Abstract
Genome-wide association studies (GWAS) have identified many single nucleotide polymorphisms (SNPs) associated with complex traits, but have explained little of the underlying genetic heritability of many of these traits. To help guide future studies, we assess the crucial question of what additional utility future GWAS will have in detecting additional SNP associations and explaining heritability, by taking into account the new availability of larger GWAS SNP arrays, reduced genotyping costs, and imputation. We first describe the pairwise and imputation coverage of all SNPs in the human genome by commercially available GWAS SNP arrays, using the 1,000 Genomes Project as a reference. Next, we describe the findings from six years of GWAS of 172 chronic diseases, calculating the power to detect each of them taking array coverage and sample size into account. We then calculate power to detect these SNPs under different conditions using improved coverage and/or sample sizes, from which we estimate percentages of SNP associations previously detected and detectable by future GWAS under each condition. Overall, we estimated that previous GWAS have detected less than one fifth of all GWAS-detectable SNPs underlying chronic disease. Furthermore, increasing sample size has a much larger impact than increasing coverage on the potential of future GWAS to detect additional SNP-disease associations and heritability.
Keywords: Genetic association studies, single nucleotide polymorphisms, complex disease, heritability
INTRODUCTION
Genome-wide association studies (GWAS) have identified many common single nucleotide polymorphisms (SNPs) that are associated with complex human traits. Some of these findings have generated and supported hypotheses about the biological mechanisms underlying these traits, such as helping to elucidate the role of zinc transporters in Type 2 Diabetes [Sladek et al., 2007]. Others have led to improvements in the treatment of complex diseases, for example by helping to predict patient response to statin therapy for myopathy [The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine Collaborative Group, 2008]. Despite these advances, criticisms of the methodology have increased with the realization that in the first several years of their popularity, GWAS have explained a limited amount of the genetic heritability of most complex traits [Visscher et al., 2012].
There are several hypotheses for why GWAS have failed to explain this “hidden” genetic heritability of complex traits. One is that GWAS do not usually address polygenic effects due to epistasis or the cumulative contribution of variants that do not reach genome-wide significance [Gibson, 2010; Yang et al., 2010]. It is also possible that we have overestimated the genetic heritability of many complex traits (e.g. schizophrenia [Girard et al., 2011; Xu et al., 2011]), so we are trying to detect some “phantom” heritability [Zuk et al., 2012]. Another prevailing hypothesis is that the heritability of common traits, especially complex diseases, is more dependent on uncommon (1–5% minor allele frequency [MAF]) and rare (<1% MAF) SNPs than assayed by early GWAS, which were originally designed based on the common disease-common variant hypothesis and underpowered to detect associations with these variants [Pritchard, 2001; Reich and Lander, 2001]. Finally, the focus on keeping false positive findings to a minimum through replication studies and Bonferroni (or similar) corrections for multiple testing may have resulted in a large number of false negative findings that could help explain some heritability [Sebastiani et al., 2009; Williams and Haines, 2011].
By now, many powerful GWAS have been performed for a wide variety of complex traits. As a result, the range of effect sizes and SNP frequencies has expanded to include uncommon SNPs and low effect sizes, as shown in the Catalog of Published Genome-Wide Association Studies [http://www.genome.gov/gwastudies]. Thus, we now have the opportunity to determine the impact of further increasing power on detecting additional associations between these traits and this range of “GWAS-detectable” SNPs. Some of the ways in which we can achieve more power for individual GWAS are through improved microarray coverage, imputation methods, and by using larger sample sizes. The latest microarrays capture many of the uncommon variants than earlier arrays designed based on the common SNPs identified by the International HapMap Project [International HapMap Consortium, 2003]. Arrays designed in the last two years cover a larger portion of all SNPs in the human genome by directly measuring more common and uncommon variants, and by measuring SNPs in linkage disequilibrium (LD) with more regions of the genome. This is possible in part due to ongoing efforts to identify novel, less common SNPs by the 1,000 Genomes Project [The 1,000 Genomes Project Consortium, 2010].
Microarrays have also improved coverage of variants in populations with non-European ancestry. For example, the latest of the Illumina Omni family of microarrays [http://www.illumina.com/], the Omni5-Quad, directly measures nearly five million SNPs and captures variation down to approximately 1% MAF in populations of Asian and African, as well as European ancestry. The latest of the Affymetrix Axiom family of microarrays [http://www.affymetrix.com/] provide population-specific arrays, which optimize coverage of SNPs down to 1–2% MAF in Asians, Africans, and Latinos in addition to Europeans [Hoffmann et al., 2011a; Hoffmann et al., 2011b]. GWAS have also increasingly used genotype imputation methods to improve the coverage of variants not directly measured by microarrays, leading to novel associations [Marchini and Howie, 2010]. Finally, GWAS have become and will continue to become more powerful due to dramatic reductions in genotyping costs, allowing for much larger sample sizes than previously feasible.
Individual GWAS are unlikely to detect all the variants that explain the genetic heritability of complex traits. In most cases, this will require the use of other methods designed for detecting polygenic effects and associations with uncommon and rare SNPs. Full genome or exome sequencing currently provide promise in these areas, and so do GWAS meta-analyses [Begum et al., 2012; Yang et al., 2012]. However, full genome and exome sequencing in large cohorts may remain prohibitively challenging for some time. GWAS meta-analyses may be the most cost-effective way to explain additional heritability in previously well-studied populations and diseases, but do not allow for inferences about others. In some cases, more powerful individual GWAS will be required to detect associations that can explain additional heritability.
Several recent reviews have shown that despite their limitations, GWAS have continued to detect novel SNP associations over the past few years [Hindorff et al., 2009; Witte, 2010; Visscher et al., 2012]. Though we expect this trend to slow and eventually end, future GWAS may still contribute many additional associations that could further improve our understanding of these traits and their heritability. Our goal is to determine the extent to which increased GWAS power can lead to additional GWAS-detectable SNP associations. To achieve this, we first describe the range of SNP frequencies and effect sizes reaching genome-wide significance over the past six years from individual GWAS of nearly all of the complex, chronic diseases studied thus far. In addition, we describe the coverage levels of the GWAS arrays used to detect these associations. We then evaluate the impact that improved array coverage and/or larger sample sizes could have on detecting additional independent associations. Finally, we test the degree to which these SNP associations might contribute to the explained genetic heritability of several well-studied complex diseases. These results can help guide decisions about the most appropriate future research projects.
METHODS
COVERAGE OF 1,000 GENOMES PROJECT DATA BY ARRAYS
We calculated the maximum pairwise correlation (r2) between each SNP in the 1,000 Genomes Project (1kGP) phase 1 low coverage pilot data (June 2011 release [http://www.1000genomes.org/]) for each population group and the SNPs on each array within a one-megabase window using LdCompare [Hao et al., 2007]. Table I lists the arrays included in this study. The 1kGP data contains SNP genotypes on 179 founders of European (60 Utah residents with Northern and Western European ancestry), Asian (60 Han Chinese from Beijing, China and Japanese from Tokyo, Japan), and African (59 Yoruba from Ibadan, Nigeria) ancestry. We then plotted the proportion of SNPs in the 1kGP data that correlate with at least one SNP on each array at thresholds of r2 > 0.01 to 0.99 (by 0.01 increments) against the corresponding r2 threshold for each population ancestry group. We calculated the area under the curve (AUC) for each array as a summary of coverage using cubic spline integration. We also calculated imputation coverage for each array using pre-phased 1kGP data and leave-one-out cross-validation with the Impute2 v.2.2.2 software [Howie et al., 2009].
Table I.
GWAS SNP microarrays with abbreviated names used in this study and the number of SNPs on the array.
| Array manufacturer | Manufacturer’s array name | Abbreviated array name | Number of SNPsa |
|---|---|---|---|
| Affymetrix | GeneChip Human Mapping 100K | Affy100K | 109,365 |
| Human SNP Array 5.0 | Affy5.0 | 443,816 | |
| GeneChip Human Mapping 500K | Affy500K | 500,568 | |
| Human SNP Array 6.0 | Affy6.0 | 934,968 | |
| Illumina | HumanHap250S (v1) | Illm250K | 241,847 |
| HumanHap300 (v1 or v2) | Illm300K | 317,503 | |
| HumanCNV370 (Duo or Quad) | Illm370K | 353,202 | |
| HumanHap550 (v1 or v3) | Illm550K | 555,352 | |
| Human610-Quad (v1) | Illm610K | 599,021 | |
| Human Omni1-Quad | Illm 1M | 1,070,858 | |
| Human Omni2.5 | Illm2.5M | 2,450,000 | |
| Human Omni5-Quad | Illm5M | 4,500,000 | |
The number of SNPs on each array includes only those pre-selected by the manufacturer (not any custom SNPs that can be added to some arrays). In cases where there were several versions of an array (v1, v2, etc.), the number of SNPs reflects the union of SNPs from all versions.
SELECTION AND SUMMARY OF PREVIOUS GWAS FINDINGS
We obtained previously published GWAS data from the National Human Genome Research Institute’s (NHGRI) Catalog of Published GWAS [http://www.genome.gov/gwastudies]. We selected studies for inclusion in this analysis if they were individual GWAS (i.e. not meta-analyses) using a case-control design with unrelated individuals, published between July 1, 2006 and June 30, 2012, and reported one or more autosomal SNP association with a chronic (non-infectious) disease at the p<5×10−8 level. In addition, each study must have used one of the Affymetrix or Illumina SNP microarrays, and it must have replicated findings in a second stage with an independent cohort from the one in the first stage.
We gathered prevalence data for each of the diseases studied from online sources such as the Centers for Disease Control and Prevention [http://www.cdc.gov/] and the Surveillance Epidemiology and End Results database [http://seer.cancer.gov/]. Diseases with a prevalence of >10% in the population under study were excluded so that each odds ratio (OR) could be considered an approximation of a risk ratio in power calculations. We removed identical SNPs that were associated multiple times with the same disease, or that were in LD (r2>0.5) with other SNPs associated with the same disease. In these cases, we kept only the most recently published association, because we wanted to reduce possible biases from the winner’s curse effect [Capen et al., 1971]. We used the web-based tool SNAP [Johnson et al., 2008] to calculate pairwise LD between SNPs associated with the same disease and within 500 kilobases of one another, using the 1kGP data as the reference. Finally, we removed SNPs with missing effect sizes or missing risk allele and frequency information.
We categorized diseases into 11 major disease categories based on the 2012 version of the National Library of Medicine’s Medical Subject Headings (MeSH) vocabulary [http://www.nlm.nih.gov/mesh/2011/mesh_browser/MBrowser.html]. We grouped diseases falling into more than one MeSH category according to the one that best represents the etiology of the disease (e.g., we categorized stroke as a cardiovascular disease rather than a nervous system disease). Not all diseases currently suspected or confirmed to be autoimmune (e.g. Celiac disease [Sollid et al., 2005]) were included with other diseases of the immune system in MeSH, so we categorized these with other diseases involving the same organ or system (e.g. Celiac disease was categorized as a digestive system disease). We combined MeSH categories for eye diseases, otorhinolaryngologic diseases, and stomatognathic diseases into a “sensory organ disease” category, since there were few GWAS of diseases in these specific categories. We provide descriptive statistics of the studies and SNPs associated within each disease category.
POWER CALCULATIONS
We calculated the power to detect each SNP-disease association using methods for multi-stage GWAS described by Skol et al. [2006] and their CaTS power calculation software [http://www.sph.umich.edu/csg/abecasis/CaTS]. This method takes into account the following factors: effect size, MAF, disease prevalence, and sample size (the number of cases and controls genotyped, and the proportion of the sample used in the initial stage of each GWAS). More details about this method can be found in Skol et al. [2006]. The key assumptions we made in order to implement this method were: 1) the genotyped SNPs were in Hardy Weinberg Equilibrium (HWE); 2) the risk allele frequency in control subjects is approximately equal to the overall risk allele frequency in the population when disease prevalence is low (<10%); 3) the reported ORs are approximately equal to risk ratios when the disease prevalence is low; 4) the reported ORs are per copy of the risk allele, i.e. a multiplicative model applies.
We also took into account SNP microarray coverage of 1kGP SNPs in each population of European, Asian, or African ancestry using a method previously described by Jorgenson and Witte [2006]. For each SNP i, this formula is:
where PNr2 is the joint power for both stages, calculated using the Skol et al. [2006] method, but with the following changes to their formula: N (the number of cases and controls combined) is multiplied r2, and then PNr2 is multiplied by wr2, the proportion of SNPs in the 1kGP data correlated at ≥r2 with at least one SNP on the array, for each possible r2. For direct comparability between studies and arrays, we assumed a type I error rate of α = 5×10−8 for the overall genome-wide significance, and α = 5×10−5 for the first stage. For the 52 GWAS that imputed genotypes, we assumed coverage was similar to the array with the closest number of SNPs reported as passing quality control (QC) in the first stage (e.g. if a study reported 2.5 million SNPs passing QC with imputation, the Illumina Omni2.5 coverage levels were assumed to apply).
PERCENT OF SNP ASSOCIATIONS AND HERITABILITY IN PREVIOUS AND FUTURE GWAS
We estimated the total expected number of independent SNP associations detectable by GWAS within each MeSH disease, effect size, and MAF category in the range of previously observed associations, calculated as:
for i of m SNPs in the category, where PPi was the power to detect SNP i in previous GWAS. For each of the GWAS test conditions shown in Table II, we calculated the percent detectable within disease, effect size, and MAF categories as:
where PTi was the power to detect SNP i in each GWAS test condition. Effect size and MAF categories were chosen a priori to be 1.0≤OR<1.5 versus OR≥1.5, and 1%<MAF≤10% versus 10%<MAF≤30% versus 30%<MAF≤50%.
Table II.
GWAS conditions tested in this study.
| GWAS Condition | Array Used | Sample Sizea |
|---|---|---|
| Previous GWAS | Same as that of previous GWAS | Equal previous GWAS |
| Imputed | Same as that of previous GWAS + Imputationb | Equal previous GWAS |
| Illm5M | Illumina Human Omni5-Quad | Equal previous GWAS |
| All 1kGPc | Theoretical array containing all 1kGP SNPs | Equal previous GWAS |
| Nx2 | Same as that of previous GWAS | Double previous GWAS |
| Nx4 | Same as that of previous GWAS | Quadruple previous GWAS |
| Imputed, Nx4 | Same as that of previous GWAS + Imputation | Quadruple previous GWAS |
| Illm5M, Nx4 | Illumina Human Omni5-Quad | Quadruple previous GWAS |
| All 1kGP, Nx4 | Theoretical array containing all 1kGP SNPs | Quadruple previous GWAS |
Cases and controls combined.
Imputation was calculated for the arrays using the 1,000 Genomes project low coverage pilot data.
1kGP: the 1,000 Genomes Project low coverage pilot data.
We also calculated the narrow-sense genetic heritability explained by the independent SNPs associated with several chronic diseases. These diseases were selected because they have been well studied both in GWAS and because they had at least two estimates of sibling recurrence risk ratios (λs) reported in the literature. We estimate GWAS-detectable heritability as a percentage of the total heritability that GWAS could explain if we were able to detect all GWAS-detectable SNPs. Under HWE and a multiplicative/log-additive model, we calculate the contribution of each SNP i to the genetic variance of a disease according to a method described by Park et al. [2010] as:
where βi is the association between the risk allele for SNP i which has frequency di. Under this model, βl = ln(ORi). We estimated total expected heritability explained by all GWAS-detectable independent SNPs for each disease as:
For each of the GWAS test conditions shown in Table II, we calculated the percent of heritability detectable by GWAS for each disease as:
RESULTS
COVERAGE
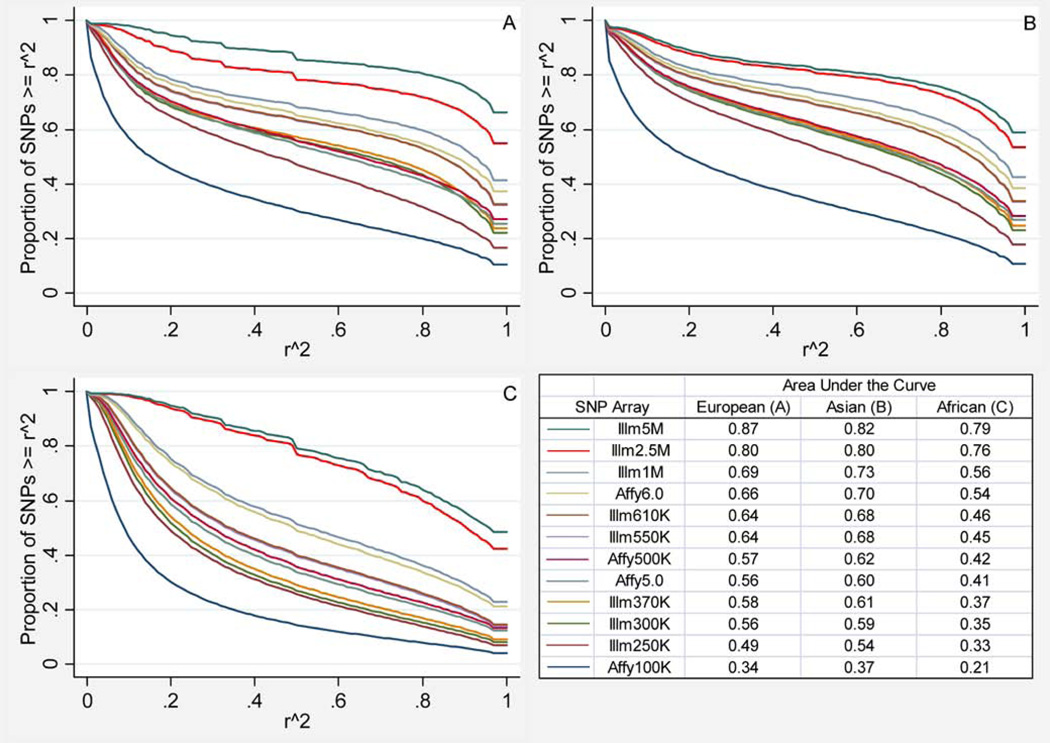
Using the 1kGP phase 1 pilot data as a reference, SNP microarrays have shown dramatic overall improvements in pairwise r2 coverage with increasing array size (Fig. 1). As expected, the smallest array (the Affymetrix GeneChip Human Mapping 100K) had the lowest level of coverage, with an AUC of 0.31 overall in populations of European, Asian, and African ancestry. 16.6% of all 1kGP SNPs were covered at an r2>0.8 threshold by this array. In contrast, the largest array (the Illumina Omni5-Quad) had an AUC of 0.82 overall, and 73.3% of all 1kGP SNPs were covered at an r2>0.8 threshold. Not surprisingly, imputation of genotypes using the 1kGP data improves coverage of most SNPs, and reduces the differences between arrays and populations (Supplementary Fig. 1). However, imputation coverage is slightly worse than pairwise coverage with the directly genotyped SNPs on the arrays when looking at a low LD threshold (e.g. r2<0.1). This is probably because these SNPs are more isolated and less common. Though we used pre-phasing to improve accuracy and efficiency, imputation of uncommon SNPs is difficult with only 60 individuals per population [Howie et al., 2012]. Array manufacturers may have intentionally targeted some of these SNPs for direct genotyping, leading to better pairwise than imputed coverage.
Fig. 1.
Coverage (maximum pairwise r2) of the 1,000 Genomes Project SNPs by microarray and population ancestry. (A) Coverage of SNPs in individuals with European ancestry. (B) Coverage of SNPs in individuals with Asian ancestry. (C) Coverage of SNPs in individuals with African ancestry. The table shows the area under the curve for each microarray and population.
As expected, pairwise coverage of the 1kGP SNPs by GWAS microarrays varies by population ancestry (Fig. 1). In general, coverage is much better in populations of European and Asian ancestry than for those with African ancestry, since LD between SNPs is lower in African ancestry populations and most GWAS and the arrays designed for them have focused more on populations with European ancestry [Bustamante et al., 2011]. Coverage also varies by MAF category in each population at r2 thresholds of 0.5, 0.8, and 0.99 (Supplementary Tables II–IV). These differences are smaller for larger arrays, especially for the Illumina Omni-5 Quad, since it was designed using the 1kGP data and has millions more directly measured SNPs than other commercial arrays. Averaged across populations, the Omni-5 Quad has only about a 2-fold difference in pairwise coverage of SNPs between the lowest (1–5%) and highest (40–50%) MAF categories compared to greater than 4-fold differences between these MAF categories for most other arrays.
PREVIOUS GWAS FINDINGS
The NHGRI Catalog of Published GWAS recorded 337 individual (not meta-analytic) studies published between July 1, 2006 and June 30, 2012 using unrelated case-control samples and reporting at least one replicated autosomal SNP association with a chronic (non-infectious) disease that was significant at the p<5×10−8 level. We selected the 329 studies that used one of the Affymetrix or Illumina SNP microarrays for which SNP content was publicly available (the other eight used Perlegen or custom arrays). We excluded five GWAS of diseases with prevalence rates of >10% in the population studied (gallstones, hypertension, hyperlipidemia, and obesity, for reasons described in the methods section), leaving 324 studies. Next, we removed nine studies with missing effect sizes in the GWAS Catalog and 25 studies with missing risk allele and frequency information, leaving 290 studies. Finally, we removed SNPs that were in LD (r2>0.5) with each other and associated with the same disease by querying the SNAP database [Johnson et al., 2008]. In these cases, we retained the most recently published association, leaving 219 studies and 729 “independent” SNPs associated with 172 chronic diseases.
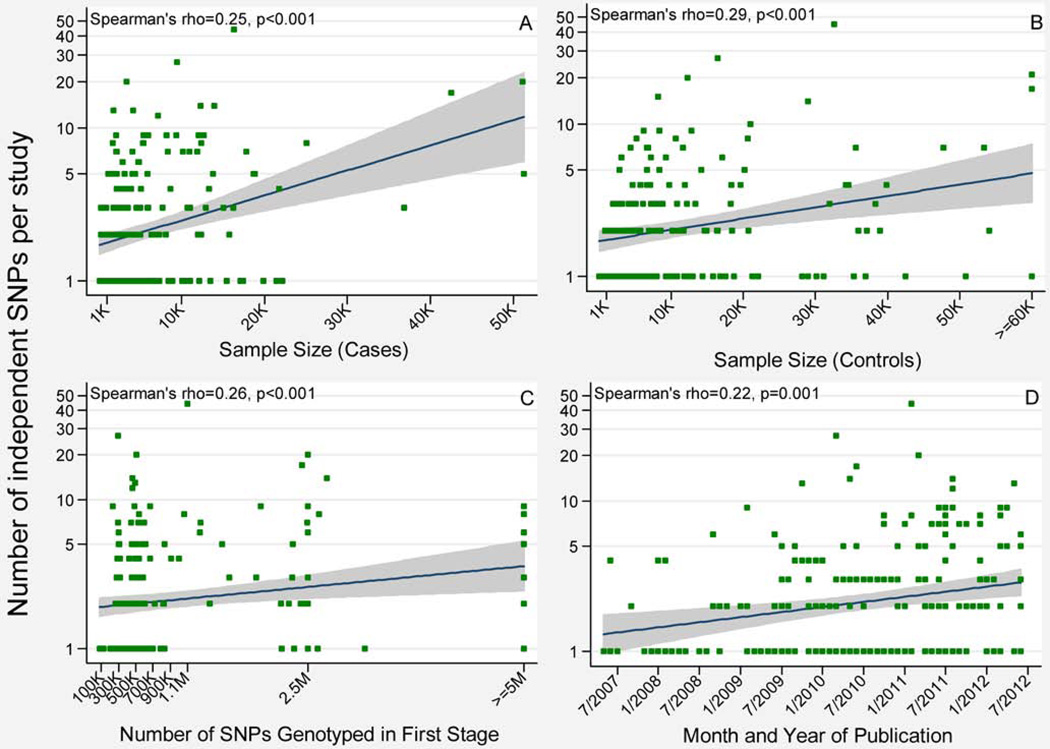
The number of independent SNPs detected per GWAS was positively and significantly correlated with study sample size (both case and control), the number of SNPs genotyped in the first stage, and publication date (Fig. 2), as previously shown [Visscher et al., 2012; Witte, 2010]. Figure 2 also shows that, as a result of the SNP selection process—whereby the most recent was chosen among those in LD and associated with the same disease—very few were included from the first year of our study period (prior to July, 2007). In addition to publication date being associated with the number of independent SNP associations reported per study, there were significant positive correlations between publication date and the number of SNPs genotyped in the first stage (Spearman’s ρ=0.38, p<0.001) as well as sample size (Spearman’s ρ=0.15, p=0.024).
Fig. 2.
The relationship between GWAS study-level characteristics and the number of significant SNP associations (p<5×10−8). The lines are predictions from linear regression models, and the gray areas are 95% confidence intervals around the regression lines. (A) The number of cases in thousands (K) versus the number of SNP associations. (B) The number of controls in thousands versus the number of SNP associations. (C) The number of genotyped SNPs passing quality control (QC) in thousands or millions (M) in the first or only stage of each GWAS versus the number of SNP associations. (D) The month and year of publication of the study versus the number of SNP associations.
We describe the characteristics of selected GWAS and SNPs in Table III, grouped by MeSH categories. This table shows that GWAS have tested a wide range of SNPs in the first stage through direct genotyping or imputation (ranging from about 80 thousand to 8 million SNPs), sample sizes (ranging from 225 to about 143 thousand cases and controls), MAFs (1%–50%), and effect sizes (OR ranging from 1.06 to 6.23). We show the individual diseases included in each MeSH category in Supplementary Table I.
Table IV shows the characteristics of selected SNPs grouped by broad categories of OR (<1.5 and ≥1.5) and MAF (1–10%, 10–30%, 30–50%). As expected, the majority (81%) of independent SNPs associated with chronic disease had low ORs (<1.5) and high MAFs (>10%). These categories explain more variance in, and have a stronger association with log-transformed sample sizes (R2=0.19, overall OR and MAF category p<0.001) than with the number of SNPs tested in the first stage (R2=0.03, p=0.002). Overall, the median sample size for detecting SNPs with OR>1.5 (6,948) was only about a third of that used to detect SNPs with ORs<1.5 (21,787), but the median number of SNPs was similar in both OR categories (507 thousand versus 562 thousand, respectively).
Table IV.
Characteristics of case-control GWAS SNPs associated with chronic diseases at p<5×10−8, by odds ratio (OR) and minor allele frequency (MAF).
| Odds Ratio | Minor Allele Frequency (MAF) |
Number of Independent SNPsa |
Median (Range) SNPs Passing QCb, (Thousands) |
Median (Range) Sample Sizec |
|---|---|---|---|---|
| 1.0≤OR<1.5 | 0.01<MAF≤0.1 | 25 | 666 (300 –7,689) | 26,005 (6,089 – 143,503) |
| 0.1<MAF≤0.3 | 250 | 570 (80 – 7,689) | 23,422 (4,644 – 143,403) | |
| 0.3<MAF≤0.5 | 312 | 541 (131 – 7,689) | 20,916 (1,607 – 143,503) | |
| OR≥1.5 | 0.01<MAF≤0.1 | 35 | 666 (282 – 7,689) | 10,769 (1,606 – 143,503) |
| 0.1<MAF≤0.3 | 68 | 501 (235 – 6,607) | 7,354 (690 – 74,544) | |
| 0.3<MAF≤0.5 | 39 | 480 (215 – 2,500) | 3,424 (225 – 39,547) | |
The sum of independent SNPs (r2<0.5) associated with each disease.
The number of SNPs passing quality control (QC) in the first GWAS stage.
The number of cases and controls in all stages.
THE IMPACT OF ARRAY COVERAGE AND SAMPLE SIZE ON FUTURE GWAS FINDINGS
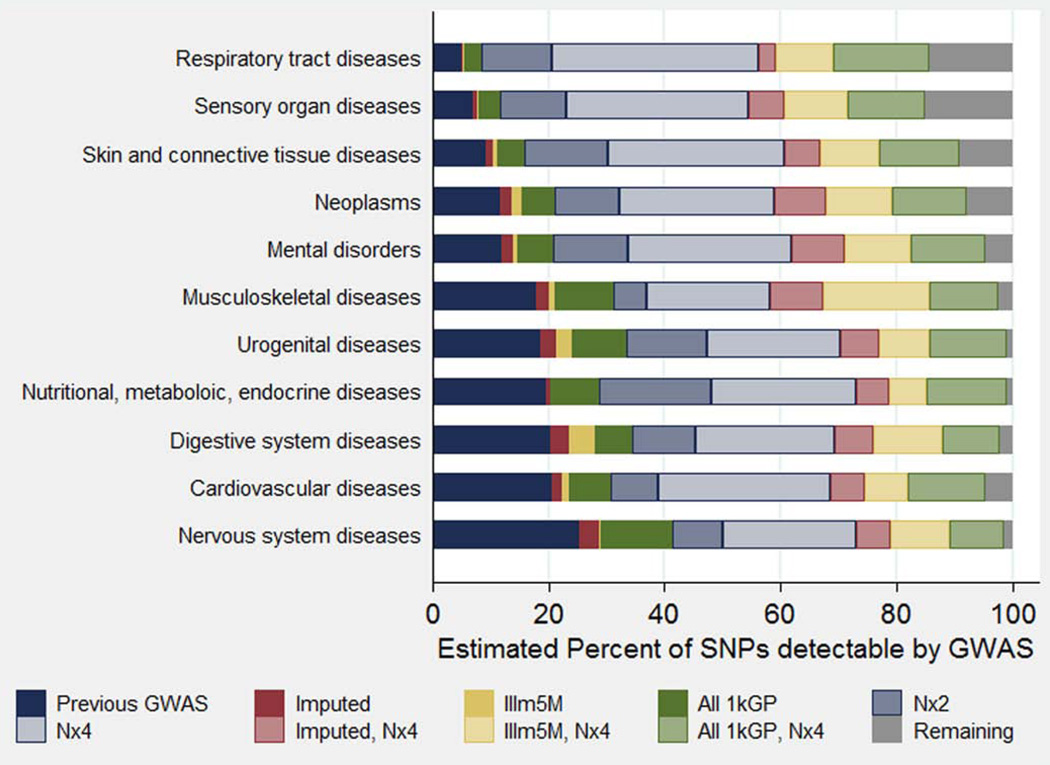
Figure 3 shows the estimated percent of SNPs detectable by GWAS under each of these conditions by disease category. These results show that we can detect many more SNPs within the range of effect sizes and MAFs observed in previous GWAS. They also show that the relative gains by increasing array coverage are much smaller than the gains by increasing sample size, whether we achieve better coverage by imputation, the Illumina Omni-5 array, or even by genotyping all 1kGP SNPs. Overall, we estimate that only 13.8% of all independent GWAS-detectable SNP associations have been detected by previous GWAS. These estimates ranged from 4.8% for respiratory tract diseases to 25.2% of nervous system diseases. With sample sizes equal to those of previous GWAS, we estimate that maximizing coverage of all 1kGP SNPs would result in the detection of still less than one quarter of all GWAS-detectable SNPs. Specifically, we estimate that 15.6% are detectable with previous arrays plus imputation, 17.0% with Illumina’s Omni5-Quad array, and 23.2% with perfect coverage of all 1kGP SNPs. In contrast, doubling or quadrupling the sample sizes without changing the coverage of previous GWAS, we estimate that we could detect 34.7% or 62.3%, respectively, of GWAS-detectable SNPs in future studies. The relative impact of increasing array coverage is slightly similar in the context of quadrupled sample sizes, rising to 69.2% with imputation, 80.2% with the Omni5-Quad array, and 93.9% with direct genotyping of all 1kGP SNPs. The percent of remaining SNPs requiring more than quadruple the sample size of previous GWAS and perfect SNP coverage ranged from 0.9% for urogenital diseases to 15.1% for sensory organ diseases.
Fig. 3.
Estimated percent of SNP associations detected by each GWAS condition tested, by disease category. Disease categories are ranked by the percent of associations detected by previous GWAS. Sensory organ diseases include the following MeSH categories: Eye diseases, Otorhinolaryngologic diseases, and Stomatognathic diseases.
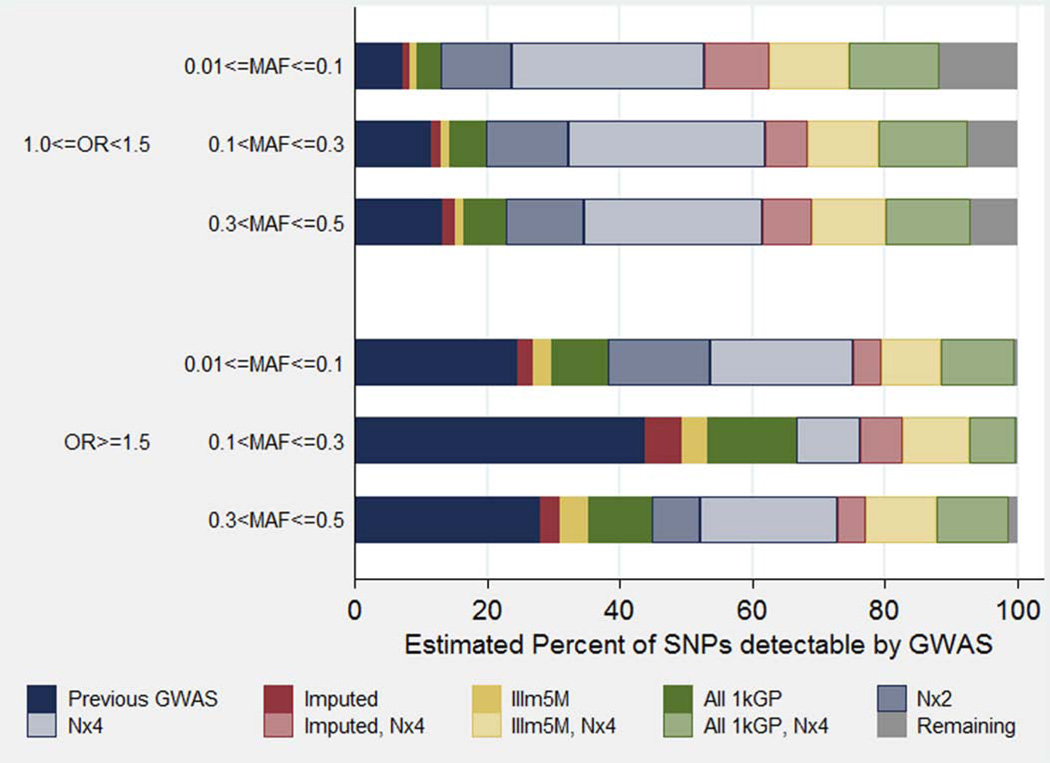
Figure 4 shows the estimated percent of SNPs detectable by GWAS under each of these conditions by OR and MAF categories. We estimate that a small percentage of the SNPs with MAFs of 1–10% have been detected by previous GWAS, as expected from the low frequencies in Table IV. This also shows that the impact of increasing sample size is greater for SNPs with weak associations (OR<1.5) than for those with stronger associations relative to the percent of SNPs detected by previous GWAS. Specifically, we estimated that quadrupling sample sizes for effects with OR<1.5 would increase the number of SNPs detected after future GWAS by over 400%. In contrast, quadrupling sample sizes for effects with OR>1.5 would only increase the number of SNPs detected after future GWAS by about 130%. SNPs with MAFs of 1–10% will be harder to detect than SNPs with MAFs of >10% in both OR categories.
Fig. 4.
Estimated percent of SNP associations detected by each GWAS condition tested, by odds ratio (OR) and minor allele frequency (MAF) category.
For several well-studied diseases, we show in Table V how much additional narrow-sense genetic heritability we can explain by GWAS that are more powerful. We selected these diseases because they have been the focus of multiple GWAS, and have multiple sibling recurrence risk ratios reported in the literature. As we did in estimating the percent of SNPs detectable by GWAS, we scaled these heritability estimates so that they represent a percentage of the maximum heritability that GWAS SNPs could explain, given the effect size and MAF distributions previously observed. We found that previous findings explain 7% to 40% of the genetic heritability that could ultimately be explained by GWAS. We also estimated that we can explain less than half of the heritability by only improving array coverage in future GWAS, and that sample sizes would need to be at least quadrupled in order to detect over half of the heritability of these diseases.
Table V.
Relative heritability that can be explained by SNPs detectable by GWAS
| Disease | λsa | Previous GWAS |
Imputed | Illm5M | All 1KGP | Nx2 | Nx4 | Imputed Nx4 |
Illm5M Nx4 |
All 1kGP Nx4 |
|---|---|---|---|---|---|---|---|---|---|---|
| Asthma | 2.6 | 7 | 8 | 9 | 13 | 24 | 59 | 62 | 72 | 88 |
| Breast cancer | 2.5 | 10 | 12 | 14 | 18 | 30 | 58 | 66 | 77 | 90 |
| Coronary heart disease | 3.2 | 13 | 15 | 15 | 21 | 30 | 57 | 66 | 75 | 90 |
| Crohn’s disease | 26.0 | 24 | 27 | 25 | 38 | 53 | 74 | 79 | 90 | 99 |
| Prostate cancer | 2.8 | 15 | 17 | 18 | 25 | 33 | 58 | 69 | 79 | 93 |
| Rheumatoid arthritis | 8.0 | 40 | 41 | 44 | 51 | 53 | 67 | 72 | 89 | 99 |
| Systemic lupus erythematosus | 30.0 | 12 | 13 | 16 | 21 | 33 | 62 | 66 | 76 | 91 |
| Type 2 diabetes | 3.5 | 14 | 15 | 16 | 22 | 40 | 67 | 75 | 83 | 99 |
λs: Sibling recurrence risk ratio. Source of λs for Asthma: Hemminki et al., 2007; Breast cancer: Risch, 2001; Coronary heart disease: Marenberg et al., 1994; Crohn’s disease: WTCCC, 2007; Prostate cancer: Risch, 2001; Rheumatoid arthritis: Harney and Wordsworth, 2002; Systemic lupus erythematosus: Harley et al., 2008; Type 2 diabetes: Das and Elbein, 2006.
DISCUSSION
In this study, we calculated pairwise and imputation coverage for nearly all GWAS SNP microarrays used for studying nearly all of the chronic diseases that have been recorded in the NHGRI Catalog of Published GWAS [http://www.genome.gov/gwastudies] in the past six years. Our pairwise coverage statistics show that GWAS SNP arrays have improved with increasing size, and that this is most dramatic with the latest SNP arrays that rely on the 1kGP data and more diverse populations. As expected, the biggest improvements are seen in coverage of uncommon SNPs (1%<MAF≤5%) in populations of European, Asian, and African ancestry. The use of the latest genotype imputation methods enhances the coverage of all 1kGP SNPs by GWAS arrays, and also narrows the coverage gaps between arrays and populations. To our knowledge, we are the first to publish these coverage statistics for nearly all commercial GWAS arrays available.
We also show that previous GWAS have detected significant associations between chronic disease and SNPs with a wide range of MAFs (down to 1%) and a wide range of effect sizes (detecting increased odds of disease of only 6% per risk allele). This range has not been observed in other reviews of GWAS findings, probably because they did not include results from the most recent and powerful GWAS. In general, the overall trends of GWAS that we observe agree well with those of previous studies using earlier data [Iles, 2008; Hindorff et al., 2009; Witte, 2010; Visscher et al., 2012]. For example, we see that significant positive correlations exist between the number of novel SNP associations detected and sample sizes, the number of SNPs tested, and publication dates. However, on average, we estimate that we have detected less than one fifth of all independent GWAS-detectable SNPs underlying chronic disease. We estimate that future more powerful individual GWAS have the potential to detect many of these additional SNP associations, and thus to explain more heritability. We found that increasing the sample size alone provides a much larger increase in GWAS-detectable SNP associations and heritability explained than improving array coverage, even with imputation or direct genotyping of all 1kGP SNPs. This is true for all major disease, MAF, and effect size categories that we studied. Even if all 1kGP SNPs were genotyped with the sample sizes used in previous GWAS, we estimate that GWAS would detect less than half of all GWAS-detectable SNPs and heritability. In contrast, quadrupling sample sizes but using the same arrays of previous GWAS would result in over 60% of SNPs and heritability detected. If it is possible to increase sample and array sizes for some diseases, future GWAS may capture most of the associations and heritability that GWAS-detectable SNPs have the potential to capture.
There are several caveats to keep in mind when interpreting these results. First, we are basing all estimates on the distribution of previously observed effect sizes and MAFs. The extremes of the true underlying distribution of SNP-disease associations are likely to be under-represented (only 8.2% of previously associated SNPs have MAFs of 1–10%), and this distribution will likely shift as we add to the number of independent SNPs associated with disease. The NHGRI Catalog of Published GWAS is also not an exhaustive source of known SNP-disease associations, and has been updated with findings from both individual and meta-analytic GWAS since mid-2012 when our collection period ended. The estimates of λs that we used for calculating heritability were also based on publications, and may continue to change somewhat over time. In addition, we calculate array coverage using a maximum pairwise approach to estimate the number of additional SNPs that remain. This may slightly underestimate coverage compared to say a multi-marker approach, and may explain why some associations were detected with seemingly low power. However, we believe that taking these issues into account would not likely change our results or conclusions that future GWAS can detect many additional SNPs and explain additional heritability, and that larger sample sizes improve their power to do so more than larger microarrays.
Another practical consideration is that in some situations, doubling or quadrupling sample sizes over those used in previous GWAS may not be realistic. For example, quadrupling the sample size over previous GWAS involving pancreatic cancer patients would require recruiting about half of all prevalent cases currently residing in the United States (based on Surveillance, Epidemiology and End Results [http://seer.cancer.gov/] and the U.S. Census [http://www.census.gov/] data). In other scenarios, increasing the sample size may be possible, but may require a less specific definition of the phenotype and more heterogeneous case samples which could in turn reduce power [Pawitan et al., 2009]. If previous GWAS have already studied a disease with very large sample sizes but poor coverage of variants in the population of interest, then it obviously makes more sense to improve coverage than to increase sample sizes to detect additional associations.
Despite the fact that individual GWAS have their limitations, we have shown that with increased power, they can detect many more SNP-disease associations. Although previous GWAS findings have explained a low amount of heritability for most diseases, they have also detected a low percentage of all the SNP-disease associations that the GWAS have the potential to detect. Even if we detect all of these GWAS-detectable SNPs, explaining all of the heritability of complex diseases will likely require other methods, especially meta-analyses and whole exome or genome sequencing. In addition, the contributions of family data, gene-gene and gene-environment interaction tests, functional validation experiments, and other approaches will be important to incorporate [Manolio et al., 2009; Pawitan et al., 2009; Cantor et al., 2010; Witte, 2010]. However, GWAS do have the potential to contribute many additional findings that may continue to add to our understanding of the biology and heritability underlying complex diseases.
Supplementary Material
Table III.
Characteristics of case-control GWAS studies and SNPs associated with chronic diseases at p<5×10−8.
| MeSHa Disease or Disorder Category | Number of Studies |
Median (Range) SNPs Passing QCb, (Thousands) |
Median (Range) Sample Sizec |
Number of Independent SNPsd |
Median (Range) Control Minor Allele Frequency |
Median (Range) Odds Ratio |
|---|---|---|---|---|---|---|
| All | 219 | 541 (80 – 7,689) | 15,722 (225 – 143,503) | 729 | 0.29 (0.01 – 0.50) | 1.26 (1.06 – 6.23) |
| Cardiovascular diseases | 22 | 2,500 (289 – 2,500) | 31,210 (225 – 143,503) | 72 | 0.25 (0.01 – 0.49) | 1.19 (1.06 – 4.82) |
| Digestive system diseases | 20 | 523 (266 – 2,466) | 25,885 (895 – 48,950) | 138 | 0.30 (0.04 – 0.50) | 1.21 (1.07 – 6.23) |
| Eye, otorhinolaryngologic, and stomatognathic diseases ("Sensory organ diseases") | 15 | 524 (299 – 6,037) | 10,462 (690 – 64,542) | 36 | 0.31 (0.01 – 0.50) | 1.38 (1.15 – 5.47) |
| Mental disorders | 6 | 1,253 (315 – 2,415) | 51,695 (2,672 – 63,649) | 16 | 0.17 (0.06 – 0.46) | 1.20 (1.12 – 1.59) |
| Musculoskeletal diseases | 20 | 1,585 (80 – 2,716) | 12,126 (1,606 – 47,926) | 64 | 0.29 (0.05 – 0.50) | 1.30 (1.10 – 3.62) |
| Neoplasms | 61 | 541 (247 – 7,689) | 12,218 (1,920 – 71,531) | 160 | 0.31 (0.02 – 0.50) | 1.29 (1.11 – 2.22) |
| Nervous system diseases | 30 | 531 (131 – 7,689) | 14,175 (1,208 – 102,338) | 84 | 0.26 (0.03 – 0.48) | 1.24 (1.09 – 5.11) |
| Nutritional, metabolic, and endocrine diseases | 12 | 2,427 (207 – 2,626) | 58,587 (792 – 141,454) | 43 | 0.32 (0.06 – 0.50) | 1.11 (1.06 – 4.05) |
| Respiratory tract diseases | 7 | 459 (215 – 550) | 35,083 (1,711 – 57,800) | 21 | 0.36 (0.14 – 0.50) | 1.20 (1.09 – 1.25) |
| Skin and connective tissue diseases | 18 | 495 (299 – 6,037) | 12,454 (3,107 – 51,423) | 72 | 0.28 (0.04 – 0.50) | 1.30 (1.11 – 2.80) |
| Urogenital diseases | 8 | 498 (303 – 2,187) | 10,769 (1,641 – 46,283) | 23 | 0.24 (0.06 – 0.50) | 1.34 (1.20 – 2.86) |
MeSH: Medical Subject Heading.
The number of SNPs passing quality control (QC) in the first GWAS stage.
The number of cases and controls in all stages.
The sum of independent SNPs (r2<0.5) associated with each disease.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01CA88164, U01CA127298, and R25CA112355.
REFERENCES
- Begum F, Ghosh D, Tseng GC, Feingold E. Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Res. 2012;40(9):3777–3784. doi: 10.1093/nar/gkr1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;13:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capen EC, Clapp RV, Campbell WM. Competitive bidding in high-risk situations. Journal of Petroleum Technology. 1971;23:641–653. [Google Scholar]
- Das SK, Elbein SC. The Genetic Basis of Type 2 Diabetes. Cellscience. 2006;2:100–131. doi: 10.1901/jaba.2006.2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Hints of hidden heritability in GWAS. Nat Genet. 2010;42:558–560. doi: 10.1038/ng0710-558. [DOI] [PubMed] [Google Scholar]
- Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;10:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Hao K, Di X, Cawley S. LdCompare: rapid computation of single- and multiple-marker r2 and genetic coverage. Bioinformatics. 2007;23(2):252–254. doi: 10.1093/bioinformatics/btl574. [DOI] [PubMed] [Google Scholar]
- Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythema- tosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney S, Wordsworth BP. Genetic epidemiology of rheumatoid arthritis. Tissue Antigens. 2002;60:465–473. doi: 10.1034/j.1399-0039.2002.600601.x. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Li X, Sundquist K, Sundquist J. Familial risks for asthma among twins and other siblings based on hospitalizations in Sweden. Clinical and Experimental Allergy. 2007;37:1320–1325. doi: 10.1111/j.1365-2222.2007.02737.x. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian African American and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011a;98:422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011b;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles MM. What can genome-wide association studies tell us about the genetics of common disease? PLoS Genet. 2008;4:e33. doi: 10.1371/journal.pgen.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. The International HapMap Project. Nature. 2003;18:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Witte JS. Coverage and power in genomewide association studies. Am J Hum Genet. 2006;78:884–888. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Marenberg ME, Risch N, Berkman LF, Floderus B, Defaire U. Genetic susceptibility to death from coronary heart disease in a study of twins. New England Journal of Medicine. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan Y, Seng KC, Magnusson PK. How many genetic variants remain to be discovered? PLoS One. 2009;4:e7969. doi: 10.1371/journal.pone.0007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- Sebastiani P, Timofeev N, Dworkis DA, Perls TT, Steinberg MH. Genome-wide association studies and the genetic dissection of complex traits. Am J Hematol. 2009;84(8):504–515. doi: 10.1002/ajh.21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;22:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Sollid LM, Jabri B. Is celiac disease an autoimmune disorder? Curr Opin Immunol. 2005;17:595–600. doi: 10.1016/j.coi.2005.09.015. [DOI] [PubMed] [Google Scholar]
- The 1,000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine Collaborative Group et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;21:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five Years of GWAS Discovery. Am J Hum Genet. 2012;13:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SM, Haines JL. Correcting away the hidden heritability. Ann Hum Genet. 2011;75(3):348–350. doi: 10.1111/j.1469-1809.2011.00640.x. [DOI] [PubMed] [Google Scholar]
- Witte JS. Genome-wide association studies and beyond. Annu Rev Public Health. 2010;31:9–20. doi: 10.1146/annurev.publhealth.012809.103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;7:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–378. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.