Abstract
Facioscapulohumeral muscular dystrophy (FSHD: MIM#158900) is a common myopathy with marked but largely unexplained clinical inter- and intra-familial variability. It is caused by contractions of the D4Z4 repeat array on chromosome 4 to 1–10 units (FSHD1), or by mutations in the D4Z4-binding chromatin modifier SMCHD1 (FSHD2). Both situations lead to a partial opening of the D4Z4 chromatin structure and transcription of D4Z4-encoded polyadenylated DUX4 mRNA in muscle. We measured D4Z4 CpG methylation in control, FSHD1 and FSHD2 individuals and found a significant correlation with the D4Z4 repeat array size. After correction for repeat array size, we show that the variability in clinical severity in FSHD1 and FSHD2 individuals is dependent on individual differences in susceptibility to D4Z4 hypomethylation. In FSHD1, for individuals with D4Z4 repeat arrays of 1–6 units, the clinical severity mainly depends on the size of the D4Z4 repeat. However, in individuals with arrays of 7–10 units, the clinical severity also depends on other factors that regulate D4Z4 methylation because affected individuals, but not non-penetrant mutation carriers, have a greater reduction of D4Z4 CpG methylation than can be expected based on the size of the pathogenic D4Z4 repeat array. In FSHD2, this epigenetic susceptibility depends on the nature of the SMCHD1 mutation in combination with D4Z4 repeat array size with dominant negative mutations being more deleterious than haploinsufficiency mutations. Our study thus identifies an epigenetic basis for the striking variability in onset and disease progression that is considered a clinical hallmark of FSHD.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD: MIM#158900) progressively and often asymmetrically affects the facial and upper extremity muscles. Recently, the prevalence of FSHD has been estimated at 12;100 000 in the Dutch population (1). There exists considerable inter- and intra-familial clinical variability with ∼20% of clinically affected individuals becoming wheelchair dependent whereas a similar proportion of gene carriers remains asymptomatic (2,3). Two genetically distinct FSHD forms have been identified, FSHD1 and FSHD2, where FSHD1 is estimated to be ∼50× more common than FSHD2.
The common form FSHD1 is caused by contraction of the D4Z4 repeat array on chromosome 4q, with each D4Z4 unit being 3.3 kb in size. For genetic testing, the size of the D4Z4 repeat array is determined by Southern blot analysis of EcoRI-digested genomic DNA using probe p13E−11 (4). The polymorphic D4Z4 repeat array varies between 11 and 100 units (43–340 kb EcoRI fragment) in the population, whereas FSHD1 individuals have one array of 1–10 units (10–40 kb; the EcoRI restriction sites are located 5.7 kb proximal and 1.1 kb distal to the distal D4Z4 unit hence a 10.1 kb EcoRI fragment for the shortest FSHD allele) (4,5). There is a rough and inverse correlation between residual repeat size and disease severity with carriers of a 1–6 D4Z4 unit repeat array being average to severely affected (6), whereas in familial carriers of a 7–10 unit allele, clinical variability and non-penetrance are much more prominent (7–10). The D4Z4 repeat array contraction leads to a less repressive local chromatin structure, marked by CpG hypomethylation in the promoter region of the DUX4 retrogene embedded in the D4Z4 unit, and a greater probability of aberrant DUX4 expression in skeletal muscle (11–13). D4Z4 chromatin relaxation only results in stable DUX4 expression when the D4Z4 repeat array contraction occurs in cis with a polymorphic DUX4 polyadenylation signal (PAS) present on a FSHD-permissive chromosomal background (4A). A similar D4Z4 repeat array is located on the equally common chromosome 4B variant and on chromosome 10, but contractions of the array on these locations typically do not result in stable DUX4 expression and disease owing to the absence of the DUX4-PAS (Fig. 1A) (14–19). DUX4 is a transcription factor normally expressed in the luminal cells of the testis, and its expression in muscle activates germline and early stem cell programs eventually resulting in muscle cell death (12,20).
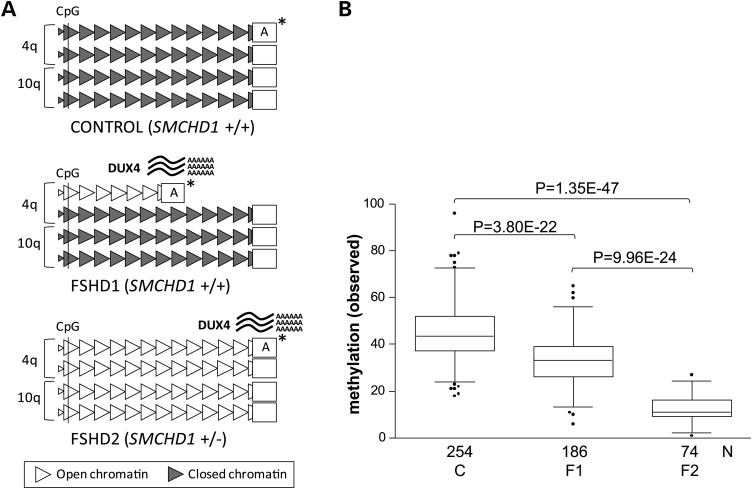
Figure 1.
Overview of the genetic and epigenetic status at D4Z4 in control and FSHD. (A) Representation of the genetic and epigenetic status at the D4Z4 repeat arrays on chromosomes 4q and 10q in the control, FSHD1 and FSHD2 individuals. Normal-sized D4Z4 arrays (10–100 units) are depicted as 12 D4Z4 units (triangles), and the contracted array (1–10 units) in FSHD1 is depicted as 6 units. Gray-colored D4Z4 units represent a repressive chromatin state, which is partially lost at the contracted array in FSHD1 and at all four arrays in FSHD2 (white colored units). The chromatin relaxation is marked by a loss of CpG methylation at D4Z4, which is measured simultaneously on chromosomes 4 and 10 in the proximal D4Z4 unit (marked by CpG). The chromatin relaxation is associated with derepression of the DUX4 gene and stable DUX4 transcription from the permissive 4qA chromosomes (marked by asterisk). (B) Box plot showing the cumulative D4Z4 methylation level on chromosomes 4q and 10q D4Z4 arrays in 254 controls (C), 186 FSHD1 individuals (F1) and 74 carriers of an SMCHD1 mutation (F2) with significant differences between all groups (p values are indicated). The reduction in D4Z4 methylation in FSHD1 is caused by the presence of minimally one contracted repeat array. The box extends from the 25th to 75th percentiles, and the line in the middle of the box is plotted at the median, the whiskers are at the 2.5th and 97.5th percentiles.
FSHD2, the uncommon form of FSHD, is caused by heterozygous mutations in the structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) gene (21,22). The SMCHD1 gene on chromosome 18p consists of 48 exons and encodes for a protein containing a putative ATPase and hinge domain. SMCHD1 is a member of the conserved family of SMC proteins involved in chromatin repression. In mice, Smchd1 has been shown to be involved in the establishment and maintenance of DNA methylation of a subset of CpG islands on the inactive X chromosome (Xi), of repetitive sequences, and of monoallelically expressed autosomal genes (21). SMCHD1 binds to the D4Z4 repeat array in somatic cells, and reduced SMCHD1 binding to the D4Z4 repeat array has been reported in individuals with FSHD2. In SMCHD1 mutation carriers, all D4Z4 repeat arrays from chromosomes 4 and 10 are hypomethylated (Fig. 1A and B) (22,23). Together, these data are consistent with a role for SMCHD1 keeping D4Z4 and DUX4 in a repressive chromatin structure in somatic tissue. Although the reduced D4Z4 methylation in FSHD2 individuals was found in peripheral blood mononuclear cells (PBMCs), fibroblasts and myoblasts, the expression of DUX4 was, as in FSHD1, only observed in skeletal muscle biopsies and in differentiated myoblasts (12). SMCHD1 can also act as a modifier in FSHD1, as individuals with both an FSHD1 allele and an SMCHD1 mutation were found to be more severely affected than relatives with only one of the two pathogenic lesions (24).
The partial loss of D4Z4 methylation in FSHD1 and FSHD2 has been demonstrated by Southern blot analysis using several methylation-sensitive restriction enzymes and, more recently, by bisulfite sequencing and methylated DNA immunoprecipitation (MeDIP) analysis at D4Z4 (11,25,26). These studies have shown that the different approaches revealed similar patterns of D4Z4 methylation, where D4Z4 hypomethylation in FSHD is universal across muscle, fibroblasts and PBMCs (11,22,26). One of the most commonly used methylation-sensitive restriction site to measure D4Z4 methylation is the FseI site as it is highly predictive to FSHD (13). The FseI site is located approximately 150 bp upstream of the DUX4 transcriptional start sites in every D4Z4 unit but is most often studied in the most proximal D4Z4 unit (14,15,27). Previously, we showed that D4Z4 methylation at this site is lower in shorter D4Z4 repeat arrays compared with longer arrays (28). Comparable correlations between repeat size and methylation were found when studying other methylation-sensitive restriction sites in the proximal D4Z4 unit (11). Hypomethylation of the proximal D4Z4 unit has also been shown to be representative for the entire array (11) and highly informative as it was instrumental in the identification of the FSHD2 gene defect (22). Moreover, reduced D4Z4 methylation in FSHD coincides with changes of several other chromatin modifications at D4Z4 (29,30), such as the chromatin compaction score (the ratio between the histone 3 modifications H3K9me3 and H3K4me2 at D4Z4), which differs significantly between controls and FSHD2, and between controls and FSHD1 (29). For all of these reasons, and because D4Z4 methylation analysis by FseI digestion can be done on large cohorts of individuals, in contrast to the more elaborate alternatives, we choose to study the CpG methylation at D4Z4 in primary PBMCs as a sign of reduced chromatin compaction.
In this study, we established the SMCHD1 mutation spectrum in a large cohort of FSHD2 individuals. We also analyzed the correlation between the degrees of D4Z4 CpG methylation in PBMCS from >500 individuals relative to D4Z4 repeat array size. We developed a D4Z4 repeat size-corrected methylation score and found a significant correlation between D4Z4 methylation at the FseI site and the clinical severity in both FSHD1 and FSHD2.
RESULTS
Identification of SMCHD1 mutations in FSHD2
We identified 60 families with one or more individuals with FSHD2 (Supplementary Material, Fig. S1), of which 41 have not been analyzed for SMCHD1 mutations previously (22). Affected individuals from these families: (i) have a phenotype consistent with FSHD, (ii) carry at least one permissive 4qA chromosome for the DUX4 mRNA, (iii) have >10 D4Z4 units on the FSHD-permissive 4qA allele and (iv) have a combined CpG methylation level on chromosomes 4 and 10 D4Z4 that is below the previously defined threshold of 25% for FSHD2 (22). The cohort of 60 families consisted of 16 familial cases (9 paternal and 7 maternal transmissions), 6 cases in which both parents show normal D4Z4 methylation levels and 38 cases where the inheritance could not be determined owing to limited availability of biological samples from other family members. Pedigrees of the 8 largest FSHD2 families are shown (Supplementary Material, Fig. S1). In 51 of the 60 families, we have identified an SMCHD1 mutation, of which 45 mutations were unique (Fig. 2 and Supplementary Material, Table S1). In total, we identified 83 carriers of an SMCHD1 mutation with an average D4Z4 methylation of 12.1% (SD 5.5%) and 45 unaffected relatives without mutation and with an average D4Z4 methylation of 46.8% (SD 14.1%). These values are consistent with those previously reported in FSHD2 individuals and in the control population, respectively (22). A mutation hotspot was identified at the 3′ splice site of exon 25, containing three different partially overlapping splice-site mutations in six unrelated families (Supplementary Material, Fig. S2). We identified 9 families (15%) with a total of 17 affected individuals for which we could not find a mutation in SMCHD1 despite an average D4Z4 DNA methylation of 16.5% (SD 3.5%), and we excluded the presence of exonic deletions in them.
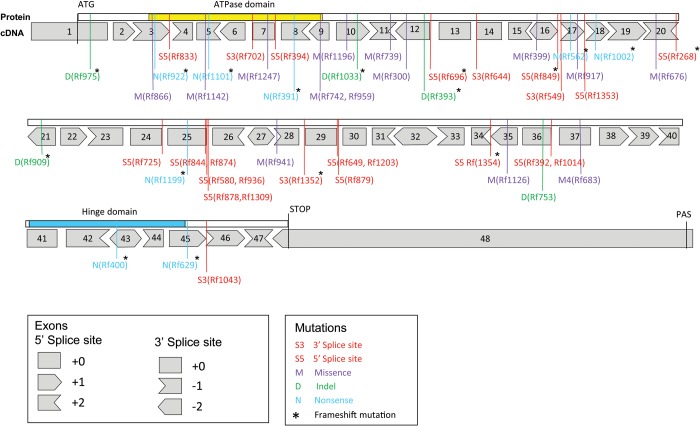
Figure 2.
SMCHD1 mutation spectrum in FSHD2. In 51 families carrying an SMCHD1 mutation, we identified 5 heterozygous insertion–deletion (indel) mutations (10%), 8 heterozygous nonsense mutations (15.5%), 14 heterozygous missense mutations (27.5%) and 24 heterozygous splice-site mutations (47%). Forty-five of the 51 SMCHD1 mutations were unique. In families Rf742 (from South of France) and Rf959 (from North of Spain), we identified the same missense mutation in exon 9. An identical splice-site mutation in exon 36 was found in two Dutch families (Rf392 and Rf1014). We found identical splice-site mutations in intron 29 in families from South Korea (Rf1203) and from the USA (Rf649). The highest incidence of shared mutations was found at the 5′ splice-site of intron 25, where three overlapping deletion mutations are identified in 6 families suggestive for a mutation hotspot (Supplementary Material, Fig. S2). All 48 SMCHD1 exons are indicated with boxes. The position of the 5′ and 3′ splice sites with respect to the ORF is also indicated and illustrated in the box below. The localization of the ATPase domain and the Hinge domain is indicated above the exon boxes. For all mutations, the nature of the mutation is indicated (M, D, N, S3 or S5, see box below) as well as the family number. An asterisk marks the mutations that disrupt the ORF.
Analysis of the SMCHD1 mutations
We identified 5 small heterozygous insertion–deletion (indel) mutations (10%), 8 heterozygous nonsense mutations (15.5%), 14 heterozygous missense mutations (27.5%) and 24 heterozygous splice-site mutations (47%) (Fig. 2). Transcription analysis showed that all nonsense and indel mutations and the splice-site mutations that were predicted to disrupt the ORF showed significant reduced levels of the mutant transcript in comparison with the wild-type transcript (Supplementary Material, Fig. S3). This is suggestive of an SMCHD1 haploinsufficiency mechanism for these types of mutations. For all tested missense mutations and those splice-site mutations that showed retention of the ORF, the mutant transcript was transcribed at similar levels to the wild-type allele, consistent with dominant negative mutations (Supplementary Material, Fig. S3). Based on these findings, SMCHD1 mutations were grouped into putative haploinsufficiency (disrupting the ORF: D-ORF) or dominant negative (preserving the ORF: P-ORF) mutations. The differences in transcript levels between D-ORF and P-ORF mutations were independent of the tissue type as they were found in PBMCs, fibroblasts and myoblasts and in some cases in different tissues obtained from the same individual (Supplementary Material, Fig. S3 and Table S1).
Correlation between D4Z4 repeat array size and methylation
We observed highly variable D4Z4 methylation values between individuals carrying different SMCHD1 mutations and hypothesized a possible correlation between the nature of the mutation and the methylation level (Fig. 1B). Because D4Z4 methylation is also repeat size dependent (11,28), we first analyzed the relationship between the combined D4Z4 methylation level on chromosomes 4 and 10 and the total number of D4Z4 units on these chromosomes in 254 controls and 186 FSHD1 individuals and 74 carriers of an SMCHD1 mutation (Supplementary Material, Table S2). This analysis showed a significant correlation (P < 0.001) between the cumulative logarithmic values of all four D4Z4 arrays and the methylation level for all three conditions (Fig. 3A). The FseI methylation levels in this study are the derivative of the D4Z4 methylation levels at all four individual repeats. Therefore, it is to be expected that in FSHD1 individuals with similarly sized pathogenic repeat arrays, those with longer arrays on the remaining three chromosomes have higher FseI methylation levels than those with shorter alleles on the other chromosomes. Subset analysis of our FSHD1 individuals indeed demonstrates the expected differences in FseI methylation indicating that the methylation at the individual repeats is independent from the other repeats (Supplementary Material, Fig. S4).
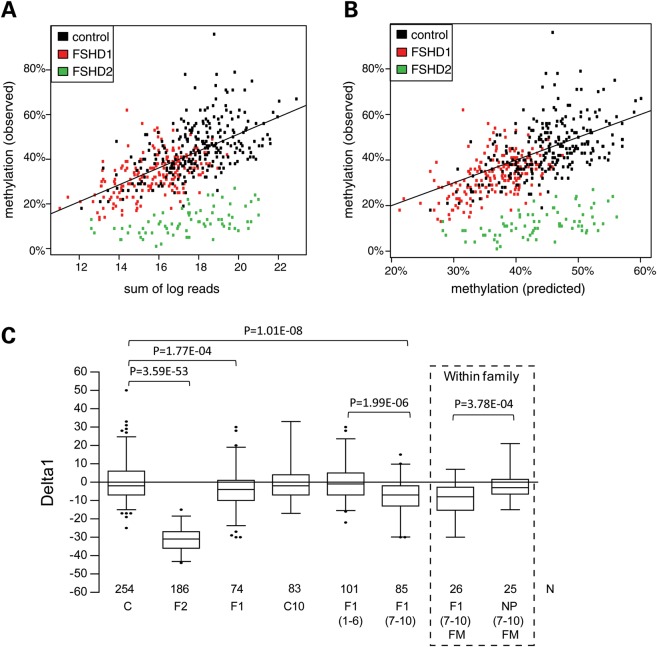
Figure 3.
Methylation and repeat array size correlation. (A) Scatterplot showing correlation between the combined D4Z4 methylation level on chromosomes 4 and 10 (OM) and the cumulative repeat array sizes (sum of log reads) for all samples examined in Figure 1B (controls in black; FSHD1 individuals in red and SMCHD1 mutation carriers in green). A logarithmic transformation of the repeat array sizes (units) clearly shows a significant correlation (P < 0.001) between the cumulative log2 values of the four D4Z4 repeat arrays and the methylation level for all three conditions. (B) Scatterplot comparison between OM at D4Z4 and PM from individuals from Figure 3A. The high correlation between both values in controls and FSHD1 individuals shows that it is possible to predict the D4Z4 methylation levels based on the combined size of the four D4Z4 repeat arrays. The OM in SMCHD1 mutation carriers is highly reduced but still correlates with the PM. (C) Boxplot showing the Delta1 for the different conditions. The average Delta1 in controls (C) is almost 0, whereas this value for SMCHD1 mutation carriers (F2) is −31.3% (P = 3.59E − 53). The average Delta1 in FSHD1 individuals (F1) is significantly (P = 1.77E − 04) reduced by 3.6%. The reduction in the average Delta1 was not significant in a subset of the control group (C10) that were selected on the basis of a contracted repeat array on chromosome 10. Further partition of the FSHD1 group based on repeat size showed that the reduced average Delta1 is specific for carriers of a 7–10 unit repeat array [F1(7–10), Delta1 −7.6%, P = 1.01E − 08), whereas the average Delta1 in carriers of a 1–6 unit repeat is almost 0 [F1(1–6)] (P = 0.96). From 26 affected carriers [F1(7–10)FM] of the 7–10 unit FSHD group, we identified 25 family members [NP(7–10)FM] that were non-penetrant carriers of the same FSHD allele (boxed). While the F1(7–10)FM group has a significantly reduced average Delta1 (−9.5%, P = 3.78E − 04) compared with their unaffected family members in group [NP(7–10)FM], the average Delta1 of the latter group was not different from controls. Sequence analysis for all individuals from the F1(7–10) group with Delta1 values below −14 failed to identify a mutation in SMCHD1 (Supplementary Material, Table S2). P-values are shown for groups that show a significant Delta1 reduction compared with the controls (C) and for the most relevant intergroup comparisons. The box extends from the 25th to 75th percentiles, and the whiskers are at the 2.5th and 97.5th percentiles.
The log transformation was based on exploratory data analysis of earlier data (11), which suggested a better fit for the log-transformed repeats than for the linear repeats. Biologically, a logarithmic effect of repeats makes sense, because the effect on methylation of going from 5 to 6 repeats is expected to be much stronger than going from 37 to 38 repeats. Indeed, applying the log transformation of the methylation data from this study showed a better fit than without log transformation. A fitted model was then designed based on controls that allowed us to calculate the predicted methylation (PM) level for any individual based on their total number of D4Z4 units (Fig. 3B and Materials and Methods). While, in general, males have been shown to be more severely affected than females (10), we did not find a significant gender effect on the D4Z4 methylation. The model enabled us to establish a Delta1 score, that is, the experimentally observed methylation (OM) level minus the PM level in controls. Delta1 is, by definition, close to zero in situations where the methylation level is only determined by repeat array size [see controls in Fig. 3C: Delta1 −0.2% (±10.0 SD)]. In SMCHD1 mutation carriers, the Delta1 is highly negative (−31.3%, P = 3.59E − 53), suggesting a strong contribution of the SMCHD1 mutation to D4Z4 hypomethylation (Fig. 3C).
Reduced Delta1 in FSHD1 individuals carrying 7- to 10-unit FSHD alleles
Analogous to controls, we expected the average Delta1 in FSHD1 individuals to be close to zero, as we anticipated that the repeat size of the FSHD allele was the only factor causing the reduction in D4Z4 methylation at chromosomes 4 and 10 (Fig. 1B). However, we observed an average Delta1 of −3.6% in FSHD1 individuals, which is significantly reduced compared with controls (P = 1.77E−04). To exclude that the reduction of Delta1 in FSHD1 individuals results from a selection bias having selected for individuals carrying on average 1 more short repeat array than control individuals, we calculated the Delta1 value in 83 control individuals carrying at least 1 short repeat array on chromosome 10 (designated C10 in Fig. 3C) and found an average Delta1 of −1.3% (±8.7% SD), which is not significantly different from the value found in all controls.
The reduced Delta1 in FSHD1 shows that, for some FSHD1 individuals, D4Z4 methylation is more reduced than might be expected based on the sizes of the D4Z4 array. This suggests that methylation reducing factors are at hand other than hypomethylation as a consequence of D4Z4 repeat array contraction. We anticipated that the biggest contributors to the reduced Delta1 in FSHD1 might be the individuals carrying an FSHD allele of 7–10 units because they show the greatest clinical variability. Therefore, we subdivided the FSHD1 group into 101 individuals with FSHD alleles of 1–6 units [F1(1–6)] and 85 individuals with alleles of 7–10 units [F1(7–10)] and confirmed that the reduced Delta1 was indeed restricted to the carriers with an array of 7–10 units (Fig. 3C and Supplementary Material, Fig. S5). These affected carriers of an array of 7–10 units had an average Delta1 of −7.6 (P = 1.01E−8) despite being excluded from carrying an SMCHD1 mutation. We did not find a significantly reduced Delta1 in FSHD1 individuals with a disease repeat array of 1–6 D4Z4 units. Finally, we also determined the average Delta1 in a group of 25 non-penetrant carriers of an 7–10 unit FSHD1 allele [NP(7–10)FM] that were family members of 26 affected individuals from the F1(7–10) cohort and observed an average Delta1 that did not differ from controls (Fig. 3C). In contrast, the difference between the average Delta1 in affected F1(7–10)FM and unaffected NP(7–10)FM family members in this group is highly significant (P = 3.78E − 04). These findings strongly suggest that FSHD1 individuals with an array of 7–10 units are epigenetically more susceptible to disease presentation than familial mutation carriers that remain unaffected.
D4z4 methylation in FSHD2 depends on repeat array size and nature of the SMCHD1 mutation
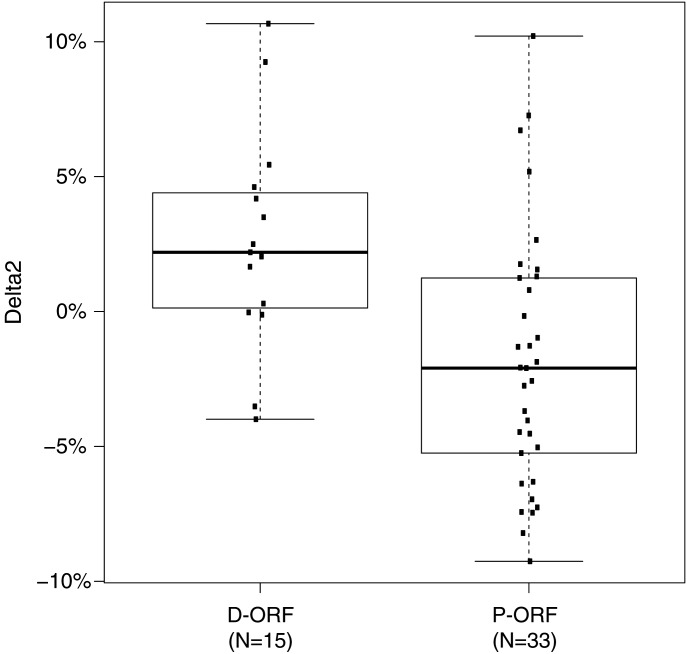
In SMCHD1 mutation carriers, we found a significant correlation between D4Z4 methylation and the sum of all D4Z4 units, but the D4Z4 unit number-dependent increase in D4Z4 CpG methylation is approximately half to that of controls (Fig. 3A). Because of the broad spectrum of SMCHD1 mutations in FSHD2 and the highly variable methylation values, we hypothesized a possible correlation between the nature of mutation and the methylation level. To accurately analyze the effect of SMCHD1 mutations on D4Z4 methylation, we fitted a second model to predict the methylation in only SMCHD1 mutation carriers (Fig. 3A and B and Materials and Methods). We defined the difference between OM and PM in this model as Delta2. This Delta2 was subsequently used to study possible correlations between the nature of mutation (D-ORF versus P-ORF) and the D4Z4 CpG methylation level. Notably, we found significantly (P = 0.014) lower Delta2 values for P-ORF mutations (mean −1.8) than for D-ORF mutations (mean 2.7), suggesting that SMCHD1 P-ORF mutations are more deleterious than D-ORF mutations (Fig. 4).
Figure 4.
Comparison of methylation between D-ORF and P-ORF SMCHD1 mutations. Boxplot showing a significantly (P = 0.014) lower Delta2 for P-ORF mutations than for D-ORF mutations. In total, we identified 33 different families with 42 carriers of a P-ORF mutation and 15 families with 32 carriers of a D-ORF mutation. For multiple carriers within one family, an average Delta2 was used. The box extends from the 25th to 75th percentiles, and the whiskers are at 1.5× interquartile range (IQR).
We also found that, for 33 P-ORF mutations, the position of the mutation within the SMCHD1 locus had a significant (P = 0.0014) impact on the level of D4Z4 CpG methylation with mutations positioned to the N-terminus of the protein having a greater effect on D4Z4 methylation than those toward the C-terminus of SMCHD1 (Supplementary Material, Fig. S6A). This effect was not seen for 15 D-ORF mutations. The N-terminus of SMCHD1 contains a predicted ATPase domain, and mutations in this domain seem more deleterious based on their very low Delta2 (Supplementary Material, Fig. S6A). Without the ATPase domain, however, the association remained significant (P = 0.0001) for P-ORF mutations, suggesting that other yet undefined domains or functionalities in SMCHD1 exist (Supplementary Material, Fig. S6B).
Correlation between D4Z4 methylation and disease severity
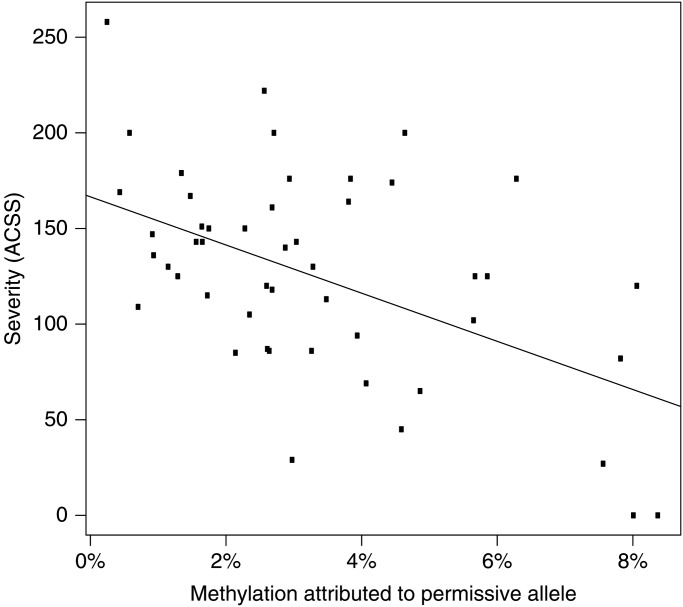
Because a rough correlation has been reported between residual pathogenic array repeat size, methylation and disease severity in FSHD1 individuals (6,11,28), we questioned whether D4Z4 methylation levels in FSHD2 individuals correlated with disease severity. We anticipated that the clinical severity in FSHD2 depends largely on the size of the permissive D4Z4 array, not the sum of all four arrays. This assumption was based on our previous finding that, in most FSHD2 individuals, the array size of the permissive allele is shorter than the average size in the control population (23), which we confirmed in this study (Supplementary Material, Fig. S7). We calculated the methylation contribution of the shortest permissive allele based on the OM and Delta2 (Materials and Methods). Using the age-corrected clinical severity scores (28) of 49 SMCHD1 mutation carriers, we observed a significant correlation (P = 0.0013) between the calculated methylation at the shortest permissive allele and severity (Fig. 5).
Figure 5.
Correlation between D4Z4 methylation and clinical severity in FSHD2. Graph showing a significant correlation of −52.7% (P = 0.0013) between the calculated methylation at the shortest permissive allele and the age-corrected clinical severity score (ACSS). Included are 49/74 carriers of an SMCHD1 mutation for which we obtained an ACSS and in which we identified the presence of an FSHD-permissive allele. Non-penetrant individuals younger than 25 years were excluded from this correlation.
DISCUSSION
We here report on evidence for epigenetic susceptibility to disease presentation in FSHD1 and FSHD2. In FSHD1, we showed that affected carriers of a disease array of 7–10 D4Z4 units, but not familial non-penetrant mutation carriers, have a greater reduction of D4Z4 CpG methylation than might be expected based on the size of the pathogenic D4Z4 repeat array. In FSHD2, this epigenetic susceptibility to disease presentation and severity depends on both the repeat array size and the nature of the SMCHD1 mutation.
We found a correlation between the sum of all D4Z4 units on chromosomes 4 and 10 and the D4Z4 CpG methylation level, demonstrating that for any given D4Z4 repeat array, its FseI methylation level is depending on the size of the array. Our analysis also confirmed that the methylation level of a D4Z4 array is independent from the sizes of the other arrays. This allowed us to introduce a new epigenetic parameter, the Delta1 score, which measures the difference between the expected D4Z4 CpG methylation based on the total number of D4Z4 units and the OM level at the FseI site in D4Z4. The highly reduced Delta1 score in SMCHD1 mutation carriers demonstrates the importance of SMCHD1 in maintaining D4Z4 methylation levels in somatic cells. Because Delta1 is independent from repeat array size, in contrast to the currently used measure of CpG methylation at D4Z4, the Delta1 score might serve as a more accurate diagnostic measure to identify FSHD2-based hypomethylation as it discriminates between D4Z4 hypomethylation owing to the presence of one or more contracted array (FSHD1) versus mutations in SMCHD1 (FSHD2), with the caveat that it requires complete D4Z4 repeat array size information.
Several studies have reported a rough and inverse correlation between D4Z4 repeat array size and clinical severity in FSHD1 families (6–10), yet this was mainly observed in affected individuals carrying a D4Z4 array of 1–6 units. In this study, we showed that a logarithmic conversion of the sum of all D4Z4 units from chromosomes 4 and 10 correlates with D4Z4 methylation at the FseI site. This finding suggests that the hypomethylation at the shortest FSHD alleles (1–6 units) is more profound than that in the 7–10 unit FSHD corroborating our earlier study that analyzed the methylation specifically at the FSHD allele (28). This probably reflects a chromatin organization of arrays in the size range of 1–6 D4Z4 units that is already sufficient to de-repress DUX4 and cause disease. This is supported by the average Delta1 value of 0 in the affected individuals with arrays of 1–6 units, like in control individuals. However, affected FSHD1 individuals carrying a disease array of 7–10 D4Z4 units, but not familial non-penetrant mutation carriers, have a greater degree of D4Z4 CpG hypomethylation than might be expected based on the sizes of the D4Z4 arrays on chromosomes 4q and 10q in these individuals. This suggests that this group of disease-presenting individuals have a greater epigenetic susceptibility to disease presentation than their non-penetrant relatives, although it is also possible that the disease process itself contributes to this difference. This latter possibility seems, however, unlikely because we did not measure a reduced Delta1 in individuals with arrays of 1–6 D4Z4 units. Interestingly, carriers of an upper range of FSHD allele sizes (7–10 units) show the highest clinical variability with ∼20% of them being asymptomatic (8). This D4Z4 repeat array size range has also been observed at a frequency of 1–2% in the Caucasian healthy control population (31–33), and we have noticed population differences in the distribution of the FSHD1 repeat sizes (34). Thus, it is possible that affected individuals in this D4Z4 repeat array size range may carry polymorphisms in chromatin modifiers of D4Z4, like SMCHD1, which determine the CpG methylation and chromatin structure at all four D4Z4 repeats. This corroborates on our previous observation that SMCHD1 mutations can aggravate the disease presentation and progression in FSHD1 families.
It is intriguing that methylation analysis at the single FseI site in D4Z4 yields results similar to more comprehensive methylation analyses such as bisulfite sequencing or MeDIP (25,26). None of the D4Z4 amplicons studied by the latter techniques overlap with the FseI site (Supplementary Material, Fig. S8). In a direct comparison of bisulfite sequencing with Southern blotting for another reported methylation-sensitive restriction site, BsaA1, a similar trend in the degree of hypomethylation was observed (13,26). Moreover, this direct comparison between PBMC DNA and myoblast DNA identified similar patterns of D4Z4 methylation between the two tissues (26). A recent study by Gaillard and colleagues also identifies D4Z4 hypomethylation more distal to the FseI site in PBMC DNA of FSHD individuals, but not within DUX4 itself (25). Interestingly, at the 5′end of DUX4, they also observe differences in DNA methylation between affected and non-affected members of FSHD1 families. The FseI site maps to the DUX4 promoter (15), and the presence of such predictive methylation-dependent site within the DUX4 promoter warrants further studies.
We analyzed 60 FSHD2 families for mutations in SMCHD1 that were selected by clinical assessment and D4Z4 DNA hypomethylation. In 51 families, we identified an SMCHD1 mutation. Therefore, in our cohort, 85% of FSHD2 families can be explained by mutations in SMCHD1, consistent with our previous report (22). The mutation spectrum is biased toward splice-site mutations (47%). Closer examination of the SMCHD1 mutation spectrum separated them into two classes: P-ORF mutations in which the SMCHD1 open reading frame is preserved and D-ORF mutations in which de open reading frame is disrupted. Further transcriptional analysis suggests that D-ORF mutations represent haploinsufficiency mutations, whereas the P-ORF are consistent with dominant negative mutations. To study the effect of different mutations on D4Z4 CpG methylation in FSHD2, the Delta2 score was introduced specific for SMCHD1 mutation carriers. The significant difference in the average Delta2 between D-ORF and P-ORF mutations strongly suggests that P-ORF mutations are more deleterious than D-ORF mutations because, according to the current disease model, DUX4 promoter methylation levels at permissive 4A alleles correlate with the transcriptional activity of DUX4. Importantly, our study suggests that the methylation level at the permissive allele and the disease severity is determined by a combination of the nature of the SMCHD1 mutation and the repeat array size of this allele: a permissive alleles carrying a smaller sized D4Z4 repeat array in combination with P-ORF mutations in general result in a greater disease severity than with D-ORF mutations, whereas for permissive alleles carrying a larger sized D4Z4 repeat array, a P-ORF SMCHD1 mutation is likely necessary to cause disease. As a consequence, the main cause for clinical variability between FSHD2 families is likely the nature of the mutation, whereas intrafamilial differences are mainly caused by the variation in repeat array size of the FSHD-permissive allele.
SMCHD1 is a member of the SMC protein family, and SMC proteins form functional dimers (35). Although SMCHD1 dimer formation has not yet been demonstrated, P-ORF mutations in SMCHD1 might result in malfunctioning dimers, possibly explaining our observations. Interestingly, we found a significant linear correlation between the position of the mutation and the degree of hypomethylation. This observation remains enigmatic because we have ruled out that this correlation is caused by mutations in the putative ATPase domain of SMCHD1. Future studies will be necessary to explain this observation although the strong correlation between mutation position and D4Z4 methylation may assist further studies of SMCHD1 structure and function.
Mutations in other SMC genes cause Cornelia de Lange syndrome (CdLS). Recently, a CdLS mutation screen identified 24 different mutations in SMC1A, all without interrupting the ORF (36). Although it was suggested that D-ORF mutations might not be tolerated or lead to a different phenotype, our results suggest that D-ORF SMC1A mutations probably result in haploinsufficiency, which might be less deleterious to dimer formation and result in a milder phenotype.
Our study goes beyond differentiating between dominant negative and haploinsufficiency mutations by providing evidence that the balance between D4Z4 repeat size and the activity of epigenetic modifiers such as SMCHD1 determines the likelihood of somatic DUX4 expression, and hence the probability of developing FSHD. With this concept of epigenetic susceptibility, we are uncovering the basis of the marked variability in disease onset and progression, a feature that has been considered a clinical hallmark of FSHD from its description in 1885 (37).
MATERIALS AND METHODS
Subjects
The 254 control individuals were collected via the Dutch Bloodbank in Leiden or selected from our FSHD families. The FSHD1 group consisted of 186 affected FSHD1 individuals, of which 101 individuals from 74 different families carried a 1- to 6-unit FSHD allele and 85 individuals from 62 families carried 7–10 unit FSHD allele. Within 13 families of the latter group, we identified 25 non-penetrant carriers of a 7–10 unit FSHD allele, all older than 26 years and showing a similar age distribution to their 26 affected family members (Supplementary Material, Fig. S9). The FSHD2 cohort consists of 60 independent families with 1 or more individuals who were diagnosed with FSHD2 based on previously established clinical, genetic and epigenetic criteria (22). These families were recruited from the United States (19), Netherlands (10), France (6), Italy (1), United Kingdom (6), Spain (4), Denmark (3), Germany (3), Canada (2), Bulgaria (1), Finland (1), Slovenia (1), Switzerland (2) and South Korea (1). Clinical assessment was performed with a standardized clinical form available at the website of the Fields Center for FSHD Research (www.urmc.rochester.edu/fields-center/) after informed consent. The mutation in a subset of these individuals has been recently reported with the discovery of the disease gene but has not been studied in a genotype–phenotype correlation analysis (22).
D4Z4 repeat sizing, haplotype analysis and methylation analysis
All three cohorts were analyzed for D4Z4 repeat size, genetic background and CpG methylation at D4Z4 (Supplementary Material, Table S2). Genomic DNA (gDNA) was isolated from PBMCs. The sizing of the D4Z4 repeats on chromosomes 4 and 10 was done by pulsed field gel electrophoresis (PFGE) as described previously (38). Haplotype analysis was done by hybridization of PFGE blots with probes A and B in combination with PCR-based SSLP analysis according to previously described protocols (38).
Repeat array sizes ranging between 20 and 60 kb (4–16 units) were confirmed using a modified PFGE program. For this analysis, EcoRI-digested gDNA was separated by PFGE at 8.5 V/cm in two identical cycles of 10 h, with a switch time increasing linearly from 1 s at the start to 2 s at the end of each cycle and by using a 5-kb ladder as internal DNA size standard (Biorad, 170–3624). Methylation of the D4Z4 repeat was established at the FseI restriction site in the proximal unit of the arrays on chromosomes 4 and 10 simultaneously as described previously (22). Detailed step-by-step protocols are freely available from the Fields Center website (www.urmc.rochester.edu/fields-center/).
Mutation analysis
Mutation analysis for SMCHD1 was performed by Sanger sequencing on index cases for all 48 coding exons using intronic primers at a position of at least 50 nucleotides from the splice donor or acceptor sites; the 5′ and 3′untranslated regions were not included. The SMCHD1 genomic sequence was obtained from Ensemble (build 37) [GRCh37:18:2655286:2805615]. Primers were designed using primer3, and primers sequences are listed in Supplementary Material, Table S3 To predict the pathogenicity of the variants we identified, we used the computer software SIFT (http://sift.jcvi.org/) and GVGD align (http://agvgd.iarc.fr/agvgd_input.php).
Southern blot-based deletion screen for SMCHD1
To screen for possible deletions in the SMCHD1 locus, we used a Southern blot-based hybridization method. Five microgram gDNA was digested with restriction enzymes EcoRV or BamHI (MBI Fermentas) according to the manufacturer's instructions. DNA was separated in a 16 h run on a 0.85% agarose gel (MP agarose [Roche]) by PFGE at 8.5 V/cm in two identical cycles, with a switch time increasing linearly from 1 s at the start to 2 s at the end of each cycle. The run was performed in 0.5× TBE buffer supplemented with 150 ng/ml ethidium bromide at 23°C. Southern blotting and hybridization conditions have been described elsewhere (38). Southern blots were hybridized overnight at 65°C with an SMCHD1 cDNA probe that recognizes all 48 exons and washed 3× for 10 min in 1× SSC and 0.1% SDS at 65°C. Blots were exposed 16–24 h to phosphor imager screens and analyzed with the Image Quant software program (Molecular Dynamics).
RNA analysis
RNA was isolated from PBMCs, fibroblast or myoblast cultures, depending on availability. Primary PBMCs were isolated from Heparinized blood by Ficol gradient and stored in Cell Culture Freezing Medium (Gibco) in liquid nitrogen. Prior to RNA isolation, PBMCs were cultured in PBmax medium (Gibco) for 5 days. RNA isolation and cDNA preparation was done as described previously (22). To study the consequences of the mutation on RNA stability or pre-mRNA processing, exonic primers were designed in the one or two exons flanking the exon that contains the mutation. The primers used for RNA analysis are listed in Supplementary Material, Table S4.
Statistical analysis
To test the impact of the mutation on D4Z4 methylation, we analyzed the relationship between the combined D4Z4 methylation level (observed methylation = OM) on chromosomes 4 and 10 and the cumulative size of the repeat arrays (in units) in 254 control individuals, 186 FSHD1 individuals and 74 SMCHD1 mutation carriers. For each sample, we determined the size of both D4Z4 arrays on chromosome 4q (4S and 4L, for the shortest and longest array, respectively) and similarly on chromosomes 10q (10S and 10L, respectively). For modeling the relationship between methylation and the repeat array sizes, we used linear mixed models throughout, in which a random effect per family was included to take within-family correlation of methylation status into account. These mixed models were fitted in R (version 3.0.2, www.r-project.org) using the glmmPQL function from the MASS library. Reported P-values are Wald tests if they concern a single parameter or likelihood ratio tests if they concern multiple parameters. They were calculated using the same function. Because the relationship between methylation and repeat lengths showed clear non-linearity, we applied a logarithmic transformation (Base 2) to the repeat measurement. To avoid instability owing to the logarithmic transformation near 0, we set the transformed repeat lengths to 2 for all values in the range smaller than 4 units [log2(4) = 2].
According to these models, the PM for control, FSHD1 and FSHD2 individuals can be calculated by the formula: [intercept + F1 × log2(4S) + F2 × log2(4L) + F3 × log2(10S) + F4 × log2(10L) + gender]. The PM in controls (PM1) can be calculated using the following values: intercept, −15.92; F1, 3.70; F2, 4.47; F3, 2.48; F4, 2.77; gender, −0.45. For calculation of the PM in FSHD2 individuals (PM2), the following values were used: intercept, −13.71; F1, 1.60; F2, 1.93; F3, 1.08 F4, 1.20; gender, −0.19. The Delta1 is the difference between the PM1 and the OM. The Delta2 is the difference between the PM2 and the OM based on the model for carriers of an SMCHD1 mutation.
Conceptually, we can split the OM and PM into four parts, each corresponding to one of the individual D4Z4 repeat arrays. We split PM as [intercept/4 + F1 × log2(4S) + gender/4] + [intercept/4 + F2 × log2(4L) + gender/4] + [intercept/4 + F3 × log2(10S) + gender/4] + [intercept/4 + F4 × log2(10L) + gender/4], which allows us to calculate the PM contributed by each of the four individual alleles. Assuming that the excess methylation Delta2 is equally distributed over the alleles, the methylation attributed to the shortest permissive allele (either 4S or 4L) in FSHD2 individuals can be estimated by the sum of the PM at this allele and Delta2 divided by four.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the US National Institutes of Health (NIH) (National Institute of Neurological Disorders and Stroke (NINDS) P01NS069539, and National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) R01AR045203), the Muscular Dystrophy Association (MDA; 217596), the Fields Center for FSHD Research, the Geraldi Norton and Eklund family foundation, the FSH Society, The Prinses Beatrix Spierfonds (W.OR12-20), Spieren voor Spieren, The Friends of FSH Research, and European Union Framework Programme 7 agreement 2012-305121 (NEUROMICS).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all individuals with FSHD and family members for their participation. We thank Dr Amanda Mason and Dr Lucia Clemens-Daxinger for critical reading.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Deenen J.C., Arnts H., van der Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L., van Engelen B.G. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology. 2014;83:1056–1059. doi: 10.1212/WNL.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padberg G.W. The Netherlands: Leiden University; 1982. Facioscapulohumeral disease. PhD thesis (http://hdl.handle.net/1887/25818. ) [Google Scholar]
- 3.Statland J.M., Tawil R. Facioscapulohumeral muscular dystrophy: molecular pathological advances and future directions. Curr. Opin. Neurol. 2011;24:423–428. doi: 10.1097/WCO.0b013e32834959af. [DOI] [PubMed] [Google Scholar]
- 4.Lemmers R.J.L., de Kievit P., van Geel M., van der Wielen M.J., Bakker E., Padberg G.W., Frants R.R., van der Maarel S.M. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann. Neurol. 2001;50:816–819. doi: 10.1002/ana.10057. [DOI] [PubMed] [Google Scholar]
- 5.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 6.Lunt P.W., Jardine P.E., Koch M.C., Maynard J., Osborn M., Williams M., Harper P.S., Upadhyaya M. Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD) Hum. Mol. Genet. 1995;4:951–958. doi: 10.1093/hmg/4.5.951. [DOI] [PubMed] [Google Scholar]
- 7.Goto K., Nishino I., Hayashi Y.K. Very low penetrance in 85 Japanese families with facioscapulohumeral muscular dystrophy 1A. J. Med. Genet. 2004;41:e12. doi: 10.1136/jmg.2003.008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricci G., Scionti I., Sera F., Govi M., D'Amico R., Frambolli I., Mele F., Filosto M., Vercelli L., Ruggiero L., et al. Large scale genotype-phenotype analyses indicate that novel prognostic tools are required for families with facioscapulohumeral muscular dystrophy. Brain. 2013;136:3408–3417. doi: 10.1093/brain/awt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tawil R., Forrester J., Griggs R.C., Mendell J., Kissel J., McDermott M., King W., Weiffenbach B., Figlewicz D. Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. The FSH-DY Group. Ann. Neurol. 1996;39:744–748. doi: 10.1002/ana.410390610. [DOI] [PubMed] [Google Scholar]
- 10.Tonini M.M., Passos-Bueno M.R., Cerqueira A., Matioli S.R., Pavanello R., Zatz M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD) Neuromuscul. Disord. 2004;14:33–38. doi: 10.1016/j.nmd.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 11.de Greef J.C., Lemmers R.J., van Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R., van der Maarel S.M. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 12.Snider L., Geng L.N., Lemmers R.J.L.F., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., Miller D.G. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS. Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 14.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O., et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 16.Hewitt J.E., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 17.Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 18.Lemmers R.J., Wohlgemuth M., Frants R.R., Padberg G.W., Morava E., van der Maarel S.M. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., Tapscott S.J. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M., Apedaile A., Hilton D.J., Dunwoodie S.L., Brockdorff N., et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 22.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R., et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Greef J.C., Lemmers R.J., Camano P., Day J.W., Sacconi S., Dunand M., van Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L., et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacconi S., Lemmers R.J., Balog J., van der Vliet P.J., Lahaut P., van Nieuwenhuizen M.P., Straasheijm K.R., Debipersad R.D., Vos-Versteeg M., Salviati L., et al. The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am. J. Hum. Genet. 2013;93:744–751. doi: 10.1016/j.ajhg.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaillard M.C., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E., Vovan C., Chaix C., Broucqsault N., Morere J., et al. Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology. 2014;83:733–742. doi: 10.1212/WNL.0000000000000708. [DOI] [PubMed] [Google Scholar]
- 26.Hartweck L.M., Anderson L.J., Lemmers R.J., Dandapat A., Toso E.A., Dalton J.C., Tawil R., Day J.W., van der Maarel S.M., Kyba M. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology. 2013;80:392–399. doi: 10.1212/WNL.0b013e31827f075c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R., et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Overveld P.G., Enthoven L., Ricci E., Rossi M., Felicetti L., Jeanpierre M., Winokur S.T., Frants R.R., Padberg G.W., van der Maarel S.M. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol. 2005;58:569–576. doi: 10.1002/ana.20625. [DOI] [PubMed] [Google Scholar]
- 29.Balog J., Thijssen P.E., de Greef J.C., Shah B., van Engelen B.G., Yokomori K., Tapscott S.J., Tawil R., van der Maarel S.M. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics. 2012;7:579–584. doi: 10.4161/epi.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A., et al. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS. Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scionti I., Greco F., Ricci G., Govi M., Arashiro P., Vercelli L., Berardinelli A., Angelini C., Antonini G., Cao M., et al. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2012;90:628–635. doi: 10.1016/j.ajhg.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Overveld P.G., Lemmers R.J., Deidda G., Sandkuijl L., Padberg G.W., Frants R.R., van der Maarel S.M. Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum. Mol. Genet. 2000;9:2879–2884. doi: 10.1093/hmg/9.19.2879. [DOI] [PubMed] [Google Scholar]
- 34.Lemmers R.J., van der Wielen M.J., Bakker E., Frants R.R., van der Maarel S.M. Rapid and accurate diagnosis of facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2006;16:615–617. doi: 10.1016/j.nmd.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 36.Mannini L., Cucco F., Quarantotti V., Krantz I.D., Musio A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 2013;34:1589–1596. doi: 10.1002/humu.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landouzy L., Dejerine J. De la myopathie atrophique progressive: myopathie héréditaire, sans neuropathie, débutant d'ordinaire dans l'enfance, par la face. Revue. de Médecine. 1885;5:253–366. [Google Scholar]
- 38.Lemmers R.J.L.F., van der Vliet P.J., van der Gaag K.J., Zuninga S., Frants R.R., de Knijff P., van der Maarel S.M. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete duplication events in human evolution. Am. J. Hum. Genet. 2010;86:364–377. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.