Abstract
Many new disease genes can be identified through high-throughput sequencing. Yet, variant interpretation for the large amounts of genomic data remains a challenge given variation of uncertain significance and genes that lack disease annotation. As clinically significant disease genes may be subject to negative selection, we developed a prediction method that measures paucity of non-synonymous variation in the human population to infer gene-based pathogenicity. Integrating human exome data of over 6000 individuals from the NHLBI Exome Sequencing Project, we tested the utility of the prediction method based on the ratio of non-synonymous to synonymous substitution rates (dN/dS) on X-chromosome genes. A low dN/dS ratio characterized genes associated with childhood disease and outcome. Furthermore, we identify new candidates for diseases with early mortality and demonstrate intragenic localized patterns of variants that suggest pathogenic hotspots. Our results suggest that intrahuman substitution analysis is a valuable tool to help prioritize novel disease genes in sequence interpretation.
INTRODUCTION
High-throughput sequencing generates large amounts of genomic data for clinical application and discovery of human disease genes. Interpretation of sequence results, however, remains a major challenge. Exome sequencing reveals variants in many genes, and it is critical to distinguish pathogenic variants from others. Previous gene annotation and inheritance patterns are important for interpretation, but variants often reside in functionally uncharacterized genes. Since sequence data from control populations reveal significant deleterious variant burden, understanding this variation could help in interpreting individual gene effects and their importance in disease phenotypes. Loss-of-function (LoF) variants, for example, are widespread in populations, though the frequencies of individual variants are rare (1–3). Effects of these variants could be tempered by carrier status or by the lack of impact for the phenotype. Missense variants are very common and pose additional challenges for exome interpretation. Although a variety of prediction methods assess the potential effects on function (4,5), variants in genes that lack disease annotation are often difficult to interpret. Variants predicted to disrupt protein function could play a role in disease or not, depending on the gene's function. Deleterious variants could persist in the population if they affect genes that do not alter reproductive fitness. Additionally, they could arise in critical genes where the variants would be subject to selection and would not persist in the population. Few studies have examined how to rank disease genes resulting from exome analyses for diagnostic interpretation (6). Here, we focus on interpreting exome data particularly for severe disease states. These are particularly important to recognize for disease prediction and for their clinical implications.
The ratio of non-synonymous to synonymous substitution rates (dN/dS ratio) (7) has been used to indicate how evolutionary selection pressure affects protein-coding sequence, particularly between species (8,9). A low ratio may indicate a negative selection, whereas a high ratio may indicate a positive selection. An interspecies ratio between human and primates has been used to indicate selection and is used in algorithms to predict the role of individual genes (10). Interestingly, far fewer studies have used the substitution ratio within one species to help predict effects (11), and this may be due to questions about the ratio's applicability within species (12). However, with the growing amount of human sequencing data, one can test the utility of this ratio broadly.
To assess the population presence of genomic variants and potentially deleterious burden, we can integrate data from control population exome studies with each human gene–disease association in Online Mendelian Inheritance In Man (OMIM, http://www.omim.org) and use these data to develop a measure of potential pathogenicity. The presence of deleterious variants in a given population may depend on whether the particular gene affected leads to adult or childhood disease, or whether the variant is present in a carrier status. To test this in a scenario where potential deleterious mutations may have a strong effect, we analyzed X-chromosome variants and integrated OMIM phenotypic data to understand the effects of exome variants in the NHLBI Exome Sequencing Project (ESP). We hypothesized that rare variants for X-linked diseases that affect reproductive fitness may be depleted in adult populations, particularly those causing a significant pediatric disease. LoF variants in genes that persist in populations may be tolerated due to individual carrier status, benign effects on protein or a gene's low effect on fitness. We present findings to assist in exome variant interpretation using a per-gene analysis and an intragenic variant distribution pattern. We also analyze all X-chromosome coding genes and their variants in the NHLBI ESP to identify new candidates for early-onset disease-associated genes.
RESULTS
Bulk variant analysis
We examined all variants on each chromosome, combining all 6503 exome results in the NHLBI GO ESP (http://eversusgs.washington.edu/EVS/) (1), and we analyzed predicted variant effects. Non-synonymous (missense) variants accounted for 34.6% of the total repertoire of variants, whereas synonymous variants accounted for 21.1% (Supplementary Material, Table S1). Splice, frameshift and stop variants, likely important in protein function [putative LoF variants], accounted for 2.3%, and the remaining fraction was comprised of intronic and other types. We examined the repertoire of these variants for each chromosome, recognizing each chromosome has a different size, coding gene density and region. The X-chromosome had a low overall ratio of non-synonymous to synonymous variant diversity relative to the autosomes (lower than all the autosomes but chromosome 22; Fig. 1A). The ratio of LoF variants versus synonymous variants was the lowest for the X-chromosome compared with the autosomes (Fig. 1B). A relative depletion of functional variants on the X-chromosome could indicate effects of selective pressure. Since ESP data are largely from adult individuals (1), we predicted that further variant analyses with these individuals could help identify patterns of variant retention and potential loss. We therefore focused our subsequent studies on the X-chromosome (Fig. 2).
Figure 1.
Putative LoF variants are relatively depleted on the X chromosome in exome data analysis. (A) Ratio of total non-synonymous variants versus total synonymous variants for each chromosome. P-value = 4.61e–05 (Wilcoxon signed ranked test). (B) Ratio of total LoF variants versus total synonymous variants for each chromosome. P-value = 4.01e–05 (Wilcoxon signed ranked test). Variants were analyzed from ESP. X-chromosome is indicated in red and autosomes in black.
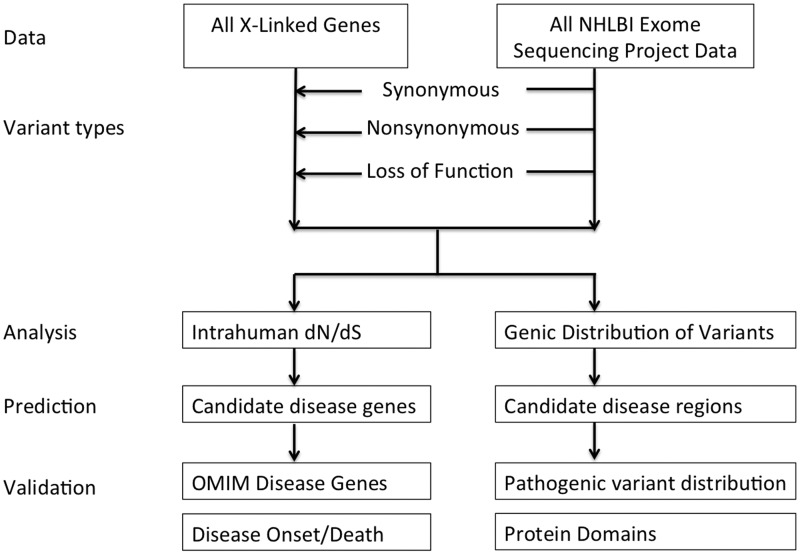
Figure 2.
Flow chart. All variants from ESP were filtered using all X-linked genes and by variant type, analyzed for prediction and validation.
We examined all variants in X-chromosome protein-coding genes in the ESP database and found that synonymous, intronic and missense variants were the most abundant and accounted for 93.5% of the total repertoire of exome variants on X-chromosome (Table 1). Stop, frameshift and splice-site variants, potentially causing LoF for the affected genes, were considered putative LoF variants (2) and were depleted on X, comprising only 1.5% collectively, lower than the average ratio of 2.3% for LoF variants from all the other chromosomes. Among all the variant sites on the X-chromosome, 28.8% (14 032 out of 48 678) occurred in OMIM disease genes, but the vast majority of variants were in genes not characterized for disease. Interestingly, there was a marked depletion of LoF variants in the OMIM disease genes compared with other variants, supporting the potential deleterious nature of LoF variants (Table 1, χ2 test, P-value <2.2e–16).
Table 1.
Variants in all genes compared with those in OMIM disease genes on X-chromosome
| Function | Number of variants |
Ratio (%) | |
|---|---|---|---|
| All genes | OMIM disease genes | ||
| Coding-synonymous | 11 186 | 3180 | 28.43 |
| Coding-other | 510 | 179 | 35.10 |
| utr-3 | 1113 | 244 | 21.92 |
| utr-5 | 821 | 163 | 19.85 |
| Intron | 17 683 | 6079 | 34.38 |
| Missense | 16 624 | 4078 | 24.53 |
| Stop-gained | 233 | 12 | 5.15 |
| Stop-lost | 10 | 2 | 20.00 |
| Splice-3 | 56 | 11 | 19.64 |
| Splice-5 | 62 | 14 | 22.58 |
| Frameshift | 380 | 70 | 18.42 |
| Total putative LoF | 741 | 109 | 14.71 |
| Total variants | 48 678 | 140 32 | 28.83 |
Non-synonymous/synonymous substitution ratio for disease gene prediction
We hypothesized that certain disease-related genes may be undergoing strong selective pressure on the X-chromosome, particularly severe diseases that affect reproductive fitness. Such genes would be highly constrained for functional variation, would lack LoF mutations and would also have a relatively lower number of non-synonymous variants in the population. Given the large number of missense variants of unclear function that can accompany sequence data, we tested whether the ratio of non-synonymous to synonymous substitution rates (dN/dS) for each gene could help identify genes under selection by human diseases. The total number of possible synonymous and non-synonymous sites varies for each gene. Thus, to compare across genes, we normalized the observed number of specific variants by the total possible number of either non-synonymous variants or synonymous variants to generate a non-synonymous ratio (dN) or a synonymous ratio (dS), respectively (13). Total possible synonymous and non-synonymous sites were predicted using DnaSP (14), and we calculated the number of non-synonymous (N) and synonymous (S) variants for each canonical transcript for each X-linked gene using bulk data from the NHLBI ESP variant server. Consistent with loss of non-synonymous versus synonymous variants, the non-synonymous ratio (dN) distribution of genes was lower than the synonymous ratio (dS) distribution of genes (Supplementary Material, Fig. S1). By calculating the dN/dS per gene, where a low dN/dS ratio could indicate increased selective pressure on a particular gene, we tested this ratio and then generated a ranked list of potential disease candidate genes.
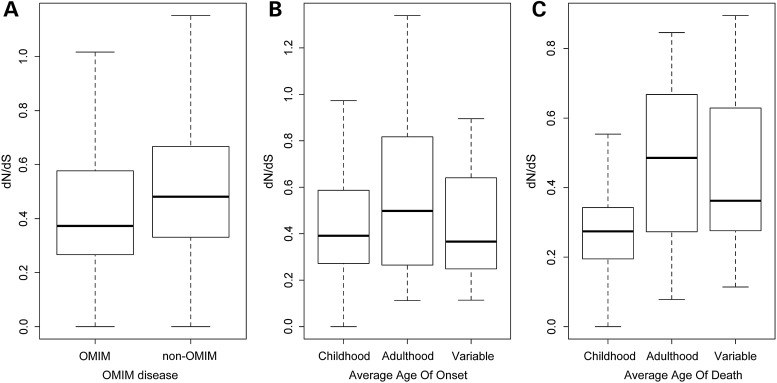
To test the utility of the dN/dS ratio to predict disease candidate genes, we first compared the dN/dS ratio for OMIM-annotated disease genes and genes lacking disease association on the X-chromosome. We defined OMIM disease genes as genes that have annotated disease phenotypes in the OMIM database. All other genes that were not annotated in disease by OMIM were defined as non-OMIM disease genes. OMIM disease genes had a significantly lower dN/dS ratio compared with non-disease genes (Fig. 3A, P-value = 6.38e−05, two-sample Wilcoxon test), supporting the potential utility of this ratio in gene prioritization.
Figure 3.
Comparison of dN/dS ratio by disease characteristics. dN/dS ratio of X-linked genes that are (A) OMIM disease genes (OMIM) or genes not yet annotated in disease (non-OMIM). P-value = 6.38e–05 (two-sample Wilcoxon test). (B) Disease genes with different average age of disease onset: childhood, adulthood and variable. P-value = 0.6834 (Kruskal–Wallis Rank Sum test). (C) Disease genes with different average age of death: childhood, adulthood and variable. P-value = 0.02972 (Kruskal–Wallis Rank Sum test).
To test whether the dN/dS score might indicate genes associated with disease severity, we compared the dN/dS ratio for genes that are associated with diseases of different onset and death. We reasoned that diseases affecting children or adults (and their severity) might affect the presence or absence of mutations in a given sequenced population. We again considered our dN/dS calculations from the ESP population, which almost entirely consists of adult variant data, and hypothesized that this ratio would be altered further in childhood disease genes. We considered all X-linked disease genes and the typical time for disease onset or death for each condition. We considered childhood, adulthood or variable categories using data from Orphanet (http://www.orphadata.org) (15). Genes for childhood-onset diseases and genes for adult-onset diseases were not significantly different in their dN/dS ratios (Fig. 3B, P-value = 0.6834, Kruskal–Wallis Rank Sum test). However, disease genes associated with childhood death had significantly lower dN/dS compared with those associated with adulthood death diseases (Fig. 3C, P-value = 0.02972, Kruskal–Wallis Rank Sum test). These data support the potential importance of the calculated dN/dS.
We also compared dN/dS to other currently available measures for ranking putative disease genes, including median combined annotation-dependent depletion (CADD) scores (16), median conservation scores [phyloP (17) and phastCons (18)] and combined network scores (19). These scores use orthogonal information, different from the population variant data used in dN/dS analysis. Lower dN/dS is significantly associated with higher median CADD scores, higher median conservation scores and higher network scores, all of which indicate dN/dS is useful in predicting disease association (Supplementary Material, Fig. S2).
In comparing prediction methodologies, we compared the area under the curve (AUC) from receiver operating characteristic (ROC) curves of the dN/dS ratio reflecting the ability to predict X-linked OMIM disease genes with other predicting measures. The dN/dS ratio performs better compared with most individual scoring parameters from the gene-based combined network score (19) (Supplementary Material, Fig. S3). dN/dS, as a single score, however does not perform better than combined score approaches, and this is expected as combined scores result from many different individual scores (Supplementary Material, Fig. S4). Since dN/dS uses population variant data that the other approaches do not have, it should be a helpful addition to existing prediction methods.
We therefore used a logistic regression model to combine dN/dS, CADD score and combined network score together to get an all combined probability score (Supplementary Material, Fig. S4 and Table S2). We did not include conservation scores because the CADD score already includes these. We found that this all combined score performs better than any of the above scores (Supplementary Material, Fig. S4). OMIM disease genes have significantly higher all combined scores compared with non-OMIM genes (Supplementary Material, Fig. S5, P-value <2.2e–16, two-sample Wilcoxon test). The all combined score therefore takes advantage of both orthogonal information and the population variant data and provided the best scoring methodology in predicting putative disease genes.
The human dN/dS calculated from a large adult population therefore may be an indicator for selective pressure, which in turn can help prioritize whether a particular gene is more likely to be related to disease. We ranked all X-linked genes by the dN/dS ratio (Supplementary Material, Table S2) and examined the genes with a lowest dN/dS ratio and considered these candidate genes for potential early/severe human phenotypes (Table 2). Of the 24 genes with the lowest ratio, one-third (8/24) were already annotated with disease in OMIM and included several notable conditions—cornelia de lange syndrome [MIM 300590] (SMC1A) (39), Ogden syndrome [MIM 300855] (NAA10) (40), Wieacker-Wolff syndrome [MIM 314580] (ZC4H2) (41) and genes implicated in intellectual disability [MIM 300161] (26), (PQBP1) (42–45). We manually reviewed the remaining functionally uncharacterized genes for potential disease association and found additional candidate genes with clinical relevance. For example, CLCN4 has been suspected in epilepsy as a de novo variant by exome sequencing (37). Given this gene prioritization based on dN/dS and our results, we propose that this method can assist in interpretation of genes not yet functionally characterized for disease.
Table 2.
Genes with the lowest dN/dS ratios on X-chromosome
| Gene | RefSeq | dN/dS | OMIM disease | Phenotype or potential function | References |
|---|---|---|---|---|---|
| EIF1AXa | NM_001412 | 0.000 | No | Implicated in uveal melanoma | (20) |
| FAM127Aa | NM_001078171 | 0.000 | No | Unknown | |
| MTCP1a | NM_001018025 | 0.000 | No | T cell proliferation; contiguous gene deletion with Moyamoya disease | (21,22) |
| PABPC1L2A | NM_001012977 | 0.000 | No | Unknown | |
| PRPS1 | NM_002764 | 0.000 | OMIM | MIM301835, 311070, 304500, 300661 | |
| RPL39a | NM_001000 | 0.000 | No | Ribosomal protein, 60S subunit | (23) |
| UBE2Aa | NM_003336 | 0.000 | OMIM | MIM300860 | |
| DDX3X | NM_001356 | 0.023 | No | Translation regulation | (24,25) |
| RAB39B | NM_171998 | 0.044 | OMIM | MIM3007744 | |
| ZDHHC9 | NM_016032 | 0.052 | OMIM | MIM300799 | |
| EIF2S3 | NM_001415 | 0.053 | No | Intellectual disability | (26) |
| HMGB3 | NM_005342 | 0.062 | No | Regulates early development and hematopoietic stem cells | (27,28) |
| RPS4X | NM_001007 | 0.063 | No | Ribosomal protein, 40S subunit | (29) |
| NUDT11a | NM_018159 | 0.063 | No | Diphosphoinositol polyphosphate phosphohydrolase, expressed in testes | (30) |
| KLHL15 | NM_030624 | 0.070 | No | Ubiquitin-proteasome system | (31) |
| NAA10 | NM_003491 | 0.074 | OMIM | MIM300855 | |
| PQBP1 | NM_001032383 | 0.078 | OMIM | MIM309500 | |
| HNRNPH2 | NM_001032393 | 0.082 | No | RNA-binding protein, splice regulator, affects hTERT splicing | (32) |
| OGT | NM_181672 | 0.091 | No | Glycosylation-related cell adhesion and disease | (33,34) |
| RAB9B | NM_016370 | 0.094 | No | Contiguous gene deletion with PLP1 | (35,36) |
| CLCN4 | NM_001830 | 0.094 | No | Possible epilepsy | (37) |
| SMC1A | NM_006306 | 0.094 | OMIM | MIM300590 | |
| PRPS2 | NM_001039091 | 0.096 | No | Synthesis of purines and pyrimidines | (38) |
| ZC4H2 | NM_018684 | 0.097 | OMIM | MIM314580 |
These genes have a coding sequence length of <500 bp.
We also calculated population-specific dN/dS ratios for African-American (dN/dS_AA) and European-American (dN/dS_EA) populations (Supplementary Material, Table S2). We found that both dN/dS_AA and dN/dS_EA ratio are lower for OMIM disease genes compared with non-OMIM genes (Supplementary Material, Fig. S6). Since many genes at the top of Table 2 were small genes, we analyzed the relationship between the coding sequence length and the dN/dS ratio. Although the median dN/dS ratio did not change with different coding sequence length, there was greater variation for small genes (Supplementary Material, Fig. S7) as small genes tended to have lower numbers of synonymous/non-synonymous variants. We noted this especially for coding sequences <500 bp, and therefore results for such small genes should be viewed with that potential variability in mind.
Intragenic variant distribution patterns
To assess intragenic variant patterns, we assessed the distribution of the missense and synonymous variants in ESP across the coding length of all X-linked genes. We found that many genes have missense variants that tend to cluster around certain coding regions and be depleted in other regions, while synonymous variants are evenly distributed. Figure 4A shows density plots of six representative disease-associated genes with such informative distribution patterns, suggesting specific coding regions that may be more sensitive to missense alteration. We also plotted all the pathogenic variants from NCBI ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) across the coding regions of these genes (Fig. 4B). Interestingly, we found that pathogenic variants tend to appear at coding regions that are relatively depleted in missense variants. On the other hand, in regions that have relatively high numbers of missense variants, there are fewer pathogenic variants. Figure 4C shows the protein domain structures of these genes. We found that protein domains also correlate with the regions of fewer missense variants in the population and with more pathogenic variants.
Figure 4.

Pathogenic variants occur at protein locations that have the least non-synonymous variant density. From top to bottom: (A) density plots for synonymous and non-synonymous variants along the coding sequence of six representative genes. X-axis shows the relative position of the synonymous and non-synonymous variants in the coding sequence. Green: synonymous. Red: non-synonymous. (B) Histograms show the pathogenic missense variants along the coding sequences. (C) Domain structures: dark gray rectangles show the protein domains and light gray bars show the whole protein. ATRX: NM_000489; CDKL5: NM_003159; F8: NM_000132; HCFC1: NM_005334; KDM5C: NM_004187; MECP2: NM_001110792.
Most X-linked genes lack disease annotation and variant pathogenicity information, so we similarly examined the distribution of the missense and synonymous variants in these genes using ESP. In many genes, localized regions of missense variant depletion were identified, and these also correlated with protein domain prediction (Supplementary Material, Fig. S8). From these results, we propose that specific gene regions with fewer population missense variants are more likely to harbor pathogenic variants and be deleterious in humans.
LoF variants
Although putative LoF mutations (e.g. stops, splice-site mutations) are less common than missense variants, their presence may indicate variant tolerance, and so we analyzed the pattern of LoF on the X chromosome. The 744 putative LoF variants from ESP involved genes at multiple locations on the X-chromosome and did not depend on parameters such as genic GC content; however, when we examined the distribution of LoF variants by gene size, they occurred more in larger-sized genes compared with smaller ones (data not shown). Since variants affecting X-linked disease genes could undergo sex-differential selection in the population, we examined the male and female distribution of variants as well (Supplementary Material, Table S3). Suggestive of a more detrimental phenotype in males, males had fewer variants in four of five categories of putative LoF variants (splice-3, splice-5, stop-gain and stop-lost), with stop gains particularly depleted. While some of the differences might be explained by fewer male samples when compared with female samples in ESP, stop gains were even further depleted (12 stop gains in females versus 3 in males). Alternatively, in exome data, sometimes predicted variant effects were not clear, even with putative LoF variants. For example, by analyzing the position of variants in genes, we found a predilection for stop-gain variants at the beginning of genes (Supplementary Material, Fig. S9). Some of the stops near the beginning of genes had codons that could be alternative starts sites (2 of 4 genes, MAGT1 and NYX, have alternative start sites), while some stops (6 of 12 genes) may not affect gene function as they would not be subject to Nonsense-mediated mRNA decay (NMD) (46). Therefore, not all stop variants were of clear functional consequence and would need further functional testing. Genes with LoF variants may be tolerated in the population if they do not affect reproduction. One would also predict that the same genes with LoF point mutations in the population may also be permissive to other forms of LoF such as deletion. To test this, we assessed whether genes were deleted by structural variation by analyzing OMIM genes on the X chromosome in the Database of Genomic Variants (DGVs, http://dgvbeta.tcag.ca/dgv/app/home), representing structural variation in control samples (47). About 47.4% of OMIM genes (27 of 57) with putative LoF variants were also deleted by structural variation in controls, whereas for OMIM genes without putative LoF variants, only 37.6% (44 of 117) were fully deleted (Supplementary Material, Table S4). These results suggest that LoF variant and deletions can overlap, and with further genomic data, additional patterns of tolerated LoF may emerge.
Disease genes on the X chromosome could also have particular functional characteristics that could help in deciphering genes without known function. We examined whether X-linked disease genes tend to reside in particular annotated pathways. To do this, we performed pathway analysis for OMIM disease genes compared with all the genes on the X chromosome using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt) (48). Metabolic pathways (KEGG) (49) were the most enriched among X-linked disease genes (Supplementary Material, Table S5). Interestingly, of the 20 metabolic pathway genes, only three genes, 15% (OTC, ALAS2 and ALG13), had putative LoF variants in ESP. In contrast, 57 of 174 X-linked OMIM disease genes (33%) had putative LoF, and 352 of 755 X-linked genes (47%) had putative LoF. The relative depletion of LoF variants in metabolic pathway genes support their function in disease and suggest that future consideration of other metabolic pathway genes may also reveal new disease candidates.
In summary, we used the dN/dS ratio from ESP population exome data as a method to prioritize putative disease causing genes on the X-chromosome. Supplementary Material, Table S2 summarizes dN/dS scores for all the genes on X-chromosome. We also made available an online sequence analysis dN/dS tool at http://humangenetics.ucsf.edu/sequencing-tool/.
This allows for calculation of dN/dS for any given coding sequence and the number of variants within the sequence. The output results are the total number of putative synonymous/synonymous sites and the dN/dS ratio. The dN/dS ratio from any coding gene or region within the gene of interest can be queried.
DISCUSSION
Despite advances in sequencing technologies for clinical diagnostics, it remains challenging in exome analysis to identify the key mutation for an underlying disease for a given patient. De novo mutations are suggestive, but additional gene annotations are needed to interpret contribution to disease, particularly with genes not yet associated with disease. For inherited variants, males tested by exome sequencing will have maternally inherited X-chromosome variants that may be important, but these would be of unclear functional significance without additional gene annotation. We have developed a basis to prioritize genes with a greater probability to cause disease. By studying variation in sequenced later-aged individuals, we can learn about genes important in earlier stages of life.
Analysis of non-synonymous to synonymous substitution rate of all X-linked genes in NHLBI ESP revealed that OMIM-annotated disease genes have a lower average dN/dS ratio. More importantly, lower dN/dS ratios are correlated with diseases with childhood mortality. This may be especially important for predicting serious pediatric conditions. Our study is interesting since we demonstrated correlation of dN/dS with diseases that have childhood mortality. Our data suggest that genes with a lower dN/dS ratio are more likely to cause disease, and these can be prioritized for study in subsequent gene discovery studies. We also found that putative LOF variants, although less numerous than missense variants, are depleted in disease genes overall, but individual LOF variants may also need further investigation for effect on function.
We also analyzed the distribution of the missense and synonymous variants from ESP along the coding sequences of all X-linked genes. We found that regions depleted in missense variants are highly correlated with those enriched in pathogenic variants relevant for human disease. For example, mutations in the ATRX gene cause alpha-thalassemia/mental retardation syndrome. Pathogenic variants are clustered in two regions, one in an N-terminal zinc finger domain and another one in C-terminal helicase domain (50). These two regions are depleted for missense variants in ESP, suggesting that variant patterns in ESP can be helpful in predicting where variants would be clinically significant. HCFC1 encodes a transcriptional regulator implicated in X-linked mental retardation and a cobalamin disorder. Pathogenic variants occur in the highly conserved N-terminal Kelch domain (51). We found that the same region is depleted for missense variants in ESP. Most X-linked genes do not have known pathogenic variants. Thus, the distribution of missense variants in sequenced populations could be utilized to predict regions that would harbor new pathogenic variants in these genes. Since whole exome sequencing often identifies multiple candidate variants, it is often difficult to determine pathogenicity. We demonstrate that the pattern of population variants may also be used as a tool to predict regions more likely to have pathogenic variants.
Our studies provide methods for prioritizing X-linked genes that have a greater probability of causing significant disease, and this will be useful for the interpretation of exome variants. Using this concept, interpretation of variants can be extended to include population-based genic variability. This can be used in addition to information on family structure and nucleotide-level change. In addition, as more genome-scale analyses are performed, even for non-pediatric diseases, our ability to interpret exomes for severe childhood conditions should improve.
We showed the first time that we can evaluate genes using an intrahuman variant substitution rate calculated from 6503 samples in the NHLBI ESP. Some studies have described approaches for evaluating variant pathogenicity by examining sequence (9,16,52,53). The former study (9) is based on variant data from smaller numbers of samples. Other studies (16,52,53) used different approaches to assess variant data, but they did not utilize intrahuman variant substitution ratio as shown in our study nor do they assess localized regions of proteins with variant depletion. For example, haploinsufficiency scoring (52) utilized copy number variation and several factors including interspecies conservation and did include some X-linked genes. A dN/dS ratio is not limited by previous gene annotation and can be calculated for any protein-coding gene. Our intrahuman dN/dS ratio is applicable to all X-chromosome protein-coding genes and adds direct information from growing human single-nucleotide variability data. Piton et al. (11) examined genes implicated in X-linked intellectual disability and used nucleotide substitution ratios to evaluate candidates for this phenotype. Our results provide evidence for generalization of this concept by evaluating all coding X-linked genes and assessing genes for multiple disease phenotypes. Our results using ESP population data on X-linked genes provide evidence for the importance of adult exome data for pediatric phenotypes.
These results are potentially useful for disease prediction, but our methods have several limitations due to currently available resources. The NHLBI ESP does include data from symptomatic individuals, especially from people with adult diseases such as cardiovascular disease. The ESP database does not provide information about which variants were from healthy or diseased individuals. However, most variants observed in a particular gene are likely from individuals that do not have a specific disease linked to that gene. Thus, the influence of having diseased individuals in the analysis will be greatly diluted, which makes it possible to use the ESP data for the analysis. Since our analysis is based on purifying selection, it is likely to be most relevant for early-onset conditions and not for late-onset diseases. Our analysis of clinical correlation used the disease databases OMIM and Orphanet, and databases can be incomplete given lag between gene–disease association and annotation. To help account for this, we used PubMed searches for the genes with the lowest substitution ratios and still found several candidate genes that are not disease annotated, but are expected to play an important function. The results of our studies are likely the most useful for pediatric and other early-onset diseases. Regarding the dN/dS ratio and clinical outcome, annotation of disease phenotypes including onset of disease and age of death for diseases are still incomplete; thus, correlation between dN/dS ratio and disease due to each gene will require further refinement with more clinical data and exome results.
We observed a greater variability in dN/dS in small genes, and this may reflect the limited number of variants even in ESP. Small genes with a low dN/dS ratio may indicate disease association or simply be due to the variability of the dN/dS value for small genes. We cannot exclude the possibility that some small genes with a low dN/dS ratio may not have accumulated sufficient variant data given the limited ESP data. We expect that the dN/dS ratio will be increasingly accurate with further exome data. Homozygotes in ESP may also represent outliers. Also, some known disease-associated genes might be missed by dN/dS prioritization, possibly because they have regions that are depleted for synonymous variants, as well as regions that are enriched for synonymous variants (for example, MECP2 in Fig. 4). This variation within a gene may dilute the overall consequence of dN/dS ratio as a whole gene. For large genes, further analysis of the dN/dS ratio in gene regions may be of further interest.
The intrahuman dN/dS ratio can be further applied as more variation data emerge. Our results provide evidence for the utility of intrahuman dN/dS, although the limits need further research (12). Future analysis of genomic data might identify other genes with dN/dS ratios suggestive of disease. Analysis of dN/dS is not limited to X-chromosome. It can be done for any genes of interest using our online tool link. Genes with suggestive dN/dS ratios would also be strong candidates for severe diseases that significantly affect reproductive fitness, and these are likely to be priority genes for detection in early disease screening and prognostication.
MATERIALS AND METHODS
Variant data
Exome variant data were obtained from the NHLBI GO ESP (1) [The Exome Variant Server (ESP6500SI-V2) http://eversusgs.washington.edu/EVS/; accessed on 22 March 2013]. This dataset includes results from 6503 exomes from 15 different NHLBI cohorts targeting primarily adults. Predicted variant effects were obtained from ESP using GVS Function. Effects included non-synonymous and synonymous variants and other exonic categories. Putative LoF variants were defined as: frameshift, stop gain/loss or involving 5′ or 3′ splice sites. We defined these as putative LoF because they may need validation by experiments. As there may be multiple transcripts for the same gene, we calculated the number of variants in ESP data for the canonical transcript. Canonical transcripts were retrieved from the UCSC Table Browser (http://genome.ucsc.edu/cgi-bin/hgTables). Variant data included 2203 African-American and 4300 European-American unrelated individuals' samples that were sequenced (1). The individuals had a variety of cardiac and pulmonary conditions including cardiovascular disease, chronic obstructive pulmonary disease, pulmonary hypertension, myocardial infarction, or they were postmenopausal women. These sample collections include adults with many of ages of at least 50 and 60 years of age, while some collections included down to early-aged adults. The cystic fibrosis collection was the only one that included children. Individual phenotypes are not released from the ESP database. All bulk variant data from ESP were used for analyses. Chromosome positions are based on Genome Reference Consortium Human Build 37 (GRCh37/hg19).
Substitution analyses
The intrahuman dN/dS ratio was calculated by evaluating the number of non-synonymous (N) and synonymous (S) variants for each gene on the X-chromosome using all the data from EVS and accounting for the theoretical potential number of non-synonymous and synonymous sites based on the gene sequence. dN is the ratio of the number of non-synonymous (N) variants divided by the number of total non-synonymous sites (N sites). dS is the ratio of the number of synonymous (S) variants divided by the number of total synonymous sites (S sites). The distribution of dN and dS were compared for all genes, and then dN/dS ratio was calculated for each gene and further examined.
The number of total putative N sites and S sites for each gene were calculated by DnaSP (14) using the Nei and Gojobori method (7). Briefly, all nucleotides in the coding region of each gene were examined to test whether the nucleotide changes would result in synonymous substitutions (S sites) or non-synonymous substitutions (N sites). The genetic code table shows that all substitutions at the second nucleotide positions of codons are non-synonymous, while a fraction of the nucleotide changes at the first and third positions are synonymous. To calculate the total putative synonymous sites, we assume equal nucleotide frequency and random substitution. Thus, we can calculate the number of synonymous sites (S sites) and the number of non-synonymous sites (N sites) for each codon. For example, in the case of codon TTA (Leu), N sites are 7/3, and S sites are 2/3. The total putative N/S sites for a given gene are the sum of the putative N/S sites for all the codons in the gene.
OMIM disease entries were obtained from the OMIM database using MIM Number Prefix # (phenotype description and molecular basis known). The gene list was extracted from Gene Links associated with the disease phenotypes. Disease onset or death was organized into three categories: childhood, adulthood and variable based on disease onset/death data from Orphanet (15) (http://www.orphadata.org). For each gene, its corresponding disease was examined for whether it typically caused childhood death, adult death, or did not cause death or led to death variably.
To assess genes that were deleted by structural variation in control samples, we analyzed the DGV for all OMIM genes on X chromosome. This database represents structural variation identified in healthy control samples (http://dgvbeta.tcag.ca/dgv/app/home) (47).
Pathway analysis was performed using WebGestalt (48) (http://bioinfo.vanderbilt.edu/webgestalt) using all genes on X chromosome as the reference gene set. All OMIM disease genes were used as the gene list for analysis. KEGG pathways were used for enrichment analysis, parameters included: hypergeometric statistic with multiple test adjustment using the Benjamini and Hochberg method.
CADD scores (16), combined network scores (19), phyloP scores (17) and phastCons scores (18) were retrieved using dbNSFP v2.4 (54,55). Logistic regression and ROC curves were made by R (http://www.r-project.org/). For comparing individual parameters used to predict OMIM disease genes, genetic_degree, signaling_degree, num_networks and num_interfaces are not shown due to insufficient data on X-chromosome. Histograms and plots were generated using R graphics (http://www.r-project.org) and ggplot2 package for R (http://ggplot2.org/). Protein domains were retrieved from the Superfamily database and the SMART database in Ensembl.
Statistical analysis
Statistical tests were performed using R (http://www.r-project.org). For dN/dS ratio comparisons between two categories, P-values were calculated by the two-sample Wilcoxon test (two-sided). For dN/dS ratio comparisons between three categories, P-values were calculated by the Kruskal–Wallis Rank Sum test. To compare variant numbers on X-chromosome with autosomal chromosomes, P-values were calculated by the Wilcoxon signed ranked test (two-sided). We used the χ2 test to identify variants that violate Hardy–Weinberg Equilibrium (P-value <0.05).
ADDITIONAL RESOURCES
Shieh lab tool is available at http://humangenetics.ucsf.edu/sequencing-tool/. It can be used to calculate the dN/dS ratio for any protein-coding gene or any regions within the gene of interest. The calculation is based on the Nei and Gojobori method (7).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grant funding from the US National Institutes of Health (grant no. HL092970 to J.S.).
Supplementary Material
Acknowledgements
The authors thank Yosr Bouhlal, Kevin Dumas and Joanna Phillips for helpful discussion and comments. The authors thank Yi Cheng and Karen Shuster for their help with the tool website. The authors thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for data analyses: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Tennessen J.A., Bigham A.W., O'Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue Y., Chen Y., Ayub Q., Huang N., Ball E.V., Mort M., Phillips A.D., Shaw K., Stenson P.D., Cooper D.N., et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am. J. Hum. Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M.X., Kwan J.S., Bao S.Y., Yang W., Ho S.L., Song Y.Q., Sham P.C. Predicting Mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2013;9:e1003143. doi: 10.1371/journal.pgen.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper G.M., Goode D.L., Ng S.B., Sidow A., Bamshad M.J., Shendure J., Nickerson D.A. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods. 2010;7:250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson P.N., Kohler S., Oellrich A., Wang K., Mungall C.J., Lewis S.E., Washington N., Bauer S., Seelow D., et al. Sanger Mouse Genetics Project. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 8.Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 9.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgi B., Voight B.F., Bucan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piton A., Redin C., Mandel J.L. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am. J. Hum. Genet. 2013;93:368–383. doi: 10.1016/j.ajhg.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryazhimskiy S., Plotkin J.B. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z., Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 15.Rath A., Olry A., Dhombres F., Brandt M.M., Urbero B., Ayme S. Representation of rare diseases in health information systems: the orphanet approach to serve a wide range of end users. Hum. Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 16.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khurana E., Fu Y., Chen J., Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput. Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M., Masshofer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B., et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miskinyte S., Butler M.G., Herve D., Sarret C., Nicolino M., Petralia J.D., Bergametti F., Arnould M., Pham V.N., Gore A.V., et al. Loss of BRCC3 deubiquitinating enzyme leads to abnormal angiogenesis and is associated with syndromic moyamoya. Am. J. Hum. Genet. 2011;88:718–728. doi: 10.1016/j.ajhg.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Z.Q., Du Bois G.C., Song S.P., Kulikovskaya I., Virgilio L., Rothstein J.L., Croce C.M., Weber I.T., Harrison R.W. Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies. Proc. Natl. Acad. Sci. USA. 1998;95:3413–3418. doi: 10.1073/pnas.95.7.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uechi T., Tanaka T., Kenmochi N. A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- 24.Shih J.W., Wang W.T., Tsai T.Y., Kuo C.Y., Li H.K., Wu Lee Y.H. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 2012;441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 25.Lee C.S., Dias A.P., Jedrychowski M., Patel A.H., Hsu J.L., Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borck G., Shin B.S., Stiller B., Mimouni-Bloch A., Thiele H., Kim J.R., Thakur M., Skinner C., Aschenbach L., Smirin-Yosef P., et al. eIF2gamma mutation that disrupts eIF2 complex integrity links intellectual disability to impaired translation initiation. Mol. Cell. 2012;48:641–646. doi: 10.1016/j.molcel.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemeth M.J., Kirby M.R., Bodine D.M. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc. Natl. Acad. Sci. USA. 2006;103:13783–13788. doi: 10.1073/pnas.0604006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao J.M., Li S.Q., Zhang H.W., Shi D.L. High mobility group B proteins regulate mesoderm formation and dorsoventral patterning during zebrafish and Xenopus early development. Mech. Dev. 2012;129:263–274. doi: 10.1016/j.mod.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 29.O'Donohue M.F., Choesmel V., Faubladier M., Fichant G., Gleizes P.E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidaka K., Caffrey J.J., Hua L., Zhang T., Falck J.R., Nickel G.C., Carrel L., Barnes L.D., Shears S.B. An adjacent pair of human NUDT genes on chromosome X are preferentially expressed in testis and encode two new isoforms of diphosphoinositol polyphosphate phosphohydrolase. J. Biol. Chem. 2002;277:32730–32738. doi: 10.1074/jbc.M205476200. [DOI] [PubMed] [Google Scholar]
- 31.Oberg E.A., Nifoussi S.K., Gingras A.C., Strack S. Selective proteasomal degradation of the B'beta subunit of protein phosphatase 2A by the E3 ubiquitin ligase adaptor Kelch-like 15. J. Biol. Chem. 2012;287:43378–43389. doi: 10.1074/jbc.M112.420281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Listerman I., Sun J., Gazzaniga F.S., Lukas J.L., Blackburn E.H. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013;73:2817–2828. doi: 10.1158/0008-5472.CAN-12-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F., Su K., Yang X., Bowe D.B., Paterson A.J., Kudlow J.E. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 34.Bektas M., Rubenstein D.S. The role of intracellular protein O-glycosylation in cell adhesion and disease. J. Biomed. Res. 2011;25:227–236. doi: 10.1016/S1674-8301(11)60031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubner C.A., Orth U., Senning A., Steglich C., Kohlschutter A., Korinthenberg R., Gal A. Seventeen novel PLP1 mutations in patients with Pelizaeus-Merzbacher disease. Hum. Mutat. 2005;25:321–322. doi: 10.1002/humu.9314. [DOI] [PubMed] [Google Scholar]
- 36.Torisu H., Iwaki A., Takeshita K., Hiwatashi A., Sanefuji M., Fukumaki Y., Hara T. Clinical and genetic characterization of a 2-year-old boy with complete PLP1 deletion. Brain Dev. 2012;34:852–856. doi: 10.1016/j.braindev.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Veeramah K.R., Johnstone L., Karafet T.M., Wolf D., Sprissler R., Salogiannis J., Barth-Maron A., Greenberg M.E., Stuhlmann T., Weinert S., et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia. 2013;54:1270–1281. doi: 10.1111/epi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatibana M., Kita K., Taira M., Ishijima S., Sonoda T., Ishizuka T., Iizasa T., Ahmad I. Mammalian phosphoribosyl-pyrophosphate synthetase. Adv. Enzyme Regul. 1995;35:229–249. doi: 10.1016/0065-2571(94)00017-w. [DOI] [PubMed] [Google Scholar]
- 39.Deardorff M.A., Kaur M., Yaeger D., Rampuria A., Korolev S., Pie J., Gil-Rodriguez C., Arnedo M., Loeys B., Kline A.D., et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rope A.F., Wang K., Evjenth R., Xing J., Johnston J.J., Swensen J.J., Johnson W.E., Moore B., Huff C.D., Bird L.M., et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirata H., Nanda I., van Riesen A., McMichael G., Hu H., Hambrock M., Papon M.A., Fischer U., Marouillat S., Ding C., et al. ZC4H2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability through impairment of central and peripheral synaptic plasticity. Am. J. Hum. Genet. 2013;92:681–695. doi: 10.1016/j.ajhg.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lubs H., Abidi F.E., Echeverri R., Holloway L., Meindl A., Stevenson R.E., Schwartz C.E. Golabi-Ito-Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J. Med. Genet. 2006;43:e30. doi: 10.1136/jmg.2005.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Garay I., Tomas M., Oltra S., Ramser J., Molto M.D., Prieto F., Meindl A., Kutsche K., Martinez F. A two base pair deletion in the PQBP1 gene is associated with microphthalmia, microcephaly, and mental retardation. Eur. J. Hum. Genet. 2007;15:29–34. doi: 10.1038/sj.ejhg.5201717. [DOI] [PubMed] [Google Scholar]
- 44.Rejeb I., Ben Jemaa L., Abaied L., Kraoua L., Saillour Y., Maazoul F., Chelly J., Chaabouni H. A novel frame shift mutation in the PQBP1 gene identified in a Tunisian family with X-linked mental retardation. Eur. J. Med. Genet. 2011;54:241–246. doi: 10.1016/j.ejmg.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Jensen L.R., Chen W., Moser B., Lipkowitz B., Schroeder C., Musante L., Tzschach A., Kalscheuer V.M., Meloni I., Raynaud M., et al. Hybridisation-based resequencing of 17 X-linked intellectual disability genes in 135 patients reveals novel mutations in ATRX, SLC6A8 and PQBP1. Eur. J. Hum. Genet. 2011;19:717–720. doi: 10.1038/ejhg.2010.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maquat L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 47.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 48.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Argentaro A., Yang J.C., Chapman L., Kowalczyk M.S., Gibbons R.J., Higgs D.R., Neuhaus D., Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl. Acad. Sci. USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu H.C., Sloan J.L., Scharer G., Brebner A., Quintana A.M., Achilly N.P., Manoli I., Coughlin C.R., II, Geiger E.A., Schneck U., et al. An X-linked cobalamin disorder caused by mutations in transcriptional coregulator HCFC1. Am. J. Hum. Genet. 2013;93:506–514. doi: 10.1016/j.ajhg.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.