Abstract
An analytical study of cell–cell communications between murine osteoblast-like MLO-A5 cells and bone marrow mesenchymal stem cell (BMSC)-like C3H10T1/2 cells was performed. C3H10T1/2 cells expressing green fluorescent protein (10T-GFP cells) were generated to enable the isolation of the BMSC-like cells from co-cultures with MLO-A5 cells. The mRNA expression levels of several osteogenic transcription factors (Runx2, Osterix, Dlx5, and Msx2) did not differ between the co-cultured and mono-cultured 10T-GFP cells, but those of alkaline phosphatase (ALP) and bone sialoprotein (BSP) were 300- to 400-fold higher in the co-cultured cells. Patch clamp and biocytin transfer assays revealed gap junction-mediated communication between co-cultured 10T-GFP and MLO-A5 cells. The addition of a gap junction inhibitor suppressed the increases in the expression levels of the ALP and BSP mRNAs in co-cultured 10T-GFP cells. Furthermore, the histone acetylation levels were higher in co-cultured 10T-GFP cells than in mono-cultured 10T-GFP cells. These results suggest that osteoblasts and BMSCs associate via gap junctions, and that gap junction-mediated signaling induces histone acetylation that leads to elevated transcription of the genes encoding ALP and BSP in BMSCs.
Introduction
With the exception of red and white blood cells and certain immune cells, individual cells in multicellular organisms generally adhere and attach to one another rather than exist as independent entities. In addition, intercellular communication between connected or adjacent cells plays an important role in the formation of tissues. In bone tissues, gap junctions form direct links between the cytoplasm of an osteocyte and another adjacent osteocyte or osteoblast [1–4]. Ions and small molecules (<1 kDa), including cyclic nucleotides and calcium ions, move between these cells through gap junctions and promote cell–cell communication [5–7]. Previous studies indicated that intercellular communication via gap junctions underlies both bone formation and bone resorption. In this regard, gap junctional communication between cells is reportedly involved in the transmission of mechanical and chemical signals from one area of the bone to another [8]. Gap junction-mediated cell–cell communication also contributes to the ability of cellular networks to initiate coordinated responses to external stimuli [2].
Recent studies have focused on the mechanisms involved in the formation of gap junctions between osteocytes and osteoblasts, as well as on intercellular communication between these cells and their neighbors. Hematopoietic stem cells are present in large numbers in the border region between unmineralized osteoid and bone marrow, and gap junction-mediated contact with osteoid-secreting osteoblasts is essential for hematopoietic stem cell maintenance within the osteoblastic niche [9,10]. In addition to hematopoietic stem cells, mesenchymal stem cells, which differentiate into osteoblasts and adipocytes, also reside within bone marrow; therefore, it is likely that gap junctional communication between these cells and osteoblasts contributes to the regulation of bone marrow mesenchymal stem cell (BMSC) status and differentiation. However, a few advances have been made toward elucidating the communication networks, gap junction mediated or otherwise, that link BMSCs with osteoblasts.
Here, we performed an analytical study of cell–cell communication between osteoblasts and BMSCs using the MLO-A5 murine late osteoblast cell line as the source of osteoblasts and the C3H10T1/2 murine multipotent cell line as the source of BMSCs. The MLO-A5 cell line was developed from transgenic mice in which the SV40 large T-antigen oncogene was expressed under the control of the osteocalcin (OCN) promoter [11]. MLO-A5 cells mineralize in culture and express large amounts of alkaline phosphatase (ALP), an osteoblast marker of active bone formation [11,12]; hence, the MLO-A5 cell line is considered as representing the mature osteoblasts that are responsible for triggering mineralization of osteoid to form bone. The C3H10T1/2 cell line was established from an early mouse embryo and is capable of differentiating into myotubes, adipocytes, chondrocytes, and osteoblasts [13]; therefore, C3H10T1/2 cells share quintessential characteristics with BMSCs. Co-culture of C3H10T1/2 cells with MLO-A5 cells resulted in intercellular communication across gap junctions formed between the two cell types. Furthermore, histone acetylation and the expression levels of the mRNAs encoding ALP and bone sialoprotein (BSP) were induced markedly in the co-cultured C3H10T1/2 cells, suggesting that gap junctional communication with osteoblasts facilitates the transcription of the genes encoding ALP and BSP in BMSCs.
Materials and Methods
Cells and reagents
C3H10T1/2, MC3T3-E1, 3T3-L1, Chinese hamster ovary (CHO), and HeLa cells were obtained from the Riken Cell Bank (Ibaragi, Japan). The MLO-A5 cell line was provided by Dr. Tomihisa Takahashi (Department of Anatomy, Nihon University School of Dentistry, Tokyo, Japan). The enhanced green fluorescent protein (EGFP) expression vector (pEGFP-N1) was obtained from Takara Bio, Inc. (Shiga, Japan). Antibiotics and cell culture medium were purchased from Gibco (Grand Island, NY) and Wako (Osaka, Japan), respectively. Fetal bovine serum was purchased from Japan Bioserum Co., Ltd. (Tokyo, Japan). Recombinant human bone morphogenetic protein 2 (BMP-2) and recombinant mouse Noggin were purchased from R&D Systems (Minneapolis, MN). Carbenoxolone (CBX), cycloheximide (CHX), and G418 (Geneticin®) were purchased from Sigma Chemical Co. (St. Louis, MO), and INI-0602 was purchased from Wako.
Generation of EGFP-expressing cells
C3H10T1/2 cells were plated into six-well plates at a density of 5×105 cells per well and allowed to grow under normal culture conditions (37°C in an atmosphere of 5% CO2/95% air) for 18 h. Subsequently, the cells were incubated in 2 mL of α-minimal essential medium (α-MEM) containing 6.25 μL of Lipofectamine LTX™ reagent (Invitrogen, Carlsbad, CA) and 2 μg of the EGFP expression vector. After transfection, cells with stable DNA integration (10T-GFP cells) were selected by culturing in α-MEM containing 10% fetal bovine serum, 1% penicillin–streptomycin, and 2 mg/mL G418. EGFP expression was confirmed via fluorescence-activated cell sorting (FACS) using a BD FACS Aria cell sorter (BD-Immunocytometry Systems, San Jose, CA). MC3T3-E1, 3T3-L1, CHO, and HeLa cells were stably transfected with the EGFP expression vector using the same techniques as those described for C3H10T1/2 cells.
Culture of MLO-A5 cells and EGFP-expressing C3H10T1/2, MC3T3-E1, 3T3-L1, CHO, and HeLa cells
EGFP-expressing cells and MLO-A5 cells were cultured separately in 10 cm dishes under normal culture conditions until they reached 70%–80% confluency. The culture medium was changed every 2 days.
Co-culture experiments
For co-culture experiments, 2×106 MLO-A5 cells were mixed gently with the same number of EGFP-expressing cells in a 15 mL tube. The mixed cells were seeded onto 10 cm dishes in growth medium and cultured for the indicated times. Where indicated, EGFP-expressing cells were isolated via using a BD FACS Aria cell sorter. For mono-culture experiments, each type of EGFP-expressing cell line was seeded onto 10 cm dishes at a density of 4×106 cells per dish, resulting in the same cell density as that used for co-culture experiments. The separate co-culture experiments were performed in six-well plates using a transwell culture system (Becton, Dickinson and Company, Franklin Lakes, NJ) to separate the MLO-A5 cells on the overlying filter from the underlying 10T-GFP cells. The MLO-A5 cells were plated onto the filters (pore size, 1 μm) forming the bottom of the upper chambers at a density of 106 cells per chamber, and the 10T-GFP cells were plated into the lower wells at a density of 106 cells per well. The upper chambers were then inserted into the lower wells, and the cells were cultured for 24 h.
CHX treatment
For both co-culture and mono-culture experiments, 10T-GFP cells were seeded onto 10 cm dishes in growth medium at a density of 2×106 cells per dish and cultured under normal conditions until they adhered to the dishes (∼3–4 h). The cells were then cultured in growth medium that was supplemented with or without CHX (100 μM) for 1 h and washed twice with phosphate-buffered saline (PBS). For co-culture experiments, 2×106 MLO-A5 cells were seeded onto the same dishes and the 10T-GFP and MLO-A5 cells were co-cultured in growth medium for 12 h. For mono-culture experiments, the 10T-GFP cells alone were cultured in growth medium for 12 h.
Noggin treatment
MLO-A5 cells (2×106) were mixed gently with the same number of 10T-GFP cells in a 15 mL tube. The mixed cells were seeded onto 10 cm dishes in growth medium that was supplemented with or without recombinant mouse Noggin (1 mg/mL) and cultured for 12 h. After the incubation, the 10T-GFP cells were isolated and subjected to real-time reverse transcription (RT)-PCR analysis. As a control, 4×106 10T-GFP cells were mono-cultured for 12 h in the same growth medium with or without recombinant mouse Noggin (1 mg/mL) and then subjected to real-time RT-PCR analysis.
BMP-2 treatment
MLO-A5 cells (2×106) were mixed gently with the same number of 10T-GFP cells in a 15 mL tube. The mixed cells were seeded onto 10 cm dishes in growth medium that was supplemented with or without recombinant human BMP-2 (100 ng/mL) and cultured for 24 h or 14 days. After the incubation, the cells were subjected to western blotting, real-time RT-PCR, and histochemical analyses. As a control, 4×106 10T-GFP cells seeded onto 10 cm dishes without MLO-A5 cells were used. The culture medium (with or without recombinant human BMP-2) was changed every 3 days.
CBX and INI-0602 treatments
10T-GFP cells (2×106) and MLO-A5 cells (2×106) were precultured in growth medium that was supplemented with or without CBX (100 μM) or INI-0602 for 1 h, and then co-cultured for 12 h as described earlier. As a control, 4×106 10T-GFP cells were precultured in growth medium that was supplemented with or without CBX (100 μM) or INI-0602 for 1 h, and then mono-cultured for 12 h. After culture, the cells were subjected to real-time RT-PCR analyses.
Histochemical examination
Cells were cultured under the indicated conditions, washed twice with PBS, fixed with 0.1 M cacodylate buffer (pH 7.2) containing 4% paraformaldehyde for 30 min, washed twice with PBS, and then double stained with ALP staining solution at pH 9.5 [nitroblue tetrazolium/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (NBT/BCIP) ready-to-use tablets; Roche Diagnostics, Penzberg, Germany] and Oil red O, as previously described [14]. The cultures were washed and then observed by fluorescence microscopy for Oil red O staining or phase contrast microscopy for ALP staining.
Whole-cell patch clamp recording
After 24 h of culture, dishes containing mixed or single cultures of MLO-A5 and 10T-GFP cells were transferred to a recording chamber that was continuously perfused with perfusion medium (126 mM NaCl, 3 mM KCl, 2 mM MgSO4, 1.25 mM NaH2PO4, 26 mM NaHCO3, 2 mM CaCl2, and 10 mM d-glucose). Dual whole-cell patch clamp recordings of the cultured cells were obtained using a fluorescence microscope that was equipped with Nomarski optics (BX51; Olympus, Tokyo, Japan) and an infrared-sensitive video camera (Hamamatsu Photonics, Hamamatsu, Japan). Electrical signals were recorded by amplifiers (Axoclamp 700B; Axon Instruments, Foster City, CA), digitized (Digidata 1422A; Axon Instruments), observed online, and then stored using Clampex software (pClamp 10; Axon Instruments). The pipette solution for recordings contained 70 mM potassium gluconate, 70 mM KCl, 10 mM N-(2-hydroxyethyl) piperazine-N′-2-ethanesulfonic acid, 15 mM biocytin, 0.5 mM EGTA, 2 mM MgCl2, 2 mM magnesium ATP, and 0.3 mM sodium GTP. Thin-wall borosilicate patch electrodes (2–5 MΩ) were pulled on a Flaming–Brown micropipette puller (P-97; Sutter Instruments, Novato, CA), and recordings were obtained at 30°C–31°C. The membrane currents and potentials were low-pass filtered at 5–10 kHz and digitized at 20 kHz.
Biocytin labeling
To visualize biocytin-labeled cells after whole-cell patch clamp recordings, the cells were cultured for 6 h, fixed, rinsed in 0.1 M PBS containing 0.5% Triton X-100 and 0.1 M glycine, and then incubated overnight with blocking solution containing 5 mg/μL Alexa Fluor 594-conjugated streptavidin (Molecular Probes, Eugene, OR). The cells were then rinsed in 0.1 M PBS containing 0.5% Triton X-100 and 0.1 M glycine, mounted on slides in Vectashield (Vector Laboratories, Burlingame, CA), and overlaid with coverslips. Images were obtained with a digital microscope (BZ-9000; Keyence, Osaka, Japan).
Analysis of mRNA expression
EGFP-expressing cells were mono-cultured or co-cultured with MLO-A5 cells for 1, 3, 6, 12, 24, 48, or 72 h, and then subjected to real-time RT-PCR analysis, as previously described [15]. Briefly, total RNA was isolated from cells using RNAiso Plus total RNA extraction reagent (Takara Bio, Inc.), according to the manufacturer's instructions. First-strand cDNA was synthesized at 50°C for 1 h in a 20 μL solution containing 1 μg of total RNA, 1× first-strand buffer, 50 ng of random primer, 10 mM dNTP mixture, 1 mM dithiothreitol, and 0.5 U of SuperScript® III RNase H− reverse transcriptase (Invitrogen). Subsequently, the first-strand cDNA was diluted 5-fold in sterile distilled water, and a 2 μL aliquot of the diluted cDNA was amplified by real-time RT-PCR using SYBR Premix Ex Taq™ II (Takara Bio, Inc.) and a CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA). The thermal cycling conditions comprised 40 cycles of 95°C for 5 s and 62°C for 25 s. The primer sets used for real-time RT-PCR analysis are described in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd). Each reaction was repeated thrice with cDNAs prepared from different samples of total RNA, and the expression levels of the mRNAs encoding ALP and BSP were normalized to those of β-actin.
Western blot analysis
10T-GFP cells were mono-cultured or co-cultured with MLO-A5 cells for the indicated periods. The 10T-GFP cells were then isolated and lysed with 1× sample buffer containing 50 mM Tris-HCl, 2% sodium dodecyl sulfate, 10% glycerol, and 6% 2-mercaptoethanol. Protein samples (10 μg) were separated on sodium dodecyl sulfate–polyacrylamide gels and transferred to nitrocellulose membranes. After transfer, the membranes were incubated with the indicated primary antibodies (diluted 1:500). The primary antibody against β-actin (C-11) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and the following primary antibodies were obtained from Cell Signaling Technology (Beverly, MA): anti-phosphorylated Smad1/5/8 (#9511); anti-total Smad1 (#9743); anti-phosphorylated p38 mitogen-activated protein kinase (MAPK) (#45118); anti-total MAPK (#9212); anti-acetylated histones H2A (#2576), H2B (#2574), H3 (#9649), and H4 (#2594); and anti-total histones H2A (#12349), H2B (#12364), H3 (#4499), and H4 (#2935). After washing, the membranes were incubated with secondary antibodies for 1 h and the immunoreactive bands were visualized using an enhanced chemiluminescence kit (Amersham, Arlington Heights, IL), according to the manufacturer's instructions.
Enzyme-linked immunosorbent assay
10T-GFP cells were mono-cultured or co-cultured with MLO-A5 cells in 10 cm dishes for 3 days without changing the growth medium. The culture medium was then collected and stored at −80°C until use. The levels of BMP-2 and BMP-4 in the culture medium were measured using Quantikine ELISA kits (R&D Systems), according to the manufacturer's instructions. The sensitivities of these kits for BMP-2 and BMP-4 in media were 29 and 3.68 pg/mL, respectively. Each experiment was performed using triplicate wells for each condition, and the culture medium from each well was tested in a single enzyme-linked immunoabsorbant assay (ELISA).
Luciferase assay of the ALP promoter construct
A construct containing the mouse ALP promoter in the pGL4.14 vector (ALPp/Lu plasmid) was synthesized by PCR [15]. 10T-GFP cells were plated into six-well plates at a density of 5×105 cells per well and allowed to grow under normal culture conditions for 18 h. The cells were then incubated with 2 mL of α-MEM containing 6.25 μL of Lipofectamine® and 2 μg of the ALPp/Luc plasmid. After transfection, the cells with stable DNA integration (10T-GFP-SA cells) were selected by culturing in the presence of 0.5 mg/mL hygromycin B (Sigma Chemical Co.). For transient transfection, 10T-GFP cells were transfected with the ALPp/Luc plasmid and a pRL-CMV vector (Promega, Madison, WI) by lipofection, as described earlier, to yield transiently transfected 10T-GFP-TA cells. The pRL-CMV construct contained the Renilla luciferase gene and was used to determine the transfection efficiency. After culture for 24 h, the 10T-GFP-SA or 10T-GFP-TA cells were co-cultured with MLO-A5 cells for a further 24 h before performing a luciferase assay using the Dual Luciferase Assay Kit (Promega), according to the manufacturer's instructions. In the Dlx5-forced expression experiment, 10T-GFP-SA cells were transiently transfected by lipofection with the Dlx5 expression vector or an empty pcDNA3.1 vector (mock control), and then mono-cultured or co-cultured with MLO-A5 cells for 24 h before performing luciferase assays. The firefly luciferase activities were normalized to those of Renilla luciferase that served as an internal control.
Luciferase assay of the (OSE2) × 6-Luc construct
The (OSE2)×6-Luc construct containing the (OSE2)× 6 promoter fused to the firefly luciferase gene was kindly provided by Dr. Gerard Karsenty (Research Associate, Baylor College of Medicine, Houston, TX). The pGL3-Basic plasmid (Promega) was also used as a negative control. 10T-GFP cells were seeded onto 10 cm dishes in growth medium at 70%–80% confluence and then incubated for 18 h. The cells were then co-transfected with the pRL-CMV vector and 10 μg of the (OSE2)×6-Luc or pGL3-Basic plasmid using Lipofectamine LTX® reagent (Invitrogen). At 48 h post-transfection, the 10T-GFP cells were mono-cultured or co-cultured with MLO-A5 cells for a further 24 h as described earlier, and then a luciferase assay was performed using a Dual Luciferase Assay Kit (Promega). The firefly luciferase activities were normalized to those of Renilla luciferase that served as an internal control.
Chromatin immunoprecipitation assay
10T-GFP cells were mono-cultured or co-cultured with MLO-A5 cells for 24 h and then isolated and fixed with PBS containing 1% formaldehyde for 10 min at room temperature. After two washes with PBS, the cells were resuspended in 0.5 mL of lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, and 1% NP40) containing Complete Protease Inhibitor Cocktail (Roche Diagnostics) and sonicated. The DNA fragments from the resulting soluble chromatin preparation were ∼400–500 bp in length. Immunoprecipitation was performed overnight with a primary antibody against Dlx5 (Cell Signaling Technology) or normal rabbit IgG (Cell Signaling Technology) that served as a negative control. The subsequent chromatin immunoprecipitation (ChIP) steps were performed as previously reported [16]. Finally, DNA from each sample was resuspended in 30 μL of water and RT-PCR was performed using 5 μL of the DNA solution as the template. The following primers were used to amplify the fragment containing the Dlx5 response element (ATTA element) within the ALP promoter: forward, 5′-CTG CCT GGG AAG TGG TC-3′; and reverse, 5′-CAC CTC TGA GCC TCA CAC-3′. As a negative control, the following primers were used to amplify the Dlx5-nonspecific region, which is located ∼1 kb upstream of the serum response element in the ALP promoter: forward, 5′-GAT TAC AGG TGT GTG AGT C-3′; and reverse, 5′-GCT CAG CCG GTG GAA GCA G-3′.
Bisulfite sequencing
After 24 h of culture, genomic DNA was purified from co-cultured or mono-cultured 10T-GFP cells using the PureLink™ Genomic DNA Kit (Invitrogen), according to the manufacturer's instructions. Bisulfite conversion was performed using the MethylCode Bisulfite Conversion Kit (Invitrogen) and the converted DNA was amplified by PCR. The following primer sets were used to amplify the 250 bp promoter regions of the genes encoding ALP and BSP: ALP forward, 5′-TTT GAA GTT AGG ATG AG-3′; ALP reverse, 5′-CTA CAA ACA ATC GTA AA-3′; BSP forward, 5′-ATTTAATTGAGGTTGAAT-3′; and BSP reverse, 5′-CTA TAA AAT CCT TAC CC-3′. The PCR products were cloned and expressed in Escherichia coli via TOPO® TA cloning (Invitrogen), and then reverse sequenced using M13 primers (Invitrogen).
Southwestern dot blot analysis
Southwestern dot blot analysis was performed according to the method of Schwarz et al. [17]. Briefly, after 24 h of culture, purified genomic DNA was prepared from mono-cultured or co-cultured 10T-GFP cells and transferred to a positively charged nylon membrane by dot blotting. The DNA was fixed by exposure to ultraviolet light and reacted with a monoclonal antibody against thymine dimers (Sigma Chemical Co.), followed by an ALP-conjugated anti-rabbit IgG and NBT/BCIP ready-to-use tablets (Roche Diagnostics).
Assessment of intracellular calcium ion levels
10T-GFP cells were mono-cultured or co-cultured with MLO-A5 cells for 6 h and then treated with the calcium indicator Rhod 2-AM (5 μM) (Dojindo, Kumamoto, Japan) for 20 min. Subsequently, the fluorescence intensity of Rhod 2-AM was determined by flow cytometry. As a positive control, 10T-GFP cells were pretreated with the calcium ionophore A23187 (2 μM) (Sigma Chemical Co.) for 1 h and then treated with Rhod 2-AM. To investigate the effect of calcium chelators on the expression levels of the mRNAs encoding ALP and BSP in co-cultured and mono-cultured 10T-GFP cells, the cells were mono-cultured or co-cultured with MLO-A5 cells in the presence or absence of 20 nM EGTA (Sigma Chemical Co.) or 2 μM A23187 for 3 h. The cells were then washed with PBS, cultured for another 6 h, and subjected to real-time RT-PCR analyses.
Statistical analysis
Quantifiable results are presented as the mean±standard deviation of triplicate cultures. Statistical differences between conditions were assessed using Student's t-tests. P<0.05 was considered statistically significant.
Results
Co-culture with MLO-A5 cells induces the expression of the mRNAs encoding ALP and BSP in C3H10T1/2 cells
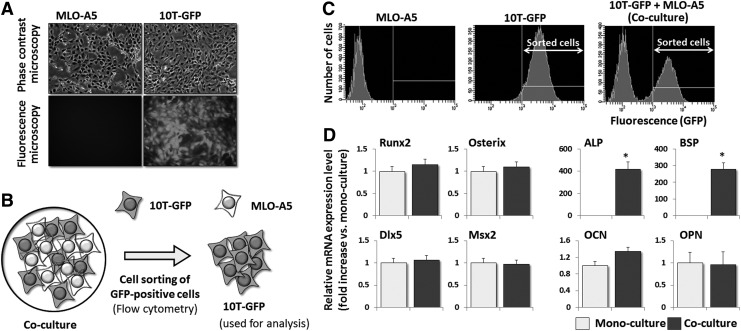
C3H10T1/2 cells expressing EGFP (10T-GFP cells) were generated and either mono-cultured or co-cultured with MLO-A5 cells, as described in the Materials and Methods section (Fig. 1A–C). The expression levels of the genes encoding osteoblast transcription factors (Runx2, Osterix, Dlx5, and Msx2) and mature osteoblast markers [ALP, BSP, osteopontin (OPN), and OCN] in the 10T-GFP cells were then assessed using real-time RT-PCR. The expression levels of the Runx2, Osterix, Dlx5, and Msx2 mRNAs were not affected by co-culture with MLO-A5 cells (Fig. 1D), and their protein levels were very low in both mono-cultured and co-cultured 10T-GFP cells (Supplementary Fig. S1); however, the expression levels of the ALP and BSP mRNAs were 300–400 times higher in co-cultured 10T-GFP cells than in mono-cultured 10T-GFP cells (Fig. 1D). The expression level of the OCN mRNA was also slightly higher in co-cultured 10T-GFP cells, while the expression level of the OPN mRNA was not affected by co-culturing (Fig. 1D).
FIG. 1.
The expression levels of the alkaline phosphatase (ALP) and bone sialoprotein (BSP) mRNAs in 10T-GFP cells are increased significantly by co-culture with MLO-A5 cells. (A) Phase contrast and fluorescence microscopy images of the 10T-GFP and MLO-A5 cells. (B) Schematic illustration of the co-culture system used in this study. Equal numbers of 10T-GFP and MLO-A5 cells were mixed, plated onto culture dishes, and co-cultured in growth medium. (C) Flow cytometry analyses of mono-cultured and co-cultured MLO-A5 and 10T-GFP cells. Co-cultured cells were sorted using a BD FACS Aria cell sorter. (D) Real-time reverse transcription (RT)-PCR analyses of the mRNA expression levels of osteogenic transcription factors (Runx2, Osterix, Dlx5, and Msx2) and osteoblast markers [ALP, BSP, osteocalcin (OCN), and OPN] in 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 24 h. The expression level of each mRNA was normalized to that in the corresponding mono-cultured cells, and the data are represented as the mean±standard deviation (SD) of n=3 replicates. *P<0.05.
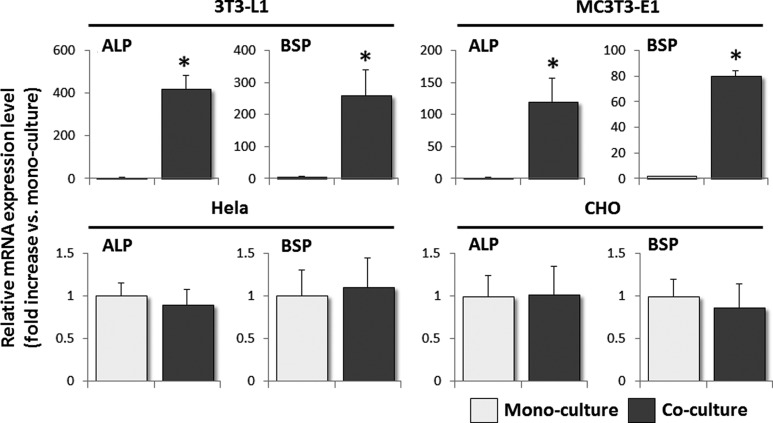
To determine whether the increases in the expression levels of the ALP and BSP mRNAs were specific to 10T-GFP cells, the same experiment was also performed using EGFP-expressing 3T3-L1, MC3T3-E1, HeLa, and CHO cells. The expression levels of the ALP and BSP mRNAs in 3T3-L1 and MC3T3-E1 cells co-cultured with MLO-A5 cells were higher than those in the corresponding mono-cultured cells, whereas co-culturing with MLO-A5 cells had no effect on the expression levels of these mRNAs in HeLa or CHO cells (Fig. 2).
FIG. 2.
The expression levels of the ALP and BSP mRNAs in 3T3-L1 and MC3T3-E1 cells, but not in HeLa and Chinese hamster ovary (CHO) cells, are increased significantly by co-culture with MLO-A5 cells. Real-time RT-PCR analyses of the expression levels of the ALP and BSP mRNAs in 3T3-L1, MC3T3-E1, HeLa, and CHO cells that were stably transfected with a enhanced green fluorescent protein (EGFP) expression vector and then mono-cultured or co-cultured with MLO-A5 cells for 24 h. The expression level of each mRNA was normalized to that in the corresponding mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05.
The expression levels of the ALP and BSP mRNAs are induced independently of BMP signaling
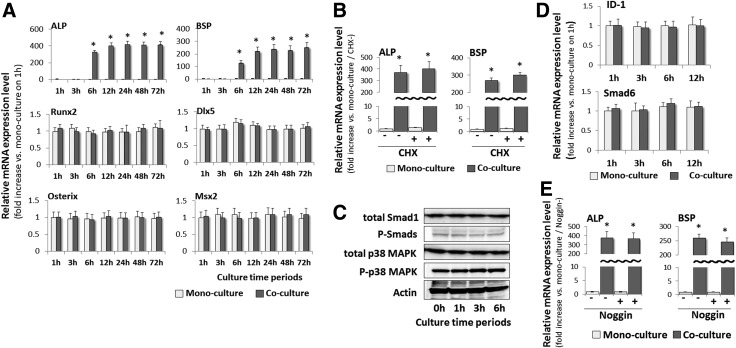
Next, the mechanisms by which the ALP and BSP mRNAs are induced in EGFP-expressing C3H10T1/2 cells were investigated. 10T-GFP and MLO-A5 cells were co-cultured for 1–72 h and real-time RT-PCR analyses were performed at various time-points (1, 3, 6, 12, 24, 48, and 72 h). The expression levels of the ALP and BSP mRNAs were markedly higher in isolated co-cultured 10T-GFP cells than in mono-cultured 10T-GFP cells after just 6 h (Fig. 3A), whereas the expression levels of the Runx2, Osterix, Dlx5, and Msx2 mRNAs were not affected by co-culture during the entire observation period (Fig. 3A). Treatment of the co-cultured 10T-GFP cells with CHX, a general protein synthesis inhibitor, had no effect on the increased levels of the ALP and BSP mRNAs (Fig. 3B).
FIG. 3.
Co-culture with MLO-A5 cells induces ALP and BSP mRNA expression in 10T-GFP cells independently of bone morphogenetic protein (BMP) signaling. (A) Real-time RT-PCR analyses of the mRNA expression levels of ALP, BSP, and osteogenic transcription factors (Runx2, Osterix, Dlx5, and Msx2) in mono-cultured and co-cultured 10T-GFP cells at various time-points. The expression level of each mRNA at each time point was normalized to that in the mono-cultured cells at the 1 h time point, and the data are represented as the mean±SD of n=3 replicates. *P<0.05 versus mono-cultured cells at the corresponding time-point. (B) Real-time RT-PCR analyses of the expression levels of the ALP and BSP mRNAs in 10T-GFP cells treated with or without cycloheximide (CHX) (10 μg/mL) for 1 h and mono-cultured or co-cultured with MLO-A5 cells for 12 h. The expression level of each mRNA was normalized to that in the non-CHX-treated mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05 versus the corresponding (with or without CHX) mono-cultured cells. (C) Immunoblot analyses of the phosphorylation levels of Smad1/5/8 and p38 mitogen-activated protein kinase (MAPK) in 10T-GFP cells. The cells were mono-cultured or co-cultured with MLO-A5 cells for the indicated times before immunoblotting with primary antibodies against P-Smads, total Smad1, phosphorylated p38 MAPK, and total p38 MAPK. The level of actin was used as an internal standard. (D) Real-time RT-PCR analyses of the expression levels of the inhibitor of DNA binding protein 1 (ID-1) and Smad6 mRNAs in 10T-GFP cells that were co-cultured or mono-cultured for the indicated times. The expression level of each mRNA was normalized to that in the mono-cultured cells at the 1 h time point, and the data are represented as the mean±SD of n=3 replicates. (E) Real-time RT-PCR analyses of the expression levels of the ALP and BSP mRNAs in 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 12 h in the presence or absence of Noggin (1 mg/mL), a BMP antagonist. The expression level of each mRNA was normalized to that in the non-Noggin-treated mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05 versus the corresponding (with or without Noggin) mono-cultured cells.
Signaling through BMPs and MAPKs initiates osteoblastic differentiation of stem cells; therefore, the roles of the BMP and MAPK signaling pathways in the stimulation of ALP and BSP mRNA expression by co-culture of 10T-GFP cells with MLO-A5 cells were investigated. Immunoblot analyses revealed that the levels of total Smad1 and phosphorylated Smad1, Smad5, and Smad8 proteins (P-Smads), which are important secondary messengers of BMP signaling, were similar in the mono-cultured and co-cultured 10T-GFP cells (Fig. 3C). Similarly, the levels of phosphorylated p38 MAPK and total p38 MAPK were also unaffected by co-culture with MLO-A5 cells (Fig. 3C). Moreover, there were no differences in the expression levels of the mRNAs encoding two direct targets of P-Smads, namely inhibitor of DNA binding protein 1 (ID-1) and Smad6 (Fig. 3D). The elevated expression levels of the ALP and BSP mRNAs in co-cultured 10T-GFP cells were also not affected by the addition of Noggin, a BMP-2 antagonist (Fig. 3E). These results indicate that BMP-2 and MAPK signaling pathways are not primarily responsible for up-regulated expression of ALP and BSP in co-cultured 10T-GFP cells.
Co-culture with MLO-A5 cells suppresses the differentiation of 10T-GFP cells into adipocytes
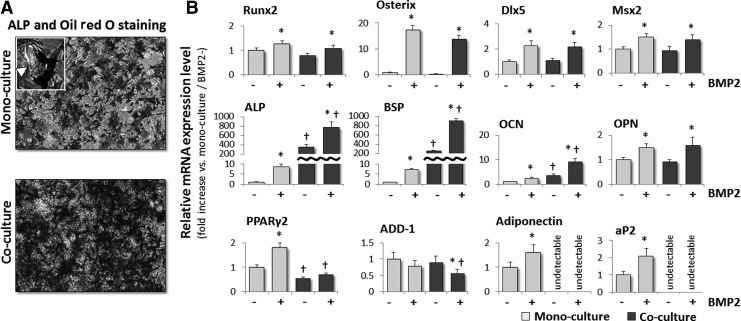
Exposure of C3H10T1/2 cells to the osteogenic cytokine BMP-2 stimulates their differentiation into osteoblasts, although some cells can also differentiate into adipocytes [18]; therefore, the effect of co-culturing with MLO-A5 cells on the differentiation of 10T-GFP cells into adipocytes versus osteoblasts was investigated. 10T-GFP cells were co-cultured with MLO-A5 cells in the absence or presence of exogenous BMP-2 for 14 days and the expression levels of osteoblast and adipocyte differentiation-related factors were then examined in isolated 10T-GFP cells. Unsorted cultures were also stained with Oil red O to observe the lipid droplets. Adipocytes containing lipid droplets were observed in the mono-cultures but not in the co-cultures of BMP-2-treated 10T-GFP cells (Fig. 4A), suggesting that the presence of the MLO-A5 cells in the co-cultures inhibited the BMP-2-induced adipocytic differentiation of the 10T-GFP cells. To test this hypothesis further, the expression levels of the mRNAs encoding four mature adipocyte markers [peroxisome proliferator-activated receptor γ2, adipocyte differentiation and determination factor 1 (ADD-1), adiponectin, and adipocyte protein 2] were also determined by real-time RT-PCR. The mRNA expression levels of all four adipogenic markers were increased by BMP-2 treatment in mono-cultured 10T-GFP cells, but not in co-cultured 10T-GFP cells. In fact, BMP-2 exposure suppressed the expression level of the ADD-1 mRNA in co-cultured 10T-GFP cells (Fig. 4B). Simultaneously, the expression levels of the ALP and BSP mRNAs in 10T-GFP cells were increased to a far greater extent by co-culture with MLO-A5 cells than by exposure to BMP-2 (Fig. 4B).
FIG. 4.
MLO-A5 cells inhibit BMP-2-induced adipocyte differentiation of 10T-GFP cells. (A) ALP and Oil red O staining of 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 14 days in the presence of BMP-2 (100 ng/mL). An NBT/BCIP staining solution was used to detect ALP and Oil red O staining was used to detect lipid droplets. The high-magnification inset shows an adipocyte with Oil red O-stained lipid droplets (arrowhead). (B) Real-time RT-PCR analyses of the mRNA expression levels of osteogenic and adipogenic markers in 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 14 days in the presence or absence of BMP-2 (100 ng/mL). The expression level of each mRNA was normalized to that in the non-BMP-2-treated mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05 versus the corresponding (mono-cultured or co-cultured) cells without BMP-2; †P<0.05 versus the corresponding (with or without BMP-2) mono-cultured cells.
Gap junctions are required for the induction of ALP and BSP mRNA expression in co-cultured 10T-GFP cells
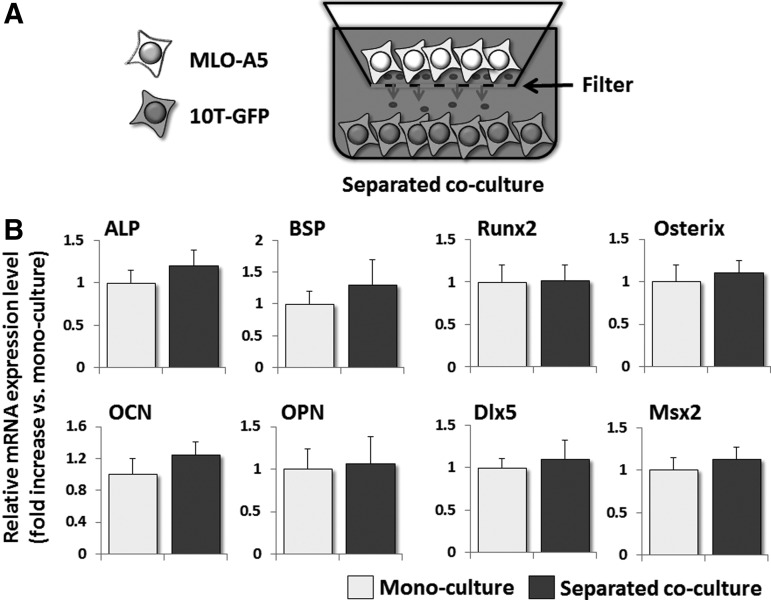
To determine whether direct contact between 10T-GFP and MLO-A5 cells is required for the induction of ALP and BSP mRNA expression in co-cultured 10T-GFP cells, 10T-GFP and MLO-A5 cells were cultured in a separated co-culture system that allowed the exchange of soluble factors between the cell types, but no direct cell–cell contact (Fig. 5A). The expression levels of osteoblast-related factors in the 10T-GFP cells were then analyzed by real-time RT-PCR. 10T-GFP cells cultured in this fashion did not show the marked increases in the ALP and BSP mRNA levels which occurred under regular co-culture conditions (Fig. 5B), suggesting that direct cell–cell contact is essential for their elevated expression.
FIG. 5.
The expression levels of the ALP and BSP mRNAs in 10T-GFP cells are not elevated by separated co-culture with MLO-A5 cells. (A) A schematic illustration of the separated co-culture system used in this study. 10T-GFP cells were plated into the lower wells of a transwell culture system, and MLO-A5 cells were plated onto filters forming the bottom of the upper chambers. (B) Real-time RT-PCR analyses of the mRNA expression levels of osteogenic markers in 10T-GFP cells that were mono-cultured or co-cultured under the separated conditions shown in (A) for 24 h. The expression level of each mRNA was normalized to that in the mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates.
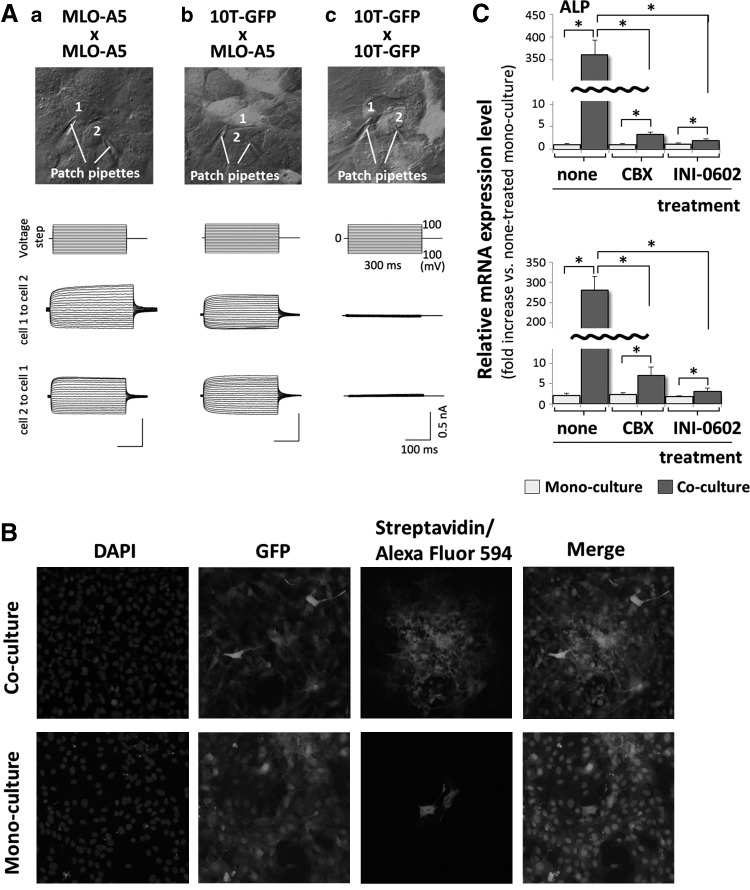
A previous study demonstrated that MLO-A5 cells express connexin43 and communicate with each other via gap junctions [19]. Based on this finding and the results described earlier, we hypothesized that co-cultured 10T-GFP and MLO-A5 cells exchange information via gap junctions. To evaluate this hypothesis, co-cultured and mono-cultured 10T-GFP cells were subjected to patch clamp assays. Passage of electric current was observed between adjacent MLO-A5 cells, as well as between adjacent MLO-A5 and 10T-GFP cells; however, no passage of electric current was detected between adjacent 10T-GFP cells (Fig. 6A). These results suggest that, during co-culture, gap junction channels are formed between neighboring MLO-A5 and 10T-GFP cells, but not between neighboring 10T-GFP cells.
FIG. 6.
ALP and BSP mRNA expression in 10T-GFP cells is triggered via gap junction-mediated intercellular communication with bone marrow mesenchymal stem cells (BMSCs). (A) Patch clamp assays showing the passage of electrical current between pairs of MLO-A5 and MLO-A5 cells (a), 10T-GFP and MLO-A5 cells (b), and 10T-GFP and 10T-GFP cells (c) that were cultured for 24 h before analysis. The cells with the attached patch pipettes are numbered. (B) Fluorescence microscopy images of biocytin-injected 10T-GFP and MLO-A5 cells. 10T-GFP cells were mono-cultured or co-cultured with MLO-A5 cells at 24 h and then, biocytin was injected into a pair of adjacent 10T-GFP and MLO-A5 cells, or a pair of adjacent mono-cultured 10T-GFP cells. The cells were cultured for another 6 h, fixed, and then stained with DAPI to detect the nuclei. EGFP expressing cells were also detected. Biocytin transfer between neighboring cells was detected using Alexa Fluor 594-conjugated streptavidin. (C) Real-time RT-PCR analyses of on the expression levels of the ALP and BSP mRNAs in 10T-GFP cells that were incubated in the presence or absence of carbenoxolone (CBX) (100 μM) or INI-0602 (100 μM) for 1 h and mono-cultured or co-cultured with MLO-A5 cells for 12 h. The expression level of each mRNA was normalized to that in the nontreated mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05.
During the patch clamp assays, the patched cells were injected with biocytin because the pipette solution for recordings contained this compound. Therefore, the presence of gap junctions was confirmed by visualizing biocytin-labeled cells. The spread of biocytin dye into neighboring cells was assessed by Alexa Fluor 594-conjugated streptavidin labeling. Dye transfer was readily observed after the injection of biocytin into co-cultured 10T-GFP/MLO-A5 cell pairs, but not after its injection into mono-cultured 10T-GFP/10T-GFP cell pairs (Fig. 6B). Furthermore, the addition of the gap junction inhibitors CBX or INI-0602 to the culture medium attenuated the co-culture-induced increases in the expression levels of the ALP and BSP mRNAs in 10T-GFP cells (Fig. 6C). These findings suggest that gap junctional communication between MLO-A5 and 10T-GFP cells regulates the expression of the ALP and BSP mRNAs in BMSCs.
Histones are acetylated in 10T-GFP cells in response to co-culture with MLO-A5 cells
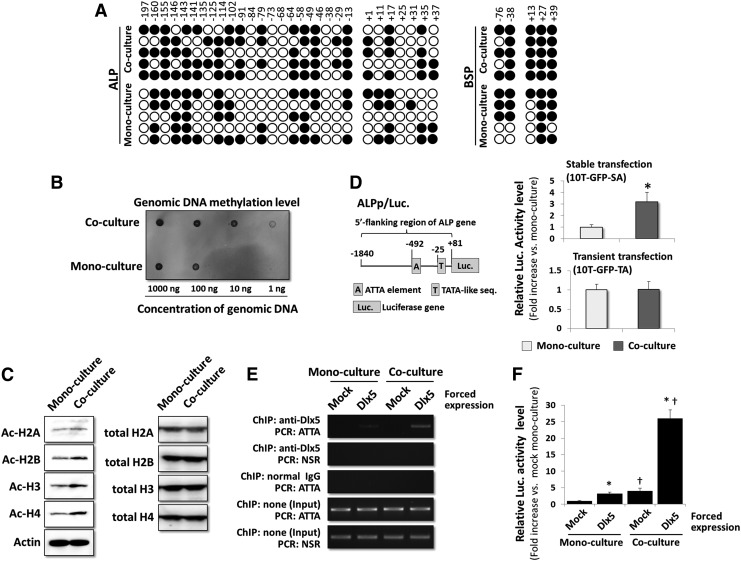
Epigenetic modifications such as genomic DNA methylation and histone acetylation regulate gene transcription; therefore, we investigated whether such modifications occur in 10T-GFP cells as a result of co-culture with MLO-A5 cells. First, the genomic methylation statuses in the vicinities (within 250 bases) of the transcription initiation sites of the genes encoding ALP and BSP were examined in mono-cultured versus co-cultured 10T-GFP cells by bisulfite sequencing. Twenty-seven CpG sites were found in the vicinity of the ALP gene transcription initiation site, but there were no significant differences between the methylation levels of these sites in the co-cultured and mono-cultured 10T-GFP cells (Fig. 7A; n=5 for each condition). By comparison, only five CpG sites were detected in the vicinity of the BSP gene transcription initiation site, and the methylation patterns at these sites were also similar in co-cultured and mono-cultured 10T-GFP cells (Fig. 7A; n=5 for each condition). Accordingly, the overall methylation status of the entire 10T-GFP genome was analyzed by Southwestern dot blot analyses using a primary antibody against methylated cytosine residues. This experiment revealed that co-cultured 10T-GFP cells had a higher level of overall genome methylation than mono-cultured 10T-GFP cells (Fig. 7B).
FIG. 7.
Chromatin remodeling in co-cultured 10T-GFP cells. (A) Bisulfite sequencing analyses of the ALP and BSP genes in 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 24 h. Genomic DNA was extracted from 10T-GFP cells and examined at positions −197 to+37 and −176 to+39 of the 5′ promoter region of the ALP and BSP gene, respectively (relative to the transcription initiation site). Methylated and unmethylated CpG sites are shown as filled and open circles, respectively. The sequences of five bacterial clones per genomic region examined are shown. (B) Southwestern dot blot analyses of 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 24 h. Genomic DNA was extracted from the 10T-GFP cells and subjected to Southwestern dot blot analysis with a primary antibody against methylcytosine residues. (C) Immunoblot analyses of the levels of histone acetylation in 10T-GFP cells that were mono-cultured or co-cultured with MLO-A5 cells for 24 h. Immunoblotting was performed using primary antibodies against acetylated and total core histones (H2A, H2B, H3, and H4). The expression levels of actin was used as an internal standard. (D) The left panel shows a schematic representation of the ALPp/Luc plasmid containing the 5′ flanking region of the ALP gene located upstream of the luciferase gene. The right panel shows the luciferase activities of 10T-GFP cells that were stably (10T-GFP-SA) or transiently (10T-GFP-TA) transfected with the ALPp/Luc plasmid and then mono-cultured or co-cultured with MLO-A5 cells for 24 h. The luciferase activity in the co-cultured cells was normalized to that in the corresponding mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05. (E) Dlx5 recruitment onto the 5′ flanking region of the ALP gene. Chromatin immunoprecipitation analyses of 10T-GFP-SA cells that were transiently transfected with a Dlx5 expression vector and then mono-cultured or co-cultured with MLO-A5 cells for 24 h. The extracted DNA fragments were analyzed by RT-PCR. ATTA, Dlx5 response element on the ALP promoter; NSR, nonspecific region on the ALP promoter; mock, control cells transfected with empty vector. (F) Luciferase activity assays showing the activities of the ALP promoter in 10T-GFP-SA cells transiently transfected with a Dlx5 expression vector and then mono-cultured or co-cultured with MLO-A5 cells for 24 h. The luciferase activities were normalized to those in the mock-transfected mono-cultured cells, and the data are represented as the mean±SD of n=3 replicates. *P<0.05 versus the corresponding (mono-cultured or co-cultured) mock-transfected cells; †P<0.05 versus the corresponding (mock-transfected or Dlx5-transfected) mono-cultured cells.
Next, the histone acetylation patterns in 10T-GFP cells were investigated. The acetylation levels of the H2A, H2B, H3, and H4 core histones were higher in co-cultured 10T-GFP cells than in mono-cultured 10T-GFP cells (Fig. 7C). Taken together, these observations suggest that DNA methylation and histone acetylation increases in co-cultured 10T-GFP cells after direct contact with neighboring MLO-A5 cells.
Remodeling of chromatin facilitates the binding of transcription factors to genomic DNA
Based on the data presented earlier, we hypothesized that remodeling of chromatin by histone acetylation features predominantly in the induction of ALP and BSP transcription in co-cultured 10T-GFP cells. This hypothesis was investigated further using an ALPp/Luc reporter plasmid containing the luciferase gene (Luc) fused to the region (∼2 kb) upstream of the ALP gene transcription initiation site (Fig. 7D). The ALPp/Luc plasmid was transiently or stably transfected into 10T-GFP cells to generate 10T-GFP-TA or 10T-GFP-SA cells, respectively; these cells were mono-cultured or co-cultured with MLO-A5 cells and then luciferase assays were performed. There were no detectable differences between the luciferase activities of the co-cultured and mono-cultured 10T-GFP-TA cells; however, the luciferase activity in the co-cultured 10T-GFP-SA cells was significantly higher than that in the mono-cultured 10T-GFP-SA cells (Fig. 7D).
The 10T-GFP-SA cells were then used to evaluate whether the increased histone acetylation resulting from co-culture of 10T-GFP cells with MLO-A5 cells facilitates the binding of transcription factors to genomic DNA. The ALPp/Luc plasmid contains a Dlx5 binding sequence (ATTA element); therefore, the 10T-GFP-SA cells were subjected to a ChIP assay using a primary antibody against Dlx5. The Dlx5 protein was only scarcely detected in 10T-GFP cells (Supplementary Fig. S1); therefore, its expression level was increased artificially by transfection of the 10T-GFP-SA cells with a Dlx5 expression vector. Antibody binding to the Dlx5 ATTA element in the ALP promoter was markedly higher in co-cultured, Dlx5-transfected 10T-GFP-SA cells than mono-cultured, Dlx5-transfected cells or mock (empty vector)-transfected cells (Fig. 7E). Luciferase assays indicated that the activity of the ALP promoter was significantly higher in Dlx5-transfected cells than mock-transfected cells, and significantly higher in co-cultured, Dlx5-transfected cells than in mono-cultured, Dlx5-transfected cells (Fig. 7F).
Discussion
The results presented here demonstrate that, as a result of co-culture with MLO-A5 cells, the expression levels of the ALP and BSP mRNAs were increased markedly in EGFP-expressing C3H10T1/2 (10T-GFP), 3T3-L1, and MC3T3-E1 cells, but not in EGFP-expressing HeLa and CHO cells. HeLa and CHO are epithelial cell lines, whereas C3H10T1/2 is a murine BMSC-like cell line and the 3T3-L1 and MC3T3-E1 cell lines are murine preadipocyte like and preosteoblast like, respectively. However, 3T3-L1 cells are able to transdifferentiate into an osteoblast lineage. Therefore, the effects of co-culture with MLO-A5 cells on the expression levels of the ALP and BSP mRNAs were restricted to cells of bone lineage.
The Runx2, Osterix, Dlx5, and Msx2 osteoblast transcription factors activate transcription of the ALP and BSP genes. However, the expression levels of these transcription factors at the protein level were very low in both mono-cultured and co-cultured 10T-GFP cells (Supplementary Fig. S1), and their mRNA levels were not increased by co-culture with MLO-A5 cells. In addition, luciferase assays using a 6×OSE2 reporter plasmid that contained six repeats of the Runx2 binding site (OSE2) located upstream of the minimal 34 bp OCN promoter revealed that transcriptional activity of Runx2 in 10T-GFP was not affected by co-culture with MLO-A5 cells (Supplementary Fig. S2). The increases in the expression levels of the ALP and BSP mRNAs were also unaffected by CHX. Taken together, these data suggest that the factor which triggers ALP and BSP gene transcription is not Runx2, Osterix, Dlx5, or Msx2, but may be a protein that is constitutively present in 10T-GFP cells under normal physiological conditions.
Osteoblast differentiation characterized by ALP and BSP expression is modulated by BMP-2 [14,15,20,21]; however, neither activation of BMP-2 signaling nor an increase in the concentration of BMP-2 in the culture medium (Supplementary Fig. S3) was observed after co-culture of 10T-GFP cells with MLO-A5 cells. Moreover, the expression levels of the ALP and BSP mRNAs were unaltered under separated co-culture conditions, which allowed the exchange of soluble factors (including cytokines such as BMP-2) between 10T-GFP and MLO-A5 cells, but no direct cell–cell contact. The concentration of BMP-2 in the growth medium of co-cultured cells was lower than 100 pg/mL (Supplementary Fig. S3), and our previous report demonstrated that BMP-2 concentrations of 10 ng/mL and lower fail to induce ALP mRNA expression in C3H10T1/2 cells [15]. Hence, the increased ALP and BSP mRNA expression levels in co-cultured 10T-GFP cells were not triggered by BMP-2 or other humoral factors.
The presence of gap junctions between MLO-A5 and 10-GFP cells was demonstrated by biocytin dye transfer and patch clamp experiments, and the use of the gap junction inhibitors CBX and INI-0602 showed that gap junctions are required for the elevations in ALP and BSP mRNA levels observed in co-cultured 10T-GFP cells. Gap junctions are channels that form between communicating cells and consist of connexin protein subunits [22,23]. Real-time RT-PCR analyses showed that MLO-A5 cells, but not 10T-GFP cells, express connexin43; however, connexins 26, 32, 37, 40, and 45 were either undetectable or only faintly expressed in both cell types (data not shown). These findings suggest that the gap junctions formed between MLO-A5 and 10T-GFP cells consist of connexin43.
Gap junctional channels regulate various cellular activities by allowing small molecular mediators, such as calcium ions, to pass from one cell to another [24–26]. Therefore, we hypothesized that calcium ions might be involved in the increased expression levels of the ALP and BSP mRNAs in co-cultured 10T-GFP cells. However, the use of a fluorescent Rhod 2-AM calcium probe revealed no significant differences between the intracellular calcium concentrations of co-cultured and mono-cultured 10T-GFP cells (Supplementary Fig. S4A). Furthermore, experiments using the calcium chelator EGTA (which decreases the level of intracellular calcium ions) and the calcium ionophore A23187 (which increases the level of intracellular calcium ions) did not suggest the involvement of calcium (Supplementary Fig. S4B, C) in this process. The activities of transcription factors containing zinc finger domains are modulated by zinc [27]; hence, changes in the intracellular concentrations of trace elements such as zinc are likely to direct critical 10T-GFP cellular functions. In addition, microRNAs (miRNAs) regulate gene expression at the post-transcriptional level and were recently shown to move between cells via gap junctions [28,29]; therefore, these small regulatory RNAs may be transported across gap junctions from MLO-A5 cells to 10T-GFP cells. The actions of miRNAs are the subject of increasing attention in a variety of fields [30–32]; in a recent study, overexpression of miR218 increased the expression levels of the ALP and BSP mRNAs in human adipose-derived stem cells [33]. Based on these reports, miRNAs may be likely candidates for the role of molecular messengers between MLO-A5 and 10T-GFP cells, although this hypothesis requires experimental validation.
Methylation of genomic DNA is an essential mechanism by which gene transcription is regulated in eukaryotes [34–37]. In addition, acetylation of the core histones that make up the nucleosome (such as H2A, H2B, H3, and H4) causes chromatin remodeling and promotes gene transcription [38–40]. Although the cytosine methylation levels in the vicinity of the ALP and BSP gene transcription initiation sites were comparable in mono-cultured and co-cultured 10T-GFP cells, the methylation level of the entire genome was higher in the co-cultured cells. Methylation of the genome generally progresses with cell differentiation [33,41]; therefore, this result may indicate that co-culture with MLO-A5 cells triggered the differentiation of 10T-GFP cells into osteoblasts. However, methylation-induced silencing of genomic DNA is inconsistent with the observed increased expression levels of the ALP and BSP mRNAs in co-cultured cells. This inconsistency may be explained as follows: First, there are no GC-rich domains and a few CpG sites upstream of the BSP gene; and second, even in 10T-GFP cells co-cultured with MLO-A5 cells, ∼40% of the cytosine residues at CpG sites are unmethylated in the regions upstream of the ALP and BSP gene transcription initiation sites. Therefore, it is possible that methylation of the genome in the vicinity of the transcription initiation sites was not of an order sufficient to suppress ALP and BSP expression.
Histone acetylation was higher in the co-cultured 10T-GFP cells than in the mono-cultured 10T-GFP cells. As such, we hypothesized that the recruitment of transcription factors to the genome, and hence ALP and BSP gene transcription, was promoted by the acetylation of core histones in co-cultured 10T-GFP cells. To evaluate this hypothesis, the ALPp/Luc plasmid was introduced into 10T-GFP cells through transient and stable transfection to generate 10T-GFP-TA cells and 10T-GFP-SA cells, respectively, which were subjected to luciferase assays. The ALPp/Luc plasmid was present in the nucleus and cytoplasm of 10T-GFP-TA cells but was integrated into the genome (unpublished observations); therefore, transcription of the luciferase gene in these cells was not affected by chromatin remodeling. On the other hand, the ALPp/Luc plasmid was integrated into the genome of 10T-GFP-SA cells and thus its transcription was affected by chromatin remodeling in the same manner as that of the endogenous ALP gene. Accordingly, luciferase activity was heightened in co-cultured 10T-GFP-SA cells but not in co-cultured 10T-GFP-TA cells. In addition, ChIP assays showed that recruitment of Dlx5 to its binding sequence (ATTA element) in the ALP promoter was augmented by the co-culture of exogenous Dlx5-expressing 10T-GFP-SA cells with MLO-A5 cells. These findings support our hypothesis that increased acetylation of histones in co-cultured 10T-GFP cells facilitates the binding of Dlx5 and possibly other transcription factors to the genome. The experiments performed with exogenous BMP-2 also support this hypothesis. Treatment with BMP-2 increased the mRNA expression levels of the Runx2, Osterix, Dlx5, and Msx2 osteoblast transcription factors to a similar extent in co-cultured and mono-cultured 10T-GFP cells. By contrast, the induction of ALP and BSP gene transcription by BMP-2 was far greater in the co-cultured cells than in the mono-cultured cells. This result may be explained by chromatin remodeling in the co-cultured 10T-GFP cells, which would lead to more efficient binding of the osteoblast transcription factors whose levels were increased as a result of BMP-2 treatment to the genome, and the subsequent induction of ALP and BSP gene transcription (Fig. 8). However, it should be noted that co-culture itself had a greater effect on the expression levels of the ALP and BSP mRNAs in 10T-GFP cells than BMP-2 exposure (∼200-fold vs. 2- to 3-fold), again suggesting that MLO-A5-derived molecular messengers play a more important role than BMP-2 in 10T-GFP cell differentiation.
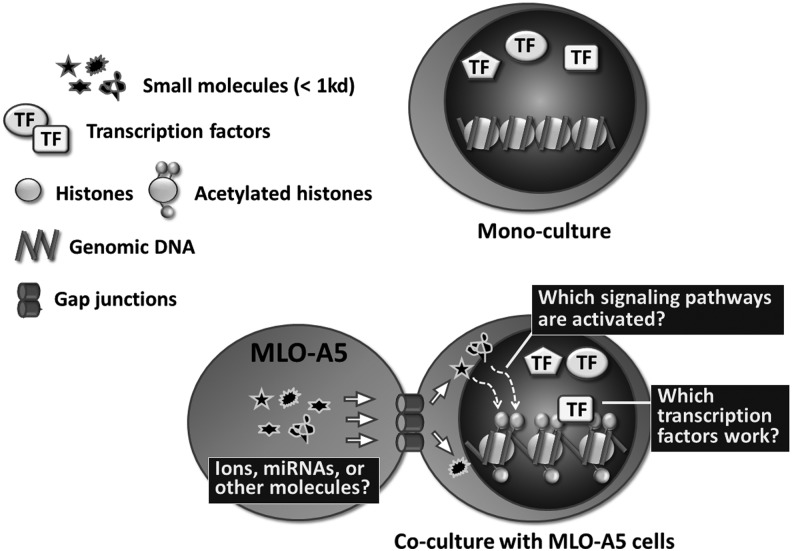
FIG. 8.
The proposed cross-talk between MLO-A5 and 10T-GFP cells. Co-cultured MLO-A5 and 10T-GFP cells may exchange small molecular messengers through gap junctions, leading to the acetylation of core histones in 10T-GFP cells. These modifications increase chromatin accessibility by relaxing chromatin condensation, enabling transcription factors to bind to the genomic DNA and modify transcription. The identities of these transcription factors and the signaling pathways they activate remain to be determined.
In conclusion, the results presented here indicate that osteoid-producing osteoblasts and BMSCs communicate via gap junctions. Furthermore, the results also suggest that chromatin remodeling and the ensuing ALP and BSP gene transcription occur in BMSCs as a result of gap junction-mediated intercellular communication (Fig. 8). To our knowledge, this study is the first demonstration of gap junction-modulated transcription in stem cells. Osteogenic differentiation of BMSCs can be induced reliably by osteogenic inducers such as BMP-2, without using the co-culture system; hence, intercellular communication may not be obligatory for this process. However, once osteoblasts are differentiated from BMSCs by these inducers, their ongoing osteogenic differentiation may be continuously induced by gap junction-mediated intercellular communication with osteoblasts, even in the absence of inducers. This feature may represent an important role of gap junctional cell–cell communication between osteoblasts and BMSCs in bone formation.
Although some questions regarding the functional roles of intercellular communication between osteoblasts and BMSCs require further exploration (Fig. 8), the findings described here have enormous implications for understanding the mechanisms of bone remodeling and BMSC maintenance and differentiation.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Yuko Nikuni-Takagaki (Department of Functional Biology, Kanagawa Dental College) for her valuable cooperation in our experiments. This work was supported in part by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology [KAKENHI (26462798) to Y.M.], and from the Dental Research Center and Sato Fund, Nihon University School of Dentistry.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Doty SB. (1981). Morphological evidence of gap junctions between bone cells. Calcif Tissue Int 33:509–512 [DOI] [PubMed] [Google Scholar]
- 2.Yellowley CE, Li Z, Zhou Z, Jacobs CR. and Donahue HJ. (2000). Functional gap junctions between osteocytic and osteoblastic cells. J Bone Miner Res 15:209–217 [DOI] [PubMed] [Google Scholar]
- 3.Sugawara Y, Ando R, Kamioka H, Ishihara Y, Honjo T, Kawanabe N, Kurosaka H, Takano-Yamamoto T. and Yamashiro T. (2011). The three-dimensional morphometry and cell-cell communication of the osteocyte network in chick and mouse embryonic calvaria. Calcif Tissue Int 88:416–424 [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Paul EM, Sathyendra V, Davison A, Sharkey N, Bronson S, Srinivasan S, Gross TS. and Donahue HJ. (2011). Enhanced osteoclastic resorption and responsiveness to mechanical load in gap junction deficient bone. PLoS One 6:e23516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tordjmann T, Berthon B, Claret M. and Combettes L. (1997). Coordinated intercellular calcium waves induced by noradrenaline in rat hepatocytes: dual control by gap junction permeability and agonist. EMBO J 16:5398–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vuyst E, Decrock E, Cabooter L, Dubyak GR, Naus CC, Evans WH. and Leybaert L. (2001). Intracellular calcium changes trigger connexin 32 hemichannel opening. EMBO J 25:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Civitelli R. (2008). Cell-cell communication in the osteoblast/osteocyte lineage. Arch Biochem Biophys 473:188–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marotti G. and Palumbo C. (2001). The mechanism of transduction of mechanical strains into biological signals at the bone cellular level. Eur J Histochem 51:15–19 [PubMed] [Google Scholar]
- 9.Boettner B. and Van Aelst L. (2009). Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol 21:684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Nieto D, Li L, Kohler A, Ghiaur G, Ishikawa E, Sengupta A, Madhu M, Arnett JL, Santho RA, et al. (2012). Connexin-43 in the osteogenic BM niche regulates its cellular composition and the bidirectional traffic of hematopoietic stem cells and progenitors. Blood 119:5144–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato Y, Boskey A, Spevak L, Dallas M, Hori M. and Bonewald LF. (2001). Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J Bone Miner Res 16:1622–1633 [DOI] [PubMed] [Google Scholar]
- 12.Barragan-Adjemian C, Nicolella D, Dusevich V, Dallas MR, Eick JD. and Bonewald LF. (2006). Mechanism by which MLO-A5 late osteoblasts/early osteocytes mineralize in culture: similarities with mineralization of lamellar bone Calcif Tissue Int 79:340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor SM. and Jones PA. (1979). Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17:771–779 [DOI] [PubMed] [Google Scholar]
- 14.Ito S, Suzuki N, Kato S, Takahashi T, Takagi M. (2007). Glucocorticoids induce the differentiation of a mesenchymal progenitor cell line, ROB-C26 into adipocytes and osteoblasts, but fail to induce terminal osteoblast differentiation. Bone 40:84–92 [DOI] [PubMed] [Google Scholar]
- 15.Mikami Y, Asano M, Honda JM. and Takagi M. (2010). Bone morphogenetic protein 2 and dexamethasone synergistically increase alkaline phosphatase levels through JAK/STAT signaling in C3H10T1/2 cells. J Cell Physiol 223:123–133 [DOI] [PubMed] [Google Scholar]
- 16.Mikami Y, Lee M, Irie S. and Honda JM. Dexamethasone modulates osteogenesis and adipogenesis with regulation of osterix expression in rat calvaria-derived cells. Cell Physiol J 226:739–748 [DOI] [PubMed] [Google Scholar]
- 17.Schwarz M, Peres G, Beer DG, Maor M, Buchmann A, Kunz W. and Pitot CH. (1986). Expression of albumin messenger RNA detected by in situ hybridization in preneoplastic and neoplastic lesions in rat liver. Cancer Res 46:5903–5912 [PubMed] [Google Scholar]
- 18.Huang H, Song TJ, Li X, Hu L, He Q, Liu M, Lane MD. and Tang QQ. (2009). BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A 106:12670–12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherian PP1, Xia X. and Jiang JX. (2008). Role of gap junction, hemichannels, and connexin 43 in mineralizing in response to intermittent and continuous application of parathyroid hormone. Cell Commun Adhes 15:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM. and Bae SC. (2000). Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol 20:8783–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, Gygi S. and Glimcher LH. (2009). TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J 28:2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dermietzel R, Traub O, Hwang TK, Beyer E, Bennett MV, Spray DC. and Willecke K. (1989). Differential expression of three gap junction proteins in developing and mature brain tissues. Proc Natl Acad Sci U S A 86:10148–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jongen WM, Fitzgerald DJ, Asamoto M, Piccoli C, Slaga TJ, Gros D, Takeichi M. and Yamasaki H. (1991). Regulation of connexin 43-mediated gap junctional intercellular communication by Ca2+ in mouse epidermal cells is controlled by E-cadherin. J Cell Biol 114:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SJ, King KR, Casali M. and Yarmush ML. (2009). DNA-triggered innate immune responses are propagated by gap junction communication. Proc Natl Acad Sci U S A 106:12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E. and Gerdes HH. (2010). Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc Natl Acad Sci U S A 107:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa M, Iwamoto T, Nakamura T, Doyle A, Fukumoto S. and Yamada Y. (2011). Pannexin 3 functions as an ER Ca(2+) channel, hemichannel, and gap junction to promote osteoblast differentiation. J Cell Biol 193:1257–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg JM. (1972). Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A 85:99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, et al. (2009). A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A 106:20794–20799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, Greco SJ, Bryan M, Patel PS. and Rameshwar P. (2011). Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 71:1550–1560 [DOI] [PubMed] [Google Scholar]
- 30.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T. and Zhang Y. (2012). MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 8:212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J, Rhee Y, Kim CH. and Lim SK. (2012). miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 27:1669–1679 [DOI] [PubMed] [Google Scholar]
- 32.Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R. and Karsenty G. (2012). miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J Cell Biol 197:509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang WB1, Zhong WJ. and Wang L. (2013). A signal-amplification circuit between miR-218 and Wnt/β-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone 58:59–66 [DOI] [PubMed] [Google Scholar]
- 34.Macgillivray AJ, Paul J. and Threlfall G. (1972). Transcriptional regulation in eukaryotic cells. Adv Cancer Res 15:93–162 [DOI] [PubMed] [Google Scholar]
- 35.Holliday R. and Pugh JE. (1975). DNA modification mechanisms and gene activity during development. Science 187:226–232 [PubMed] [Google Scholar]
- 36.Rigal M, Kevei Z, Pélissier T. and Mathieu O. (2012). DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J 31:2981–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones PA, Wolkowicz MJ, Rideout WM, 3rd, Gonzales FA, Marziasz CM, Coetzee GA. and Tapscott SJ. (1990). De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc Natl Acad Sci U S A 87:6117–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller S, Erard M, Burggraf E, Couppez M, Sautière P, Champagne M. and Van Regenmortel MH. (1982). Immunochemical detection of changes in chromatin subunits induced by histone H4 acetylation. EMBO J 1:939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abell AN, Jordan NV, Huang W, Prat A, Midland AA, Johnson NL, Granger DA, Mieczkowski PA, Perou CM, et al. (2011). MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell 8:525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson RT. (1978). Structure of chromatin containing extensively acetylated H3 and H4. Cell 13:691–699 [DOI] [PubMed] [Google Scholar]
- 41.Rossant J. and Papaioannou VE. (1984). The relationship between embryonic, embryonal carcinoma and embryo-derived stem cells. Cell Differ 15:155–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.