50 and nH values given are estimated.
Figure 2. Maximal Currents Observed for Mutants at His126.
These data were obtained from the same batch of oocytes to minimize oocyte variability. Values are relative to wild-type (100%); data = mean ± SEM, n = 3–5.
Probing the His234-Ile258 Hydrogen Bond
Crystallographic studies suggest a functionally important hydrogen bond between the side chain of His234 on helix M2 and the backbone carbonyl of Ile258 on M3 (Figure 1C). We have frequently employed a general strategy for probing hydrogen bonds to the protein backbone, using nonsense suppression methodology to incorporate α-hydroxy acid analogs of α-amino acids (England et al., 1999a, 1999b; Noren et al., 1989). Briefly, a stop codon is introduced at the position of interest, and mRNA is prepared by in vitro runoff transcription. The desired α-hydroxy acid is ligated to a suppressor tRNA bearing the anticodon corresponding to the stop codon used. The tRNA and mRNA are then coinjected into Xenopus oocytes, and the cells are incubated to allow for expression of the protein containing the α-hydroxy acid (Nowak et al., 1998; Dougherty and Van Arnam, 2014).
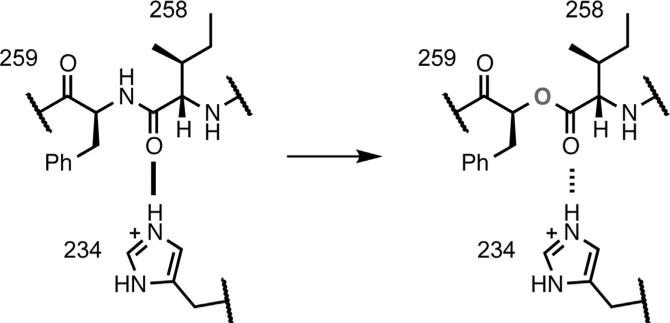
The consequences of substitution with an α-hydroxy acid are 2-fold (Figure 3). Replacement of a residue with its α-hydroxy analog results in the loss of the backbone N-H bond, eliminating hydrogen bond donation. In addition, replacement of the backbone amide with an ester diminishes the hydrogen bond-accepting ability of the carbonyl at the i–1 residue relative to the mutation. Previous studies have shown that both effects can produce substantial perturbations of protein function (Dougherty and Van Arnam, 2014). In the present case, to probe the Ile258 carbonyl, we replaced Phe259 with phenyllactic acid, the α-hydroxy acid analog of Phe.
Figure 3. Backbone Ester Mutagenesis.
Incorporation of phenyllactic acid at Phe259 introduces a backbone ester mutation, converting a strong hydrogen bond (solid line) to a weaker hydrogen bond (dashed line).
Backbone mutation to modulate the proposed M2-M3 hydrogen bond produced functional channels that expressed well (Table 1). The substitution did result in a shift in pH50 by about 1 pH unit toward a more acidic value. This suggests that weakening the M2-M3 hydrogen bond results in a relative destabilization of the open state of the channel, consistent with the model of Figure 1. Of course, by introducing a backbone mutation near the middle of the M3 αhelix, we were impacting several potential hydrogen bonds. Interestingly, in the open channel structure (Protein Data Bank [PDB] ID 3EHZ) there was a distortion of the M3 helix in the region of the Ile258 carbonyl, such that this carbonyl appeared to make a stronger hydrogen bond to the M2 His side chain (O,,,N distance, 3.0Å) than to the backbone NH of the i+3 residue in M3 (Tyr262; O,,,N distance, 3.5Å). At the same time, the NH of Phe259 (which was deleted in the α-hydroxy experiment) made a strong hydrogen bond to the i–4 residue (Gly255; O,,,N distance, 3.1Å). When we perturbed this hydrogen bond in the same manner (conversion of Ala256 to its α-hydroxy analog), we were unable to observe functional channels. Nonsense suppression can be effective at the Ala256 site, however, as we were able to rescue wild-type behavior by incorporation of Ala. Further experiments are required to tease out the role of any hydrogen bond networks in this region of the protein, although currently there is no evidence to indicate that hydrogen bonds other than those formed by His234 are actively involved in channel gating. Thus, we can say that our results are consistent with the presence of an important hydrogen bond between the backbone carbonyl of Ile258 on helix M3 and the side chain of His234 on helix M2, but we cannot rule out other interactions.
Synthesis and Incorporation of Noncanonical Histidine Analogs
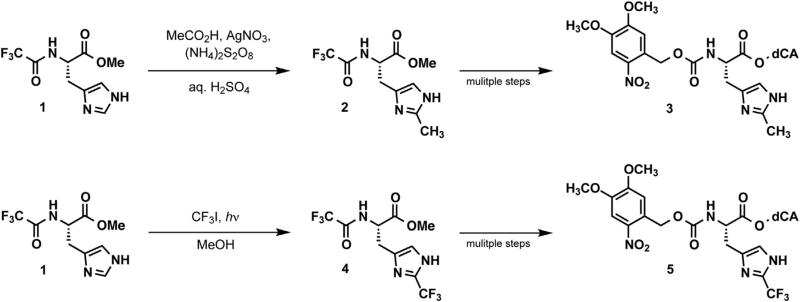
We next hoped to determine the specific molecular requirements at His234 necessary for the function of GLIC. In particular, we were curious whether GLIC could tolerate subtle changes in the size or basicity of the imidazole side chain of His234. Recall that all conventional mutations of His234 produce nonfunctional channels. We first considered 2-methylhistidine (2-CH3His), an analog that would be expected to have increased steric bulk but only a minor (~0.7 pKa unit) increase in basicity relative to histidine, based on the effect of adding a methyl group to the corresponding position on imidazole (Bruice and Schmir, 1958). For the synthesis of the desired noncanonical amino acid, we adapted a literature procedure for radical alkylation of histidine (Figure 4A; Jain et al., 1997). While the original reaction conditions resulted in only trace quantities of 2-CH3His, we found that using acetic acid as a cosolvent and increasing the concentrations of silver nitrate and ammonium persulfate provided a low, but acceptable yield of the desired product, with high regio-selectivity for the 2-position. This provided us with relatively rapid access to 2-CH3His, which could then be appropriately protected and appended to suppressor tRNA, as previously described (Noren et al., 1989).
Figure 4. Synthetic Schemes.
Preparation of (top) 2-CH3His and (bottom) 2-CF3His for ligation to suppressor tRNA.
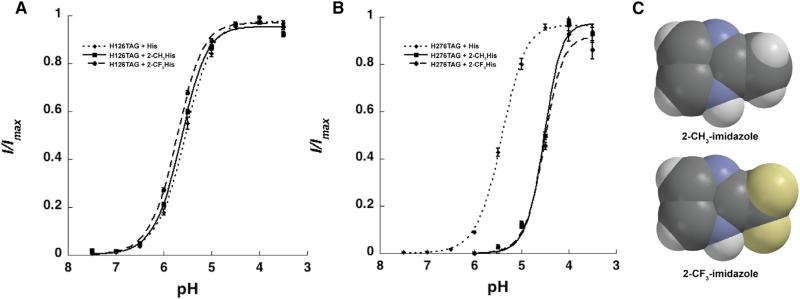
Interestingly, coinjection of H234TAG mRNA with 2-CH3His-charged tRNA gave no evidence of functional channels containing 2-CH3His, even after long incubation times and multiple injections of large quantities of material. Control experiments in which 2-CH3His was incorporated at the two other histidine sites (His126 and His276) produced functional channels, establishing that 2-CH3His can be readily incorporated via nonsense suppression (Figure 5A). Given the modest effect on His pKa caused by CH3 substitution, we considered these results to suggest that the introduction of even a minimal steric clash in the vicinity of His234 may disrupt protein folding, membrane trafficking, or the gating process.
Figure 5. Incorporation of His Analogues into Functional Receptors.
(A and B) Control experiments demonstrate robust nonsense suppression and membrane trafficking with histidine (dotted) and synthetic histidine anlaogs 2-CH3His (solid) and 2-CF3His (dashed) at His126 (A) and His276 (B). Mean normalized current responses are plotted from 12–20 oocytes per condition, with SE indicated.
(C) Space-filling models of 2-CH3-imidazole (top) and 2-CF3-imidazole (bottom), showing they are comparable in size.
See also Figure S2.
Examination of structural data from the open state of GLIC (3EHZ) suggests possible unfavorable interactions between a group installed at the 2-position of the His234 side chain and the side chains of Ile261 and Ile258 (Figure 6A). We therefore introduced additional mutations in combination with H234TAG in an effort to alleviate crowding around the substituted histidine. The I261G mutation alone produced functional channels. Concentration-response curves showed some cell-to-cell variation (Figure S1 available online), but currents were first observed upon acidification at pH 6, and we estimated pH50 to be approximately 5.0. Combining the I216G mutation with 2-CH3His incorporation at His234 did produce functional channels. The steric sensitivity of this region is emphasized by the fact that 2-CH3His did not produce functional channels in the presence of the I261A mutation (Figure 6B). For the 2-CH3His/I261G double mutant, currents were first observed upon acidification at pH 5.5, and increased roughly linearly as pH was lowered to 3.5. While the pH profiles of these currents were consistent, the oocytes did not tolerate proton concentrations high enough to reach a maximal current. As such, we were unable to obtain a reliable pH50 value for this mutant.
Figure 6. Studies of His Analogues at His234.
(A) Space-filling model of His234 and surrounding residues in the presumed open state structure (PDB ID 3EHZ).
(B) Whole-cell current responses of oocytes expressing GLIC mutants to buffer applications at pH 7.5, 6.5, 5.5, and 4.5. See also Figure S3.
To demonstrate that the mechanism of channel activation is not in some way modified by the I261G mutation, perhaps via a disruption of the M3 helix, we combined I261G with the conventional mutations H234L and H234F, and observed negligible currents (data not shown). Thus, His234 still plays an essential role in gating, even in the presence of the I261G mutation, and the observed function of the His234(2-CH3His) mutant appears to be permitted specifically by the reduction of steric bulk in the proximity of the 2-position.
Given the sensitivity to structural perturbation in the region of His234, we could not expect the subtle pKa shift associated with the addition of the methyl group to be discernible. As such, we sought a more dramatic electronic perturbation, and we chose 2-trifluoromethylhistidine (2-CF3His). As shown in Figure 5B, 2-CH3His and 2-CF3His are similar in size; however, the two have very different basicities. While the conjugate acid of 2-methylimidazole has a pKa of 7.75, the corresponding value for 2-trifluoromethylimidazole is 2.06 (Bruice and Schmir, 1958). Thus 2-CF3His is a very weak base that would almost certainly remain in the neutral form at any pH accessible in the electrophysiology assay. We therefore prepared 2-CF3His using a literature procedure (Figure 4B; Kimoto et al., 1984), and appended it to a suppressor tRNA as described for 2-CH3His.
Again, positive control experiments at His126 and His276 indicated that nonsense suppression with 2-CF3His occurs robustly (Figure 5B; Figure S2). The concentration-response curves for the 2-CF3His and the 2-CH3His mutants overlay at both sites, indicating the following: (1) any steric perturbation due to trifluoromethylation is similar to that introduced by methyl substitution at these sites; and (2) as the pKa values of these compounds are very different, the protonation state of neither His126 nor His276 affects GLIC function.
Attempts at incorporation of 2-CF3His at His234 in combination with the I261G mutation, however, gave no significant currents. Establishment that receptors were synthesized and delivered to the cell surface by, for example, immunofluorescence, was not possible due to the inherently low expression levels associated with nonsense suppression experiments in Xenopus oocytes (Figure S3). However, given that we have established that 2-CF3His is compatible with our nonsense suppression methodology, and that 2-CH3His at this site can produce functional receptors, we consider the most reasonable interpretation of the 2-CF3His results to be that the noncanonical amino acid was incorporated, but that the resulting receptors are nonfunctional because protonation cannot occur.
SIGNIFICANCE
Structural and functional data for GLIC have shown the importance of His234 in the transmembrane region of this protein. Here we have shown that even subtle steric perturbation of His234 ablates function, and this can be partially rescued by compensatory steric alterations in the adjacent M3 helix. In addition, we have produced two lines of evidence to support a key role for protonation of His234 in receptor function. First, the subtle mutation by backbone mutagenesis of the putative hydrogen bond partner to the protonated His234 alters function in the expected direction. Second, introducing a His analog that would not be expected to be protonated at pH > 2 (2-CF3His) produces nonfunctional channels. Overall, these experiments show that formation of the M2-M3 hydrogen bond is essential for stabilizing the ion-conducting state, and strongly support the in silico experiments showing that His234 must exist in the protonated form for GLIC to attain its activated conformation (Wang et al., 2012). We conclude that His234 is a sterically restricted site whose protonation is essential for the activation of GLIC.
EXPERIMENTAL PROCEDURES
Molecular Biology
The cDNA for GLIC was in the pGEMhe plasmid. Site-directed mutagenesis was performed using the Stratagene QuikChange protocol to generate the appropriate codon. For noncanonical amino acid mutants and conventional mutants generated by nonsense suppression, the site of interest was mutated to the TAG stop codon. Plasmids were linearized with the SbfI restriction enzyme, and receptor mRNA was then prepared by in vitro runoff transcription using the Ambion T7 mMessage mMachine kit.
Hydroxy or amino acid-dCA couples were enzymatically ligated to truncated 74-mer THG73 tRNA as previously described (England et al., 1999a; Nowak et al., 1998). The 74-mer tRNA was prepared using the Ambion T7MEGAshortscript kit by transcription from a modified DNA oligonucleotide template, as described in the literature, to enhance RNA transcript homogeneity (Kao et al., 1999). Crude tRNA-amino acid or tRNA-hydroxy acid product was used without desalting, and the product was confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry on a 3-hydroxypicolinic acid matrix. Deprotection of the NVOC group on the tRNA-amino acids was carried out by 5 min photolysis on a 1 kW xenon lamp with WG-335 and UG-11 filters immediately prior to injection.
Note that we were using residue numbering consistent with PDB ID 3EHZ (Figure 1; Hilf and Dutzler, 2009). In some other published structures of GLIC, residues are offset by 1.
Oocyte Preparation and RNA Injection
Stage V–VI oocytes of Xenopus laevis were harvested and injected with RNAs as described previously (Nowak et al., 1998). For nonsense suppression experiments, each cell was injected with 50–100 ng each of receptor mRNA and appropriate tRNA approximately 48 hr before recording. Mutants yielding small responses required 72 hr of incubation, with a second injection of mRNA and tRNA 48 hr before recording.
For wild-type experiments and conventional mutants, each cell received a single injection of 1–25 ng of receptor mRNA approximately 24 hr before recording. Injection volumes for each injection session were 50–100 nl per cell.
As a negative control for suppression experiments at each site, unacylated full-length tRNA was coinjected with mRNA in the same manner as charged tRNA. These experiments yielded negligible responses for all sites. Wild-type recovery conditions (injecting tRNA charged with the appropriate amino acid to regenerate a wild-type channel via nonsense suppression at a TAG stop codon) were injected alongside mutant nonsense suppression conditions as a positive control. THG73-phenyllactic acid was unprotected and was injected directly without irradiation.
Electrophysiology
Oocyte recordings were made in two-electrode voltage clamp mode using the OpusXpress 6000A (Axon Instruments). Oocyte equilibration and washes were performed with Ca-free ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES) adjusted to pH 8 with 1 N NaOH. The pH of buffers for concentration-response curves were adjusted accordingly with NaOH or HCl, and buffers with pH below 6.8 contained 5 mM MES in place of HEPES. Initial holding potential was –60 mV. Data were sampled at 125 Hz and filtered at 50 Hz. Oocytes were equilibrated for 30 s at 1 ml/min before each pH application. The pH buffer applications lasted for 15 s at 4 ml/min, followed by a 15 s waiting period to allow the cells to attain a peak current. Cells were then washed for 40 s at 3 ml/min before the following equilibration. Concentration-response data were obtained for nine buffer pHs, for a minimum of two cell batches, and for a minimum of eight cells total. The concentration-response relations for each cell were fitted to the Hill equation, Inorm = 1/(1 + (EC50/A))nH, where Inorm is the normalized current peak at [H3O+] = A, EC50 is the value of [H3O+] that elicits a half-maximum response, and nH is the Hill coefficient. The values for each cell that displayed a strong fit to the Hill equation were then averaged to give the reported values, where pH50 = –log(EC50). At pH < 4, uninjected cells gave current responses that were occasionally up to ~200 nA. As such, data were only reported from cells that gave responses above pH 4, or that gave larger, more robust currents below pH 4, and other cells were considered nonresponsive.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (NS034407 to D.A.D.) and The Wellcome Trust (WT 81925 to S.C.R.L., who is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science). We would like to thank Fiona Love, Rell Parker, and Dr. Brandon Henderson for excellent technical assistance, and Henry A. Lester for helpful discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2014.10.019.
REFERENCES
- Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Prado de Carvalho L, Cartaud J, Neyton J, Le Poupon C, Taly A, Grutter T, Changeux J-P, Corringer P-J. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007;445:116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux J-P, Delarue M, Corringer P-J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Bruice TC, Schmir GL. Imidazole catalysis. II. The reaction of substituted imidazoles with phenyl acetates in aqueous solution. J. Am. Chem. Soc. 1958;80:148–156. [Google Scholar]
- Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, Cordero-Morales JF, Chakrapani S, Roux B, Perozo E. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 2010;466:272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daCosta CJB, Baenziger JE. Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure. 2013;21:1271–1283. doi: 10.1016/j.str.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Dougherty DA, Van Arnam EB. In vivo incorporation of non-canonical amino acids by using the chemical aminoacylation strategy: a broadly applicable mechanistic tool. ChemBioChem. 2014;15:1710–1720. doi: 10.1002/cbic.201402080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret G, Van Renterghem C, Weng Y, Prevost M, Moraga-Cid G, Huon C, Sonner JM, Corringer P-J. Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc. Natl. Acad. Sci. USA. 2011;108:12143–12148. doi: 10.1073/pnas.1104494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiselé J-L, Bertrand S, Galzi J-L, Devillers-Thiéry A, Changeux J-P, Bertrand D. Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 1993;366:479–483. doi: 10.1038/366479a0. [DOI] [PubMed] [Google Scholar]
- England PM, Lester HA, Dougherty DA. Incorporation of esters into proteins: improved synthesis of hydroxyacyl tRNAs. Tetrahedron Lett. 1999a;40:6189–6192. [Google Scholar]
- England PM, Zhang Y, Dougherty DA, Lester HA. Backbone mutations in transmembrane domains of a ligand-gated ion channel: implications for the mechanism of gating. Cell. 1999b;96:89–98. doi: 10.1016/s0092-8674(00)80962-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc. Natl. Acad. Sci. USA. 2013;110:18716–18721. doi: 10.1073/pnas.1313156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutter T, de Carvalho LP, Dufresne V, Taly A, Edelstein SJ, Changeux J-P. Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. USA. 2005;102:18207–18212. doi: 10.1073/pnas.0509024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaine G, Deluz C, Grasso L, Wyss R, Tol MB, Hovius R, Graff A, Stahlberg H, Tomizaki T, Desmyter A, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf RJC, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- Jain R, Cohen LA, El-Kadi NA, King MM. Regiospecific alkylation of histidine and histamine at C-2. Tetrahedron. 1997;53:2365–2370. [Google Scholar]
- Jensen AA, Frølund B, Liljefors T, Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J. Med. Chem. 2005;48:4705–4745. doi: 10.1021/jm040219e. [DOI] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rüdisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto H, Fujii S, Cohen LA. Photochemical trifluoromethylation of some biologically significant imidazoles. J. Org. Chem. 1984;49:1060–1064. [Google Scholar]
- Lemoine D, Jiang R, Taly A, Chataigneau T, Specht A, Grutter T. Ligand-gated ion channels: new insights into neurological disorders and ligand recognition. Chem. Rev. 2012;112:6285–6318. doi: 10.1021/cr3000829. [DOI] [PubMed] [Google Scholar]
- Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- Nowak MW, Gallivan JP, Silverman SK, Labarca CG, Dougherty DA, Lester HA. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. In: Michael Conn P, editor. Methods in Enzymology. Elsevier; New York: 1998. pp. 504–529. [DOI] [PubMed] [Google Scholar]
- Nury H, Van Renterghem C, Weng Y, Tran A, Baaden M, Dufresne V, Changeux J-P, Sonner JM, Delarue M, Corringer P-J. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- Posson DJ, Thompson AN, McCoy JG, Nimigean CM. Molecular interactions involved in proton-dependent gating in KcsA potassium channels. J. Gen. Physiol. 2013;142:613–624. doi: 10.1085/jgp.201311057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost MS, Sauguet L, Nury H, Van Renterghem C, Huon C, Poitevin F, Baaden M, Delarue M, Corringer P-J. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat. Struct. Mol. Biol. 2012;19:642–649. doi: 10.1038/nsmb.2307. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Shahsavar A, Poitevin F, Huon C, Menny A, Nemecz À, Haouz A, Changeux J-P, Corringer P-J, Delarue M. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. USA. 2014;111:966–971. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixma TK, Smit AB. Acetylcholine binding protein (AChBP): a secreted glial protein that provides a high-resolution model for the extracellular domain of pentameric ligand-gated ion channels. Annu. Rev. Biophys. Biomol. Struct. 2003;32:311–334. doi: 10.1146/annurev.biophys.32.110601.142536. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J. Biol. Chem. 2007;282:15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. USA. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Lester HA, Lummis SCR. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Wang H-L, Cheng X, Sine SM. Intramembrane proton binding site linked to activation of bacterial pentameric ion channel. J. Biol. Chem. 2012;287:6482–6489. doi: 10.1074/jbc.M111.305839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.