Abstract
The Hirschsprung disease (HSCR) is a complex congenital disorder, arising from abnormalities in enteric nervous system (ENS) development. There is a gender disparity among the patients, with the male to female ratio as high as 5 : 1. Loss-of-function mutations of HSCR genes and haploinsufficiency of their gene products are the primary pathogenic mechanisms for disease development. Recent studies identified over half of the HSCR disease susceptibility genes as targets for the sex-determining factor SRY, suggesting that this Y-encoded transcription factor could be involved in sexual dimorphism in HSCR. Among the SRY targets, the tyrosine kinase receptor RET represents the most important disease gene, whose mutations account for half of the familial and up to one-third of the sporadic forms of HSCR. RET is regulated by a distal and a proximal enhancer at its promoter, in which PAX3 and NKX2-1 are the resident transcription factors respectively. We show that the SRY-box 10 (SOX10) co-activator interacts and forms transcriptional complexes with PAX3 and NKX2-1 in a sequence-independent manner and exacerbates their respective transactivation activities on the RET promoter. SRY competitively displaces SOX10 in such transcription complexes and represses their regulatory functions on RET. Hence SRY could be a Y-located negative modifier of RET expression; and if it is ectopically expressed during ENS development, such SRY repression could result in RET protein haploinsufficiency and promotion of HSCR development, thereby contributing to sexual dimorphism in HSCR.
INTRODUCTION
Hirschsprung disease (HSCR), or aganglionic megacolon, is a complex congenital disease arising from abnormalities in the enteric nervous system (ENS) development during embryogenesis (1–3). The incidence of HSCR is ∼1 in 5000 live births and varies significantly among ethnic groups, with Asians being the highest with an incidence of 1.4 per 5000 births. HSCR can be generally classified into two categories: long- (L-HSCR) and short-segment (S-HSCR) aganglionosis. There are roughly 20% L-HSCR and 80% S-HSCR among the patient population. Significantly, there is a sex difference among the S-HSCR category with male to female ratio >5 : 1 among some populations (1). Sexual dimorphism in HSCR has been a major medical phenomenon whose pathogenesis has been a long-standing dilemma in the field.
The precursors for the ENS originate from neural crest (NC) cells, which migrate from the vagal and sacral portions of the NC, populate the primitive gut and differentiate into neural ganglions and glial cells lining the gastrointestinal wall (3,4). The ENS is responsible for maintaining the peristalsis and bowel movement in the digestive system. Accordingly, HSCR is a neurocristopathy that arises from insufficiency/absence of NC cell migration and/or neural differentiation in certain segments, primarily the colon, of the gastrointestinal track during embryonic development (5,6). At present, genetic studies have identified mutations in various genes, including RET, GDNF, NRTN, EDNRB, EDN3, ECE1, SOX10, PHOX2B, ZEB2 and L1CAM and numerous susceptibility genes and loci, involved in ENS development and could potentially contribute to the complex etiologies of HSCR (3,7). Among the HSCR disease genes, mutations in the tyrosine kinase receptor RET account for ∼50% of the familial and 7–35% of the sporadic forms of HSCR (1,8). Significantly, disease penetrance depends on numerous risk factors, including the nature of the mutations, gene–gene and gene–environment interactions, and actions of associated genetic/epigenetic modifiers (3,7,9–11). In general, male siblings possess higher disease penetrance than female siblings in familial cases, suggesting the existence of a male-specific disease modifier(s) for HSCR. At present, the exact nature of such genetic modifier(s) is unknown.
The SRY gene is the sex-determining gene on the male-specific Y chromosome, responsible for switching on the testis differentiation during embryogenesis (12). It is the founder of a family of transcription factor genes, designated as SRY-box (SOX) genes (13). SRY and SOX proteins harbor a functionally interchangeable and conserved DNA/protein-binding domain, termed high-mobility group (HMG) box (14). In a recent study, the specific targets for SRY in fetal gonads at the time of sex determination in the mouse have been determined (15). In addition to various known sex-determining genes, significant numbers of genes involved in numerous neurological disorders are among the SRY targets. Gene ontology analysis showed that among the neurological disorders, the HSCR is a major one with 7 of the 13 known disease genes in the database being represented among the SRY targets (Supplementary Material, Table S1). Further analysis identified additional SRY target genes (Supplementary Material, Fig. S1), potentially involved in ENS development, and mutations of which could contribute to the overall HSCR pathogenesis (3,7). If SRY is expressed during ENS development, the binding of the Y-encoded SRY transcription factor to the promoters of these HSCR and related genes suggests that it could exert regulatory functions to the expression of these genes, thereby affecting the developmental process(es) in a male-specific manner. Hence, SRY could potentially be a male-specific genetic modifier for HSCR, predisposing males to the disease processes.
To determine the probable role of SRY in modifying the expression of HSCR genes, we have selected the RET gene for a detailed study, since RET mutations and/or haploinsufficiency are the major causes of the disease among the familial and sporadic patient populations (1,2). Our results demonstrate that SRY could be expressed in diseased tissues in HSCR patients. It binds to the promoter of the human RET gene at its distal and proximal enhancer regions by interacting with the resident transcription factors, PAX3 and NKX2-1, respectively (16–18). SRY competes with SOX10 bindings to the same transcription factors, and represses the transactivation activities of these transcriptional complexes on RET gene. Hence, SRY could inhibit RET expression, thereby generating a haploinsufficiency of the RET product(s) and suppressing its function(s) in ENS development.
RESULTS
SRY is expressed in enteric nervous tissues in HSCR patients
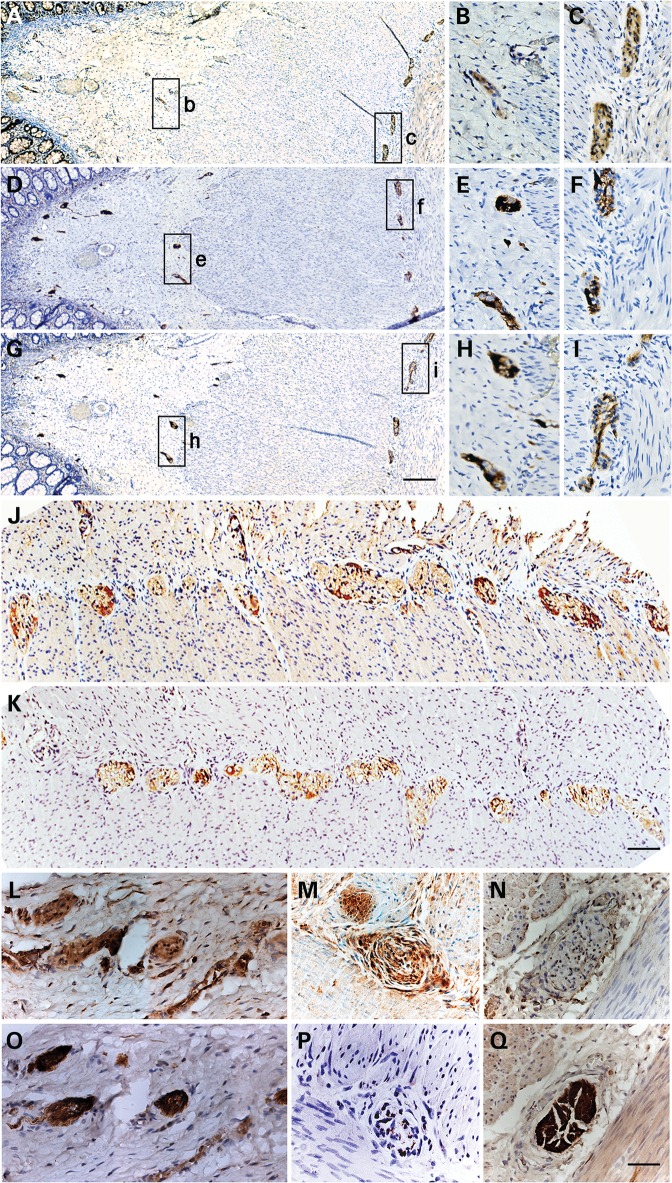
As a genetic modifier, we surmise that SRY could likely be expressed in migrating NC cells and/or ENS precursor cells, thereby affecting the over neural network development in the gut. Although the effective period of actions for any mutant gene and/or genetic modifier is likely to be at the time of NC cell migration and ENS development during embryogenesis, it is conceivable that residual SRY expression could still persist and be detectable in colon tissues of HSCR patients. Using a panel of RNA samples derived from surgical resected ganglionic colon tissues of 24 male HSCR patients with affected short segments (19) and 12 normal colon controls (10 male patients with colorectal cancer and 2 patients with imperforate anus), we had examined the SRY expression using gene-specific primers and RT-PCR analysis (Supplementary Material, Table S2). This initial experiment showed heterogeneous levels of SRY in 18 of the 24 HSCR samples, but none in the 12 normal colon controls (Supplementary Material, Fig. S2). To determine the locations and cell type(s) expressing SRY, tissue sections from archival pathological preparations of surgical biopsies from 27 male HSCR patients and 1 female HSCR patient and 2 male colorectal cancer patients were analyzed with immunohistochemistry using specific antibodies against the human SRY, and calretinin and neurofilament M as references. Calretinin and neurofilament have been used as diagnostic markers for ganglion cells in HSCR rectal biopsies (20–23). Calretinin, in particular, has been demonstrated to be reliable in delineating the boundaries between aganglionic and ganglionic portions in the bowels of these patients (23). Eight male and one female HSCR samples consisted of paired aganglionic and ganglionic sections, while the rest of the samples were derived from ganglionic parts of the biopsies. Our results showed that SRY protein was detectable only in the ganglionic parts, but not the aganglionic sections of the respective HSCR tissues while normal colon samples from colorectal cancer specimens were also negative for SRY. Similar to the RNA results, heterogeneous staining patterns were observed on 12 of the 27 male samples with visible SRY immunostaining signals, but not the female sample and normal colon controls. The SRY protein was primarily located at the myenteric plexuses (Fig. 1A, C and J) between the longitudinal and circular muscle layers, and the submucosal plexuses (Fig. 1A, B and K). These structures were intensely positive for calretinin (Fig. 1D–F) and neurofilament M (Fig. 1G–I and N). Occasionally, differences in immunostaining of SRY (Fig. 1L and M) and these neural markers (Fig. 1O and P) could be observed at selected ganglionic plexuses. Hence, although the SRY expression could be heterogeneous, it was detectable at both RNA and protein levels in the ganglionic but not in aganglionic portion of the HSCR tissues or normal control colon tissues. To a large extent, its expression pattern parallels those of ganglionic markers, calretinin and neurofilament M, in enteric ganglions. Our observations suggest that SRY could be ectopically expressed in the precursor cells of the ENS during embryonic development among selected HSCR patients. However, its expression could be extinguished as these precursors failed to differentiate into ganglionic cells in the aganglionic segment(s), but could still be residually detected in ganglionic cells in the ganglionic segment(s) of the colon in the HSCR patients.
Figure 1.
Detection of SRY, calretinin and neurofilament M expression in ganglionic tissue of HSCR patients using immunohistochemistry. (A) SRY was primarily detected in the ganglionic structures at the myenteric plexuses (box c) and the submucosal plexuses (box b), respectively, on resected tissue of a 5-week-old male patient. Boxed areas in (A) were enlarged in (B) and (C), respectively. (D) An adjacent section immunostained with a calretinin antibody. Boxes e and f were enlarged in (E) and (F), respectively. (G) An adjacent section immunostained with a neurofilament M antibody. Box h and i were enlarged in (H) and (I), respectively. The ganglionic structures were positive for SRY and the ganglionic biomarkers. (J and K) Immunostaining of SRY and neurofilament M, respectively, on myenteric plexuses on adjacent sections of resected ganglionic tissues of a 2-week-old male HSCR patient. (L and O) Immunostaining of SRY and neurofilament M, respectively, on the submucosal plexuses of a 1-year-old male patient. Selected ganglionic structures could be positive for SRY (M) and negative (P) for neurofilament M (a 7-week-old male) and vice versa (N and Q) (a 2-week-old male). Bar in (G) represents 400 µm in (A), (D) and (G); bar in (K) represents 100 µm in (B), (C), (E), (F), (H)–(K); and bar in (Q) represents 50 µm in (L)–(Q), respectively.
SRY disrupts the transactivation of PAX3-SOX10 and NKX2-1 at the distal and proximal enhancer regions of the RET gene
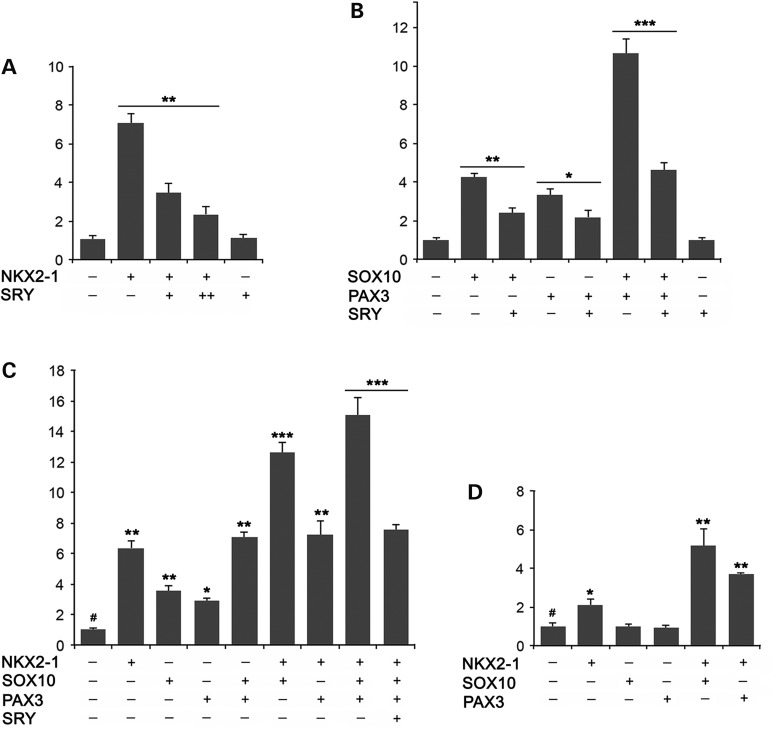
Among the HSCR disease genes, the RET gene has been the most characterized one since its mutations account for majority of the familial and many sporadic forms of the disease (1,2,24). Further, the regulation of the RET gene is conserved among various vertebrate species, ranging from humans to zebrafish, without the necessity of sequence similarity (25), thereby raising the possibility of translating SRY regulation of the RET gene from mouse to humans. Studies on the human RET promoter have identified primarily two enhancer regions, one distal and one proximal to its transcription start site. The distal enhancer region is located at ∼3.4 kb and harbors two closely located PAX- and SOX-binding sites while the proximal enhancer region harbors an NKX2-1-binding site immediately upstream of the transcription start site. Previous studies have demonstrated that PAX3 and NKX2-1 are the resident transcription factors critical for the enhancer functions of these two sites for RET expression (16–18). PAX3 interacts with SOX10, another vital transcription factor for ENS development (26), and synergistically exacerbates the transactivation of RET expression. To determine the probable function of SRY on the transactivation of RET by NKX2-1 and PAX3, we had examined its effects in a reporter system with a luciferase gene directed by a 3.7 kb promoter (−3531 to +196) of the human RET gene (27), harboring both distal and proximal enhancers (Supplementary Material, Fig. S3). Co-transfection of NKX2-1 with this reporter gene greatly exacerbated the luciferase activities in Neuro-2A cells. Inclusion of a human SRY expression vector in the transfection showed a dosage-dependent repression of the NKX2-1 transactivation, whereas SRY alone did not show any significant effect on the RET3.7-luciferase activities (Fig. 2A). Similar analyses on PAX3 and SOX10 showed that both PAX3 and SOX10 could independently stimulate the expression of the RET3.7-luciferase reporter, and could synergistically exacerbate their respective transactivation activities, when they were present in the transfected cells (Fig. 2B). Again, such transactivation activities were repressed with a co-transfected SRY transgene in the same cells.
Figure 2.
SOX10 stimulates and SRY represses NKX2-1 and PAX3 regulation of RET promoter at proximal and distal enhancer regions in Neuro-2A cells. (A) SRY represses NKX2-1 transactivation of a 3.7 kb RET promoter (Supplementary Material, Fig. S3) directed luciferase gene in a dosage-dependent manner. **P < 0.01 by Student's t-test between sample pairs. (B) PAX3 and SOX10 are capable of activating the RET-luciferase individually and synergistically, which is repressed by SRY. *P < 0.05, **P < 0.01 by Student's t-test. (C) NKX2-1, PAX3 and SOX10 activate RET3.7-luciferase additively, and are repressed by SRY. *P < 0.05, **P < 0.01 and ***P < 0.001 at each column compared with value at column 1 (#); and between sample pair. (D) PAX3 and SOX10 individually cannot, but can exacerbate the NKX2-1 activation of a 1.5 kb RET promoter (Supplementary Material, Fig. S3) directed luciferase, harboring only the proximal enhancer. *P < 0.05, **P < 0.01, when compared with value at column 1 (#). Error bars represent SDs derived from triplicate experiments.
To explore if the distal and the proximal enhancer and the respective transcription factors could collaborate with each other in their transactivation of RET promoter, NKX2-1, PAX3 and SOX10 were transfected individually and in combinations with the RET3.7-luciferase reporter in Neuro-2A cells. Our results showed that these transcriptional factors were capable of stimulating the RET promoter-directed luciferase activities in additive manners, with the highest activity in cells co-transfected with the three transcription factors and the RET-reporter, which again could be repressed by the presence of the human SRY transgene (Fig. 2C). To separate the effects of the distal enhancer from those of the proximal enhancer, a 1.5 kb promoter (−1273 to +196) harboring only the proximal enhancer region was used to direct the luciferase reporter (RET1.5-luciferase) (Supplementary Material, Fig. S3), and analyzed similarly in Nuero-2A cells. Our results showed that this short RET promoter was capable of responding to NKX2-1 stimulation, but not PAX3 and SOX10 individually. However, when either SOX10 or PAX3 was co-transfected with NKX2-1, significant exacerbations of the RET1.5-luciferase activities were observed (Fig. 2D), suggesting potential interactions and collaborations between these transcription factors and NKX2-1 on regulation of RET gene.
SRY competes with SOX10 in interactions with PAX3 and NKX2-1 and represses their transactivation activities
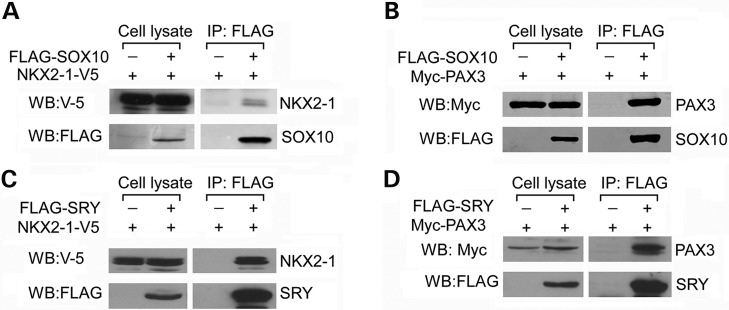
Previously SOX10 had been demonstrated to interact physically with PAX3, and bind to the putative SOX site, adjacent to the PAX site at the distal enhancer region, but its binding to the SOX site was not required for its transactivation (18). Our results confirmed these findings, and further suggested that SRY could impede on such collaborative effects of SOX10 by competing against its interactions to NKX2-1 and PAX3. To explore this possibility, we have systemically studied the SOX10 and SRY interactions with NKX2-1 and PAX3, using co-immunoprecipitation and GST pull-down assays. Co-transfection of FLAG-tagged SOX10 with either V5-tagged NKX2-1 or Myc-tagged PAX3 to the human HEK293 cells and immunoprecipitation with an FLAG (SOX10) antibody showed that both NKX2-1 and PAX3 were co-immunoprecitated with SOX10 (Fig. 3A and B), confirming the physical interactions of SOX10 with these transcription factors. Similar studies with FLAG-tagged SRY transgene showed that SRY was also capable of interacting physically with NKX2-1 and PAX3 in the transfected cells (Fig. 3C and D).
Figure 3.
Demonstration of interactions between SOX10 and (A) NKX2-1 and (B) PAX3; and between SRY and (C) NKX2-1 and (D) PAX3, respectively, in HEK293 cells by co-immunoprecipitation. HEK293 cells were transfected with epitope-tagged transcription factors, as indicated, and immunoprecipitated with an antibody against the FLAG epitope for SOX10 in (A) and (B); and SRY in (C) and (D) analyzed with western blots with antibodies against FLAG (SOX10 and SRY), V5 (NKX2-1) and Myc (PAX3). Western blots of cell lysates detected the protein expression of respective transgene in the transfected cells. WB:Myc/V5/FLAG = western blot with respective Myc/V5/FLAG epitope antibodies.
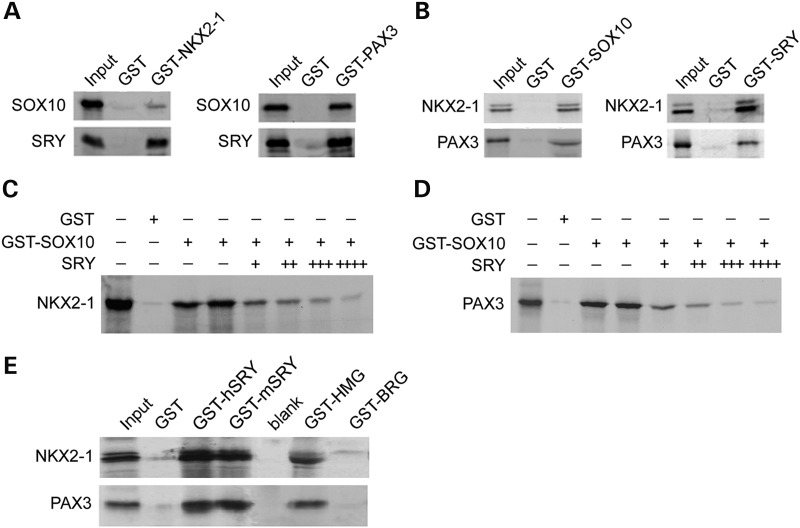
GST pull-down assay was used to confirm SOX10 and SRY interactions with NKX2-1 and PAX3. Using either a GST-NKX2-1 or a GST-PAX3 fusion protein as bait, both 35S-labeled SOX10 and SRY were being pulled down, but not a GST-alone bait (Fig. 4A), while the reverse bindings with either a GST-SOX10 or a GST-SRY fusion protein as bait, both 35S-labeled PAX3 and NKX2-1 were being pulled down with GST-binding beads (Fig. 4B). To demonstrate the competitiveness of SOX10 and SRY in their bindings to NKX2-1 and PAX3, GST-SOX10 fusion protein was used as bait in pull-down assays with either 35S-labeled NKX2-1 or PAX3 and cold SRY was added in increasing quantities to the binding reactions. Our results showed that GST-SOX10 bindings to NKX2-1 or PAX3 were gradually and competitively suppressed with increasing amounts of cold SRY protein, suggesting that SRY is capable of competing with SOX10 in its physical interactions with NKX2-1 and PAX3, respectively (Fig. 4C and D).
Figure 4.
SOX10 and SRY competitive interactions with NKX2-1 and PAX3 demonstrated by GST pull-down assays. (A) GST-NKX2-1 (left) and GST-PAX3 (right) were used as baits to pull-down individually and radioactively labeled SOX10 and SRY protein. (B) GST-SOX10 and GST-SRY pull down of labeled NKX2-1 and PAX3, respectively. GST-SOX10 pull down of labeled (C) NKX2-1 and (D) PAX3 can be competitively suppressed by additions of non-radioactive SRY in a dosage-dependent manner. (E) Mapping of the HMG box as the domain in SOX10 and SRY responsible for binding to NKX2-1 and PAX3. GST-hSRY and GST-mSRY are GST fusion proteins harboring human and mouse SRY, respectively. GST-HMG and GST-BRG are GST fusion proteins harboring the mouse HMG box and the BRG (bridge peptide between HMG box and Q-domain of the mouse SRY).
SRY is the founder of the SOX genes, which encode proteins with a conserved and special HMG box (SRY box) present in the SRY protein (13). Beside this conserved domain, the flanking sequences of the SOX proteins share very little homology with the flanking sequences of SRY (13,28,29). The competitive bindings of SOX10 and SRY to NKX2-1 and PAX3 suggested that this conserved domain between SRY and SOX10 was likely involved in the process(es). To explore this possibility, GST-fusion proteins containing the human SRY, mouse SRY, HMG box alone and the bridge (BRG) domain of the mouse SRY (30,31) were used in GST pull-down assays with 35S-labeled NKX2-1 or PAX3 proteins. Our results showed that human and mouse SRY were capable of pulling down both transcription factors (Fig. 4E). As discussed above, we postulated that the HMG box of both SOX10 and SRY was likely their binding domain to NKX2-1 and PAX3. Using a GST-HMG box fusion protein as bait, NKX2-1 and PAX3 bindings to this conserved domain were indeed confirmed with GST pull-down assay (Fig. 4E).
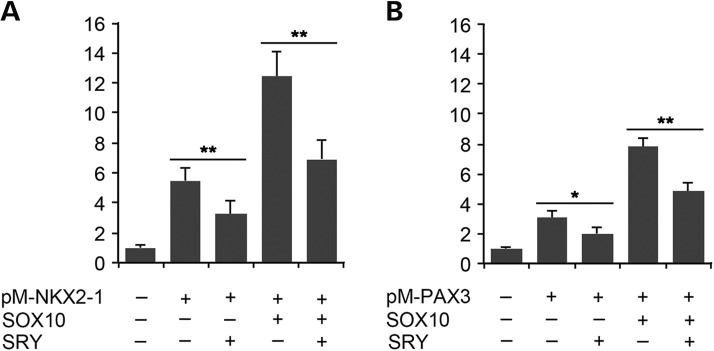
SOX10 and SRY interact with NKX2-1 and PAX3 independent of any cis-binding sequences
To determine if SOX10 stimulation and SRY repression of NKX2-1 and PAX3 transactivation of the RET promoter require their binding to specific sequence(s) adjacent to the respective NKX2-1 and PAX3 sites at the RET promoter, SOX10/SRY and NKX2-1/PAX3 interactions were studied with a mammalian two-hybrid system (32). The coding sequences for NKX2-1 or PAX3 were individually fused in-frame with the GAL4 DNA-binding domain (GAL4-BD) in the pM vector, resulting in two expression vectors, pM-NKX2-1 and pM-PAX3, respectively. The resulting hybrid genes encode fusion proteins capable of binding to the UAS sequence of the UAS-luciferase reporter via the GAL4-BD, thereby bringing the NKX2-1 or PAX3 to and activating the luciferase gene. Hence, the two-hybrid system eliminated the requirement of any binding sites for NKX2-1, PAX3 or SOX10/SRY at the responsive promoter-directed reporter gene. Transfection with either the pM-NKX2-1 or pM-PAX3 expression vectors with the UAS-luciferase reporter showed that the GAL4-BD was indeed capable bringing these two transcription factors to the UAS site, thereby transactivating the UAS-luciferase reporter in the transfected Neuro-2A cells (Fig. 5A and B). Inclusion of an SOX10 expression vector in the transfection greatly stimulated their transactivation activities, suggesting that SOX10 interacted with the respective transcription factor and synergistically stimulated the UAS-luciferase reporter. Again, inclusion of SRY in the transfections, either alone or in combination with SOX10, suppresses such transactivation (Fig. 5A and B). Our results, therefore, demonstrated that SOX10 interacted with either NKX2-1 or PAX3 without the requirement of binding to specific sequences on the responsive promoter and exacerbated their respective transcriptional activities, which were repressed by either SRY binding to or competitively displacing SOX10 in the NKX2-1 or PAX3 transcription complexes.
Figure 5.
SOX10 and SRY interact with NKX2-1 and PAX3 without the requirement of binding to any cis-sequence on the promoter, as demonstrated by the mammalian two-hybrid assay. (A) A GAL4-BD and NKX2-1 (pM-NKX2-1) fusion protein gene is capable of transactivating a co-transfected GAL4-responsive UAS-luciferase gene in Neuro-2A cells. Such transactivation is exacerbated by inclusion of an SOX10 gene, suggesting that SOX10 interacts with NKX2-1 and synergistically activates the NKX2-1 transcriptional activities. Inclusion of an SRY in these transfections represses the NKX2-1 and SOX10 transactivation functions on UAS-luciferase gene. (B) Similar assays with a GAL4-BD-PAX3 (pM-PAX3) fusion protein gene and SOX10 demonstrate that SOX10 exacerbates and SRY represses the PAX3 transcriptional activities. *P < 0.05 and **P < 0.01 by Student's t-test between respective sample pairs. Error bars represent SDs from triplicate experiments.
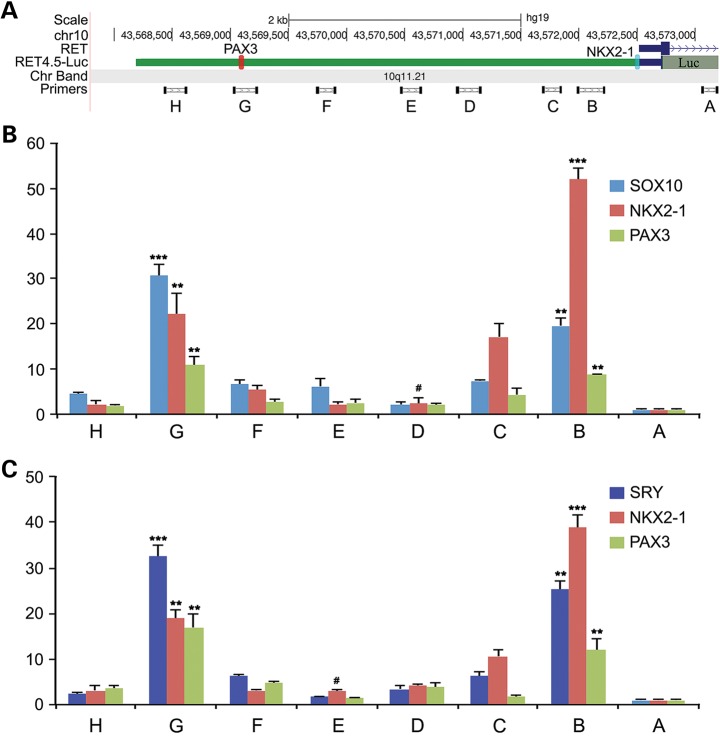
SOX10 and SRY co-localize with NKX2-1 and PAX3 at the chromatin of the enhancer regions on the RET promoter
To demonstrate that SOX10 and SRY could co-localize with the resident transcription factors at the distal and proximal enhancer regions of the RET promoter, a luciferase gene directed by a 4.5 kb promoter (−4320 to +196) of the human RET (RET4.5-luciferase) gene was constructed (Supplementary Material, Fig. S3), and co-transfected with either epitope-tagged SOX10, NKX2-1 and PAX3 or epitope-tagged SRY, NKX2-1 and PAX3 in Neuro-2A cells. The presence of NKX2-1, PAX3, SOX10 and combinations thereof stimulated the luciferase reporter activities while addition of SRY to the transfection mixtures repressed such activities (data not shown), as previously demonstrated (Fig. 2). Chromatin immunoprecipitation was performed individually with specific antibodies against the respective epitope of SOX10, NKX2-1, PAX3 or SRY. The corresponding precipitated chromatin DNAs were analyzed with quantitative PCR and specific primer pairs spanning the 4.5 kb promoter of the human RET gene (Fig. 6A). The primers in set A were located at exon 1 but outside the 4.5 kb promoter, and were absent in the mouse Neuro-2A genome, while other primer sets (i.e. B to H) were present in the transfected RET4.5-luciferase gene. PCR products with the A primer set were used as control, from which all other PCR products of the same immunoprecipitated chromatin DNAs were compared. Hence, all enrichments of precipitated chromatin DNAs at B to H locations were calculated as fold increases over the corresponding value (=1) at the A site of the RET gene (Fig. 6A). Using this system, we had analyzed the enrichment of chromatin at the specific regions along the promoter of the RET gene immunoprecipitated with the respective antibodies against these transcription factors. When the RET4.5-luciferase gene was co-transfected with SOX10, NKX2-1 and PAX3 and analyzed with chromatin immunoprecipitation, specific enrichments of chromatin immunoprecipitated with individually SOX10, NKX2-1 or PAX3 were observed with primer sets B and G, corresponding to the proximal and distal enhancer regions, respectively (Fig. 6B). Using similar strategy, the same chromatin regions at the proximal and distal enhancers of the RET promoter could also be demonstrated with SRY in place of SOX10 in the transfection to Neuro-2A cells (Fig. 6C). The co-localizations of the three transcription factors, i.e. NKX2-1, PAX3 and SOX10 or NKX2-1, PAX3 and SRY, at both the proximal and distal enhancers suggested that they could form transcriptional complexes at the respective enhancer regions.
Figure 6.
Detection of NKX2-1, PAX3, SOX10 and SRY bindings to the RET promoter using chromatin immunoprecipitation and quantitative PCR analyses with spanning primer sets. A 4.5 kb promoter (Supplementary Material, Fig. S3) harboring both the distal and proximal enhancers and directing the luciferase gene (A) was co-transfected with epitope-tagged NKX2-1, PAX3 and SOX10 (B) or SRY (C). Chromatin immunoprecipitation was conducted individually with antibodies against the respective epitope-tagged transcription factors. Specific spanning primer sets, A to H (A; Supplementary Material, Table S2) were used to detect the relative enrichments of the corresponding segments of the RET promoter using quantitative PCR. The results (B and C) showed that all transcription factors bind preferentially to regions defined by primer sets B and G, corresponding to the proximal and distal enhancers, suggesting that they could form transcriptional complex(es) involving both enhancers. **P < 0.01 and ***P < 0.001 when compared with the lowest position excluding A (i.e. D (#) in B and E (#) in C, respectively) along the promoter of RET using Student's t-test. Error bars represent SDs from triplicate experiments.
DISCUSSION
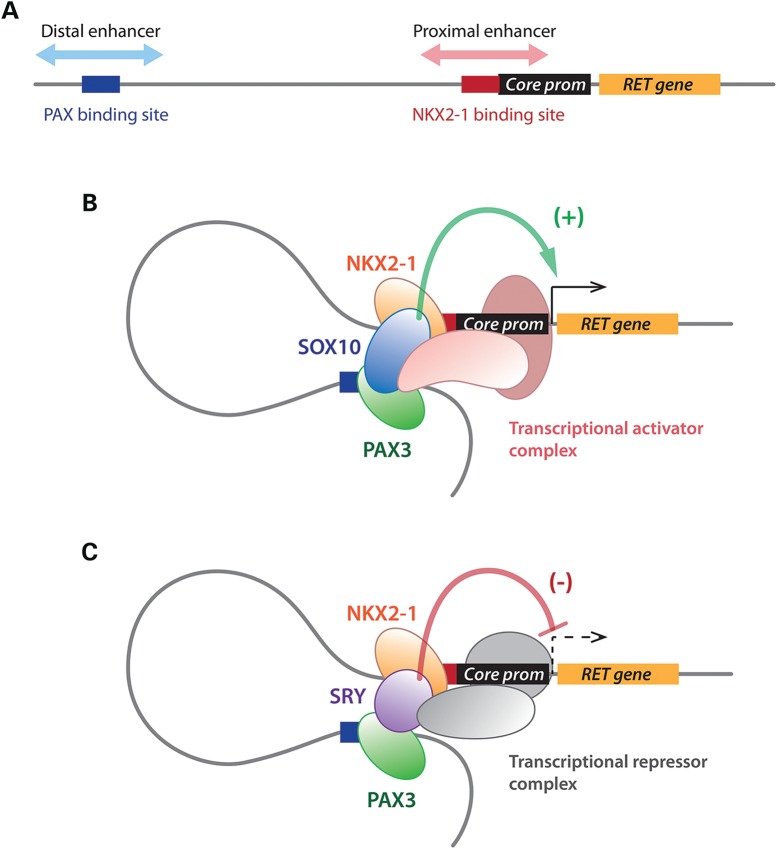
The sexual dimorphisms in incidence and disease penetrance in HSCR are unsolved mysteries in the biomedical field (1,2). Currently, the exact etiologies are unknown, despite the identification of disease-causing mutations in a sizable number of genes, involved in NC cell migration and ENS development, to be responsible for HSCR (1,2,6,7,9,33). These sexual dimorphisms could be results of the actions of genetic modifiers on the Y chromosome, which could be ectopically expressed in precursors and exert male-specific effects on the same HSCR disease genes in ENS. The Y-located SRY gene fits such criteria since it is capable of bindings to the promoters of a significant number of HSCR disease genes, and numerous ENS developmental genes (15). Our detailed analysis of SRY actions on the promoter of the tyrosine kinase receptor RET, a major HSCR disease gene, demonstrated that it could compete with the related SOX10 transcription factor at the distal and proximal enhancers and repress SOX10 interactions and stimulation of the respective resident transcription factors, PAX3 and NKX2-1, in the normal regulation of RET expression. We further demonstrated that SOX10 stimulation and SRY repression on these transcription factors were independent of their bindings to any sequence at the RET promoter, but were through direct interactions with PAX3 and NKX2-1. The observations that PAX3 and NKX2-1 were localized at both distal and proximal enhancers in vivo with either SOX10 or SRY (Fig. 6), further raised the possibility that these two enhancer regions could form a knotted structure in a transcriptional complex at the RET promoter (Fig. 7). At present, the exact composition of such transcriptional complex(es) is uncertain and might include the core elements of the preinitiation complex (PIC) (34) and other transcription factors, such as PHOX2B or HOXB5 (16,27), and additional enhancer site(s) (8) involved in NKX2-1 and PAX3 regulation of the RET gene. We surmise that such transcriptional complex could likely be gene and cell type specific (35,36), pertaining to ENS precursors and might have both temporal and spatial properties. Such multifactorial transcriptional regulatory mechanism(s) could result in gradated levels of RET expression, associated with disparities and/or polymorphisms in the functionalities of the respective component factors, during ENS development and might account partially for the variations in disease penetrance in HSCR (3,6,9). Nevertheless, the present data suggest that SOX10 is the normal and positive modifier or co-activator, while SRY is a negative modifier or repressor on the regulation of RET gene expression.
Figure 7.
Model of molecular interactions and transcriptional complex formation at the promoter of the RET gene. (A) Linear view of the locations of the distal and proximal enhancer regions harboring a PAX and an NKX2-1-binding site, respectively, on the RET promoter. (B) SOX10 interacts with both PAX3 and NKX2-1, thereby brings these two transcription factors, and hence the enhancers, into a knotted structure. At present, the exact composition of such a transcriptional complex is uncertain, and it might include additional co-factors, such as polymerase II and factors associated with the PIC, and co-activators capable of exacerbating the regulatory activities of the resident factors at the respective enhancers. (C) SRY competes and displaces SOX10 in the transcriptional complex, and exerts inhibitory functions on the transactivation of RET.
What is the probable mechanism(s) responsible for SRY in sexual dimorphisms in HSCR development? SRY is the primary male sex determinant and is normally expressed in the developing gonads at the time of sex determination (12). However, its expression has been detected in somatic tissues, such as pre and postnatal brain in humans and mouse (37–40). Transgenic mice harboring a fluorescent protein reporter gene directed by a human SRY promoter show significant transgene expression in the migrating NC cells (41), suggesting that the human SRY promoter, and hence the SRY gene, is capable of expressing in the precursors for ENS under transgenic conditions. As demonstrated in our expression analysis, both residual SRY RNA and protein were detected in colon tissues of HSCR patients, but not normal controls, suggesting that SRY could indeed be aberrantly expressed at the time of ENS development during embryogenesis and could contribute to the complex pathogenesis of HSCR. SRY and SOX10 harbor a conserved DNA/protein-binding domain, i.e. the HMG box, but diverge significantly at their flanking sequences (13). Importantly, this HMG box is functionally interchangeable between SRY and other SOX proteins (14), suggesting that they could bind to the same SOX cis-elements and/or co-activator(s) for the SOX proteins, i.e. SOX10 in the present case. The current study demonstrated that the HMG box is indeed responsible for SOX10 and SRY interactions with PAX3 and NKX2-1. Since SOX10 harbors a transactivating domain, it can co-activate the RET gene expression with these two transcription factors at the distal and proximal enhancers. On the other hand, SRY lacks such transactivating domain, and it is incapable of transactivation, which might result in inhibitory actions on RET expression by various means. First, displacement of SOX10 by SRY in the NKX2-1/PAX3/SOX10 transcription complex might produce a dominant-negative effect(s) on the overall RET transcriptional activities (42–44). Second, SRY has been demonstrated to interact with the gene silencer/repressor complex, KAP1-HP1 (31). The incorporation of SRY in the NKX2-1/PAX3 could bring this and other gene silencer complexes (45,46) to the proximity of the transcription start site, thereby repressing the RET gene. Third, SRY also interacts with a powerful chromatin modulator, PARP1 (30), which could poly(ribosyl)ate the chromatin proteins, thereby altering the protein-DNA affinity, chromatin structure and transcription factor accessibility (47,48) to the RET gene. Fourth, SRY interacts with the transcriptional complex could induce instability, leading to the dissolution of the entire complex from the RET promoter. Currently, it is uncertain which of these or other yet-to-be identified repressive mechanisms or combination thereof could play critical role(s) in SRY repression of the RET gene expression, and be responsible for RET insufficiency during ENS development.
Transgenic mouse studies demonstrated that haploinsufficiency of HSCR gene products could cause megacolon phenotypes in transgenic mice (49,50). Based on these observations, we surmise that the levels of aberrant SRY expression, and therefore the severity of its interference on the normal expression of its target genes (Supplementary Material, Table S1 and Fig. S1), involved in ENS development, could be critical in its contribution to sexual dimorphisms in the disease development. At low or moderate levels, SRY repression of HSCR disease genes could result in hypoganglionosis while high levels could induce aganglionosis in selected segment(s) of the ENS. Further, the disease processes might also depend on the number ENS developmental genes being affected, and other accessory and/or collaborative impairment events. If so, which pathway is being affected the most by aberrant SRY expression in ENS development? Among the signaling pathways in ENS (3,7), the GDNF/RET/GFRα pathway is likely to be the most impaired with key members, i.e. RET, GDNF, NRTN and GFRα2 being targets for SRY. The present study has demonstrated that SRY is capable of interfering with the normal expression of the RET gene, the most important HSCR disease gene responsible for a majority of familial and sporadic forms of the disease among the patients (1,2). We surmise that other pathways, such as EDN3/EDNRB and the hedgehog signaling pathways with members being SRY targets, could likely be affected by SRY repression and/or inappropriate activation. The overall disruption in various signaling pathways collectively in ENS development could be crucial in SRY contribution to the sexual dimorphisms in HSCR.
Sexual dimorphisms are prevalent in many human disorders, such as autoimmune disease, diabetes, cancers, schizophrenia, autism, depression, attention deficit disorder and neural degeneration, in terms of incidence, disease progression and/or treatment responses (51–60). Our recent study demonstrated that SRY and SOX9 share a significant number of common genes among their respective targets (15) and the HMG boxes of SRY and other SOX proteins are functionally interchangeable (14); hence, SRY could bind and regulate targets of other SOX transcription factors. Such scenario raises the possibility that SRY, when ectopically expressed in non-gonadal tissues, could exert male-specific effects on many developmental pathways, since the SOX proteins are key regulators in numerous cell fate specification and differentiation processes (61,62), particularly neurodevelopment. Indeed, as demonstrated in the gene ontology study, many genes involved in many neural diseases are SRY targets, and hence could potentially be affected by ectopic expression of SRY during neurodevelopment and/or disease processes. Specifically, the monoamine oxidase A (MAOA) gene is associated with over one-third of the neurological diseases with members as SRY targets (Supplementary Material, Table S1). MAOA catalyzes the deamination of monoamine neurotransmitters, such as serotonin, norepinephrine and dopamine, important for neurodevelopment, physiology, cognitive functions and related diseases (63–68). A detailed characterization of the human MAOA gene promoter demonstrated that SRY and Sp1 form a transcription complex and synergistically activate MAOA expression (58). Since MAOA gene dysfunctions and fluctuations in its enzyme activities are associated with various neuropsychiatric disorders, as discussed above, the SRY influence on MAOA expression could potentially exert a male-specific effect(s) on the contributions of MAOA dysfunctions in these neuropsychiatric diseases. We surmise that the spatial and temporal patterns and the magnitude of SRY ectopic expression are critical elements on its roles in sexual dimorphisms in neurological diseases, including HSCR. The identification of SRY targets associated with these neurological disorders (Supplementary Material, Table S1) has offered opportunities for detailed investigations on how SRY affects the normal expression of these neural targets. More significantly, elucidating the genetic, epigenetic, viral or environmental events/factors promoting SRY ectopic expression in non-gonadal tissues could be key in understanding the mechanisms by which SRY exerts its male-specific genetic modifier functions in the disease pathogeneses.
MATERIALS AND METHODS
RT-PCR analysis and immunohistochemistry
For RT-PCR analysis, RNA samples were extracted from resected colon ganglionic specimens from 24 patients of HSCR, 10 adult patients with colorectal cancer and 2 patients with imperforate anus and analyzed with RT-PCR, as previously described (19), using human SRY-specific primers (Supplementary Material, Table S2). Ribosomal 18S RNA was used as a positive control.
For immunohistochemistry, a total of 27 male and 1 female HSCR colon tissues and 2 adult male normal colons from colorectal cancer patients were analyzed in the present study. Seventeen cases were obtained from archival specimens at the Department of Pathology, University of California, San Francisco, which included eight pairs of aganglionic colon segment tissues and ganglionic colon segment tissues from 5 days to 5-month-old boys and one pair from a 3-week-old girl. Eleven cases of HSCR tissue sections were obtained from Division of Pediatric Surgery, Department of Surgery, The University of Hong Kong (19). All studies were performed under approved protocols by the respective institutional Committee on Human Research.
Immunostaining was performed with standard methods using specific antibodies, as described previously (69). The antibodies used for immunohistochemistry were: a mouse monoclonal antibody against human SRY (custommade at Ab-Mart, Shanghai, China; at 1 : 1000 dilution) and a rabbit polyclonal antibody against a recombinant human SRY protein (70) (at 1 : 250 dilution); a monoclonal antibody against the human calretinin (Clone DAK-Calret 1, Dako, Inc., Carpinteria, CA, USA, at 1 : 100 dilution), a monoclonal antibody against the human neurofilament M (Clone RMO-270, Life Technologies, Carlsbad, CA, USA; at 1 : 500 dilution). Positive stainings were visualized with a biotinylated anti-mouse IgG or anti-rabbit IgG (1 : 500) and DAB substrates, counterstained with haematoxylin, and examined and recorded with a Zeiss Imager A2 microscope and AxioCam MRc camera.
RET promoter-directed luciferase assays
The luciferase reporter plasmid (pXP1-FLWT) containing 3.7 kb of the human RET promoter, as previously described (27). It is designated as RET3.7-luciferase (Supplementary Material, Fig. S3). The 1.5 and 4.5 kb promoters were amplified with high-fidelity PCR and specific primers (Supplementary Material, Table S2) from RET-containing human genomic BAC DNA and subcloned into the pGL3 basic luciferase plasmid, resulting in RET1.5- and RET4.5-luciferase reporter vectors, respectively (Supplementary Material, Fig. S3). Human NKX2-1, SOX10 and PAX3 cDNAs were purchased from Open Biosystems, Inc. The coding sequences of the respective cDNAs were inserted in-frame into the corresponding epitope-tagged mammalian expression vectors: SOX10 and SRY in p3XFLAG-CMV-7 (FLAG-tagged) (Sigma, St. Louis, MO, USA); NKX2-1 in pcDNA3.1/V5-His vector (V5-tag) and PAX3 cDNA in pcDNA3.1/Myc expression vector (Myc-tag) (Life Technologies).
The mouse Neuro-2A cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and were cultured in DME medium supplemented with 10% FBS, at 37°C and 5% CO2. They were seeded at 1 × 105/well in 24-well plates, and were transfected with the various combinations of SOX10, NKX2-1, PAX3 and SRY using X-tremeGENE 9 reagent (Roche Biochemicals, Indianapolis, IN, USA). The β-galactosidase reporter vector, pCMV-LacZ, was included in all transfection mixtures and served as an internal control for transfection efficiency and transgene expression. Forty-eight hours after transfection, the cells were harvested and analyzed with the luciferase assay system, and β-galactosidase enzyme assay system (Promega, Madison, WI, USA) according to the manufacturer's instructions. The relative luciferase activities were calculated with respect to the β-galactosidase activity of the co-transfected LacZ gene, as described before (15).
Protein–protein interactions using co-immunoprecipitation and GST pull-down assays
Co-immunoprecipitation and GST pull-down assays were used to demonstrate protein–protein interactions between SOX10 and NKX2-1/PAX3 or SRY and NKX2-1/PAX3, as previously described (30,31). Epitope-tagged FLAG-SOX10 was co-transfected with NKX2-1-V5 and Myc-PAX3 in various combinations in human HEK293 cells. Co-immunoprecipitation was performed with EZview™ Red ANTI-FLAG® M2 Affinity Gel (Sigma) to pull down SOX10- or SRY-containing protein complexes. Other components of the SOX10- or SRY-complexes were detected by western blotting. First, a rabbit anti-FLAG antibody was used to detect either SOX10 or SRY, to confirm the presence of the initial FLAG-SOX10 or FLAG-SRY in the transfected HEK293 cells and the corresponding immunoprecipitated products. To detect the co-immunoprecipitation of NKX2-1-V5 or Myc-PAX3, a rabbit anti-V-5 or anti-Myc antibody was used respectively in parallel western blots of the immunoprecipitated proteins.
To synthesize GST fusion proteins consisting of SOX10, SRY, NKX2-1, or PAX3, the respective cDNAs were inserted in-frame in the pET41b vector. The recombinant proteins were synthesized in BL21(DE3) pLysS bacterial cells, and purified by affinity chromatography using glutathione-Sepharose resins, as previously described (30,31). To generate the radioactively labeled substrate, the corresponding cDNAs were inserted in pET28b, which were used in in vitro protein synthesized with the TnT T7 Quick Coupled Transcription/Translation System (Promega) in the presence of 35S-methionine (PerkinElmer, Waltham, MI, USA). GST pull-down assays were performed with a specific GST-fusion bait and corresponding 35S-labeled substrate, as previously described (30,31). The bound proteins were analyzed in 10–15% SDS–PAGE gels and detected by autoradiography.
Mammalian two-hybrid analysis
The interactions between NKX2-1/PAX3 and SOX10/SRY were analyzed with the mammalian two-hybrid system (Clontech Laboratories, Palo Alto, CA, USA) (32). The coding sequences for NKX2-1 or PAX3 were inserted in-frame immediately after the GAL4-binding domain (GAL4-BD) sequence in the pM vector, resulting in pM-NKX2-1 and pM-PAX3, respectively. The GAL4-binding site, upstream activator sequence (UAS), was inserted into the minimal promoter of GL2-luciferase reporter gene, resulting in the UAS-Luc reporter gene. Any GAL4-BD fusion protein would bind to the promoter of the UAS-Luc reporter, and exert any transactivation or repression of the respective transcriptional function of the fused protein. To demonstrate the interactions between NKX2-1/PAX3 and SOX10/SRY, pM-NKX2-1 or pM-PAX3 was co-transfected with the UAS-Luc, and with or without SOX10/SRY. The effects of SOX10 or SRY on NKX2-1 or PAX3 transactivation of the UAS-Luc gene were determined by luciferase assays, as described above.
Detection of transcription factor bindings on the RET promoter by chromatin immunoprecipitation
Five million Neuro-2A cells were cultured in 10-cm culture dish overnight, and co-transfected with epitope-tagged FLAG-SOX10, NKX2-1-V5, Myc-PAX3 and the RET4.5-Luc harboring a luciferase gene directed by a 4.5 kb RET promoter (Supplementary Material, Fig. S3). Parallel transfection was conducted with FLAG-SRY, substituting the FLAG-SOX10 gene in the reaction mixture. Forty-eight hours later, the cells were harvested and cross-linked with 1% formaldehyde for 10 min at room temperature. Chromatin immunoprecipitation was performed with normal mouse IgG, anti-FLAG, anti-V5 and anti-Myc antibodies, as described previously (15). Enrichment of DNA fragments was determined by quantitative PCR with primer pairs at different regions of RET promoter and exon 1. Data were normalized to normal IgG control immunoprecipitation, and then referenced as folds of enrichment to primer pair A (=1), which were located at the exon 1 sequence outside the 4.5 kb RET promoter in the RET4.5-Luc reporter.
SUPPLEMENTARY MATERIAL
FUNDING
This work was partially supported by NIH grant HD38117 and VA Merit grant 1I01BX000865 to Y.-F.C.L. Y.-F.C.L. is a Research Career Scientist of the Department of Veterans Affairs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Dong Ji Zhang and Ms. Jay So Man Ting for technical assistance, Dr Xin Yuan for a gift of the polyclonal antibody against human SRY and Professor Yuet Wai Kan for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., et al. Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarti A., McCallion A.S., Lyonnet S. In: Hirschsprung Disease, in OMMBID–The Online Metabolic and Molecular Bases of Inherited Diseases. Valle D., Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., Gibson K., Mitchell G., editors. 2014. Chapter 251, DOI:10.1036/ommbid.29, McGraw-Hill: New York, NY. [Google Scholar]
- 3.Heanue T.A., Pachnis V. Enteric nervous system development and Hirschsprung's disease: advances in genetic and stem cell studies. Nat. Rev. Neurosci. 2007;8:466–479. doi: 10.1038/nrn2137. [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Chan A.K., Sham M.H., Burns A.J., Chan W.Y. Analysis of the sacral neural crest cell contribution to the hindgut enteric nervous system in the mouse embryo. Gastroenterology. 2011;141:992–1002. doi: 10.1053/j.gastro.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Martucciello G. Hirschsprung's disease as a neurochristopathy. Pediatr. Surg. Int. 1997;12:2–10. doi: 10.1007/BF01194793. [DOI] [PubMed] [Google Scholar]
- 6.McKeown S.J., Stamp L., Hao M.M., Young H.M. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:113–129. doi: 10.1002/wdev.57. [DOI] [PubMed] [Google Scholar]
- 7.Lake J.I., Heuckeroth R.O. Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G1–24. doi: 10.1152/ajpgi.00452.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emison E.S., McCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., Portnoy M.E., Cutler D.J., Green E.D., Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 9.Borrego S., Ruiz-Ferrer M., Fernandez R.M., Antinolo G. Hirschsprung's disease as a model of complex genetic etiology. Histol. Histopathol. 2013;28:1117–1136. doi: 10.14670/HH-28.1117. [DOI] [PubMed] [Google Scholar]
- 10.Wallace A.S., Anderson R.B. Genetic interactions and modifier genes in Hirschsprung's disease. World J. Gastroenterol. 2011;17:4937–4944. doi: 10.3748/wjg.v17.i45.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang R., Torigoe D., Sasaki N., Agui T. QTL analysis identifies a modifier locus of aganglionosis in the rat model of Hirschsprung disease carrying Ednrb(sl) mutations. PLoS ONE. 2011;6:e27902. doi: 10.1371/journal.pone.0027902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svingen T., Koopman P. Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev. 2013;27:2409–2426. doi: 10.1101/gad.228080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowles J., Schepers G., Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 14.Bergstrom D.E., Young M., Albrecht K.H., Eicher E.M. Related function of mouse SOX3, SOX9, and SRY HMG domains assayed by male sex determination. Genesis. 2000;28:111–124. [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y.M., Zheng M., Lau Y.-F.C. The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Reports. 2014;8:723–733. doi: 10.1016/j.celrep.2014.06.055. [DOI] [PubMed] [Google Scholar]
- 16.Leon T.Y., Ngan E.S., Poon H.C., So M.T., Lui V.C., Tam P.K., Garcia-Barcelo M.M. Transcriptional regulation of RET by Nkx2–1, Phox2b, Sox10, and Pax3. J. Pediatr. Surg. 2009;44:1904–1912. doi: 10.1016/j.jpedsurg.2008.11.055. [DOI] [PubMed] [Google Scholar]
- 17.Lang D., Chen F., Milewski R., Li J., Lu M.M., Epstein J.A. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J. Clin. Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang D., Epstein J.A. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum. Mol. Genet. 2003;12:937–945. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- 19.Lui V.C., Samy E.T., Sham M.H., Mulligan L.M., Tam P.K. Glial cell line-derived neurotrophic factor family receptors are abnormally expressed in aganglionic bowel of a subpopulation of patients with Hirschsprung's disease. Lab. Invest. 2002;82:703–712. doi: 10.1097/01.lab.0000017364.13014.ae. [DOI] [PubMed] [Google Scholar]
- 20.Schouten W.R., ten Kate F.J., de Graaf E.J., Gilberts E.C., Simons J.L., Kluck P. Visceral neuropathy in slow transit constipation: an immunohistochemical investigation with monoclonal antibodies against neurofilament. Dis. Colon Rectum. 1993;36:1112–1117. doi: 10.1007/BF02052258. [DOI] [PubMed] [Google Scholar]
- 21.Deguchi E., Iwai N., Goto Y., Yanagihara J., Fushiki S. An immunohistochemical study of neurofilament and microtubule-associated Tau protein in the enteric innervation in Hirschsprung's disease. J. Pediatr. Surg. 1993;28:886–890. doi: 10.1016/0022-3468(93)90688-h. [DOI] [PubMed] [Google Scholar]
- 22.de Arruda Lourencao P.L., Takegawa B.K., Ortolan E.V., Terra S.A., Rodrigues M.A. A useful panel for the diagnosis of Hirschsprung disease in rectal biopsies: calretinin immunostaining and acetylcholinesterase histochemistry. Ann. Diagn. Pathol. 2013;17:352–356. doi: 10.1016/j.anndiagpath.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Morris M.I., Soglio D.B., Ouimet A., Aspirot A., Patey N. A study of calretinin in Hirschsprung pathology, particularly in total colonic aganglionosis. J. Pediatr. Surg. 2013;48:1037–1043. doi: 10.1016/j.jpedsurg.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Barcelo M., Ganster R.W., Lui V.C., Leon T.Y., So M.T., Lau A.M., Fu M., Sham M.H., Knight J., Zannini M.S., et al. TTF-1 and RET promoter SNPs: regulation of RET transcription in Hirschsprung's disease. Hum. Mol. Genet. 2005;14:191–204. doi: 10.1093/hmg/ddi015. [DOI] [PubMed] [Google Scholar]
- 25.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., McCallion A.S. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 26.Bondurand N., Sham M.H. The role of SOX10 during enteric nervous system development. Dev. Biol. 2013;382:330–343. doi: 10.1016/j.ydbio.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J., Garcia-Barcelo M.M., Tam P.K., Lui V.C. HOXB5 cooperates with NKX2–1 in the transcription of human RET. PLoS ONE. 2011;6:e20815. doi: 10.1371/journal.pone.0020815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coward P., Nagai K., Chen D., Thomas H.D., Nagamine C.M., Lau Y.F. Polymorphism of a CAG trinucleotide repeat within Sry correlates with B6.YDom sex reversal. Nat. Genet. 1994;6:245–250. doi: 10.1038/ng0394-245. [DOI] [PubMed] [Google Scholar]
- 29.Su H., Lau Y.F. Identification of the transcriptional unit, structural organization, and promoter sequence of the human sex-determining region Y (SRY) gene, using a reverse genetic approach. Am. J. Hum. Genet. 1993;52:24–38. [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Oh H.J., Lau Y.F. The poly(ADP-ribose) polymerase 1 interacts with Sry and modulates its biological functions. Mol. Cell. Endocrinol. 2006;257–258:35–46. doi: 10.1016/j.mce.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Oh H.J., Li Y., Lau Y.F. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol. Reprod. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y., Batalao A., Zhou H., Zhu L. Mammalian two-hybrid system: a complementary approach to the yeast two-hybrid system. Biotechniques. 1997;22:350–352. doi: 10.2144/97222pf02. [DOI] [PubMed] [Google Scholar]
- 33.Tam P.K., Garcia-Barcelo M. Molecular genetics of Hirschsprung's disease. Semin. Pediatr. Surg. 2004;13:236–248. doi: 10.1053/j.sempedsurg.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Levine M., Cattoglio C., Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodrich J.A., Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 2010;11:549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewing P., Chiang C.W., Sinchak K., Sim H., Fernagut P.O., Kelly S., Chesselet M.F., Micevych P.E., Albrecht K.H., Harley V.R., et al. Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 38.Czech D.P., Lee J., Sim H., Parish C.L., Vilain E., Harley V.R. The human testis-determining factor SRY localizes in midbrain dopamine neurons and regulates multiple components of catecholamine synthesis and metabolism. J. Neurochem. 2012;122:260–271. doi: 10.1111/j.1471-4159.2012.07782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer A., Mosler G., Just W., Pilgrim C., Reisert I. Developmental profile of Sry transcripts in mouse brain. Neurogenetics. 2000;3:25–30. doi: 10.1007/s100480000093. [DOI] [PubMed] [Google Scholar]
- 40.Milsted A., Underwood A.C., Dunmire J., DelPuerto H.L., Martins A.S., Ely D.L., Turner M.E. Regulation of multiple renin-angiotensin system genes by Sry. J. Hypertens. 2010;28:59–64. doi: 10.1097/HJH.0b013e328332b88d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyer A., Pilon N., Raiwet D.L., Lussier J.G., Silversides D.W. Human and pig SRY 5′ flanking sequences can direct reporter transgene expression to the genital ridge and to migrating neural crest cells. Dev. Dyn. 2006;235:623–632. doi: 10.1002/dvdy.20670. [DOI] [PubMed] [Google Scholar]
- 42.Jain S., Naughton C.K., Yang M., Strickland A., Vij K., Encinas M., Golden J., Gupta A., Heuckeroth R., Johnson E.M., Jr., et al. Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development. 2004;131:5503–5513. doi: 10.1242/dev.01421. [DOI] [PubMed] [Google Scholar]
- 43.Cosma M.P., Cardone M., Carlomagno F., Colantuoni V. Mutations in the extracellular domain cause RET loss of function by a dominant negative mechanism. Mol. Cell. Biol. 1998;18:3321–3329. doi: 10.1128/mcb.18.6.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cosma M.P., Panariello L., Quadro L., Dathan N.A., Fattoruso O., Colantuoni V. A mutation in the RET proto-oncogene in Hirschsprung's disease affects the tyrosine kinase activity associated with multiple endocrine neoplasia type 2A and 2B. Biochem. J. 1996;314:397–400. doi: 10.1042/bj3140397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Groner A.C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Denervaud N., Bucher P., Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng B., Ren X., Kerppola T.K. KAP1 represses differentiation-inducible genes in embryonic stem cells through cooperative binding with PRC1 and derepresses pluripotency-associated genes. Mol. Cell. Biol. 2014;34:2075–2091. doi: 10.1128/MCB.01729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beneke S. Regulation of chromatin structure by poly(ADP-ribosyl)ation. Front. Genet. 2012;3:169. doi: 10.3389/fgene.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas C., Tulin A.V. Poly-ADP-ribose polymerase: machinery for nuclear processes. Mol. Aspects Med. 2013;34:1124–1137. doi: 10.1016/j.mam.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lui V.C., Cheng W.W., Leon T.Y., Lau D.K., Garcia-Barcelo M.M., Miao X.P., Kam M.K., So M.T., Chen Y., Wall N.A., et al. Perturbation of hoxb5 signaling in vagal neural crests down-regulates ret leading to intestinal hypoganglionosis in mice. Gastroenterology. 2008;134:1104–1115. doi: 10.1053/j.gastro.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 50.Shen L., Pichel J.G., Mayeli T., Sariola H., Lu B., Westphal H. Gdnf haploinsufficiency causes Hirschsprung-like intestinal obstruction and early-onset lethality in mice. Am. J. Hum. Genet. 2002;70:435–447. doi: 10.1086/338712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abel K.M., Drake R., Goldstein J.M. Sex differences in schizophrenia. Int. Rev. Psychiatry. 2010;22:417–428. doi: 10.3109/09540261.2010.515205. [DOI] [PubMed] [Google Scholar]
- 52.Figueira M.L., Ouakinin S. Gender-related endocrinological dysfunction and mental disorders. Curr. Opin. Psychiatry. 2010;23:369–372. doi: 10.1097/YCO.0b013e3283399b86. [DOI] [PubMed] [Google Scholar]
- 53.Harrison P.J., Tunbridge E.M. Catechol-O-methyltransferase (COMT): a gene contributing to sex differences in brain function, and to sexual dimorphism in the predisposition to psychiatric disorders. Neuropsychopharmacology. 2008;33:3037–3045. doi: 10.1038/sj.npp.1301543. [DOI] [PubMed] [Google Scholar]
- 54.Isensee J., Witt H., Pregla R., Hetzer R., Regitz-Zagrosek V., Noppinger P.R. Sexually dimorphic gene expression in the heart of mice and men. J. Mol. Med. 2008;86:61–74. doi: 10.1007/s00109-007-0240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mendrek A., Stip E. Sexual dimorphism in schizophrenia: is there a need for gender-based protocols? Expert Rev. Neurother. 2011;11:951–959. doi: 10.1586/ern.11.78. [DOI] [PubMed] [Google Scholar]
- 56.Paus T. Sex differences in the human brain: a developmental perspective. Prog. Brain Res. 2010;186:13–28. doi: 10.1016/B978-0-444-53630-3.00002-6. [DOI] [PubMed] [Google Scholar]
- 57.Spring S., Lerch J.P., Henkelman R.M. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage. 2007;35:1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 58.Wu J.B., Chen K., Li Y., Lau Y.F., Shih J.C. Regulation of monoamine oxidase A by the SRY gene on the Y chromosome. FASEB J. 2009;23:4029–4038. doi: 10.1096/fj.09-139097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baron-Cohen S., Knickmeyer R.C., Belmonte M.K. Sex differences in the brain: implications for explaining autism. Science. 2005;310:819–823. doi: 10.1126/science.1115455. [DOI] [PubMed] [Google Scholar]
- 61.Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 62.Sarkar A., Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bortolato M., Chen K., Shih J.C. Monoamine oxidase inactivation: from pathophysiology to therapeutics. Adv. Drug Deliv. Rev. 2008;60:1527–1533. doi: 10.1016/j.addr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shih J.C., Chen K., Ridd M.J. Monoamine oxidase: from genes to behavior. Annu. Rev. Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volavka J., Bilder R., Nolan K. Catecholamines and aggression: the role of COMT and MAO polymorphisms. Ann. N Y Acad. Sci. 2004;1036:393–398. doi: 10.1196/annals.1330.023. [DOI] [PubMed] [Google Scholar]
- 66.Craig I.W. The role of monoamine oxidase A, MAOA, in the aetiology of antisocial behaviour: the importance of gene-environment interactions. Novartis Found. Symp. 2005;268:227–237. doi: 10.1002/0470010703.ch16. discussion 237–241, 242–253. [DOI] [PubMed] [Google Scholar]
- 67.Bortolato M., Godar S.C., Alzghoul L., Zhang J., Darling R.D., Simpson K.L., Bini V., Chen K., Wellman C.L., Lin R.C., et al. Monoamine oxidase A and A/B knockout mice display autistic-like features. Int. J. Neuropsychopharmacol. 2013;16:869–888. doi: 10.1017/S1461145712000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jonsson E.G., Norton N., Forslund K., Mattila-Evenden M., Rylander G., Asberg M., Owen M.J., Sedvall G.C. Association between a promoter variant in the monoamine oxidase A gene and schizophrenia. Schizophr. Res. 2003;61:31–37. doi: 10.1016/s0920-9964(02)00224-4. [DOI] [PubMed] [Google Scholar]
- 69.Li Y., Tabatabai Z.L., Lee T.L., Hatakeyama S., Ohyama C., Chan W.Y., Looijenga L.H., Lau Y.F. The Y-encoded TSPY protein: a significant marker potentially plays a role in the pathogenesis of testicular germ cell tumors. Hum. Pathol. 2007;38:1470–1481. doi: 10.1016/j.humpath.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan X., Lu M.L., Li T., Balk S.P. SRY interacts with and negatively regulates androgen receptor transcriptional activity. J. Biol. Chem. 2001;276:46647–46654. doi: 10.1074/jbc.M108404200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.