Abstract
Pathological aggregation and mutation of the 43-kDa TAR DNA-binding protein (TDP-43) are strongly implicated in the pathogenesis amyotrophic lateral sclerosis and frontotemporal lobar degeneration. TDP-43 neurotoxicity has been extensively modeled in mice, zebrafish, Caenorhabditis elegans and Drosophila, where selective expression of TDP-43 in motoneurons led to paralysis and premature lethality. Through a genetic screen aimed to identify genetic modifiers of TDP-43, we found that the Drosophila dual leucine kinase Wallenda (Wnd) and its downstream kinases JNK and p38 influenced TDP-43 neurotoxicity. Reducing Wnd gene dosage or overexpressing its antagonist highwire partially rescued TDP-43-associated premature lethality. Downstream of Wnd, the JNK and p38 kinases played opposing roles in TDP-43-associated neurodegeneration. LOF alleles of the p38b gene as well as p38 inhibitors diminished TDP-43-associated premature lethality, whereas p38b GOF caused phenotypic worsening. In stark contrast, disruptive alleles of Basket (Bsk), the Drosophila homologue of JNK, exacerbated longevity shortening, whereas overexpression of Bsk extended lifespan. Among possible mechanisms, we found motoneuron-directed expression of TDP-43 elicited oxidative stress and innate immune gene activation that were exacerbated by p38 GOF and Bsk LOF, respectively. A key pathologic role for innate immunity in TDP-43-associated neurodegeneration was further supported by the finding that genetic suppression of the Toll/Dif and Imd/Relish inflammatory pathways dramatically extended lifespan of TDP-43 transgenic flies. We propose that oxidative stress and neuroinflammation are intrinsic components of TDP-43-associated neurodegeneration and that the balance between cytoprotective JNK and cytotoxic p38 signaling dictates phenotypic outcome to TDP-43 expression in Drosophila.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease that preferentially impacts motoneurons. ALS typically follows an unrelenting course, leading to respiratory failure and death in 3–5 years. Sadly, there is no effective treatment for this disease.
Recent progress toward a treatment for ALS has been propelled by major breakthroughs in ALS genetics. Although the majority of ALS cases arise sporadically (sALS), roughly 10% of ALS cases are familial in nature (fALS), exhibiting a dominant pattern of inheritance. The first identified genetic cause of ALS, mutation of the superoxide dismutase 1 (SOD1) was identified in 1993 (1). SOD1 mutations account for ∼20% of fALS cases, impact SOD1 folding and aggregation potential, and are thought to confer various toxic gains of function (reviewed in 2). Transgenic expression of mutant SOD1 in either motoneurons or glia elicits motoneuron degeneration in mice and SOD1 transgenic rodents have been the principle mammalian model for exploring neuropathologic mechanisms in ALS for ∼20 years.
The seminal discovery that the RNA-binding protein 43-kDa TAR DNA-binding protein (TDP-43) is a major component of Ub-positive cytoplasmic inclusions in degenerating neurons of patients with sALS, or the ALS spectrum disease frontotemporal lobar degeneration (FTLD) (3), reshaped thinking about ALS pathogenesis. Subsequent discoveries that mutations in TDP-43 (4–9) and a second RNA-binding proteins, FUS/TLS (fused in sarcoma/translocated in liposarcoma), cause familial forms of ALS (10,11), pointed toward neuronal RNA metabolic defects as critical components of ALS. Another landmark discovery that hexanucleotide GGGGCC (G4C2) repeat expansions in the 3′UTR of C9ORF72 are responsible for ∼40% of fALS cases is also consistent with a critical role for altered RNA metabolism (12,13). G4C2-expanded C9ORF72 transcripts form intranuclear aggregates and may promote toxicity, in part, through sequestration of key RBPs (14–16) and ATG-independent translation of toxic dipeptides (17–19).
TDP-43 is a nuclear protein that comprises two RNA recognition motifs (RRM) and an unstructured, Gly-rich, C-terminal domain where most of the ∼50 different ALS mutations reside. The Gly-rich motif exhibits prion-like characteristics and is thought to mediate protein–protein interactions (20). Although it remains unclear how ALS-associated mutations in TDP-43 instigate disease, such mutations have been reported to increase its stability (21) and aggregation potential (22), and alter TDP-43 splicing activity in vivo (23). TDP-43 preferentially binds to (UG)n repeats and participates in mRNA splicing, including alternative exon skipping (24–26). Genome wide analyses have defined a landscape of several thousand TDP-43 regulated substrates and revealed that TDP-43 is enriched deep within large introns that are characteristic of many neuron-specific genes (24,25). TDP-43 also negatively regulates its own RNA (27), leading to a model whereby cytosolic aggregation of TDP-43 leads to increased translation of TDP-43 message, feedforward TDP-43 aggregation, and ultimately, depletion of essential TDP-43 splicing functions in the nucleus. Indeed, nuclear clearing of TDP-43 is frequently observed in degenerating motoneurons of ALS/FTLD patients (8). Although its splicing functions have garnered the most attention, TDP-43 has also been implicated in transcription regulation (28,29), microRNA processing (30) and regulation of stress granule (SG) formation (31,32). Alterations in any or all these processes may contribute to neurodegeneration in ALS or FTD.
TDP-43 neurotoxicity has been modeled in rodents, C. elegans, zebrafish and Drosophila. A common denominator of these studies is that overexpression of either wild-type or mutant TDP-43 proteins elicits ALS-like phenotypes including impaired motor function and dramatic lifespan reduction (7,33–37). In Drosophila, selective expression of TDP-43 in motoneurons leads to axonal swelling, neuromuscular junction defects, age-dependent paralysis and dramatic lifespan reduction (reviewed in 38). TDP-43 maintains a largely nuclear distribution pattern when overexpressed in Drosophila, though cytoplasmic inclusions and insoluble TDP-43 aggregates have been reported and may contribute to toxicity (22,36,39,40). In keeping with the theme of altered RNA metabolism, an intact RRM is required for TDP-43 to elicit full toxicity in Drosophila (41,42).
A number of laboratories have identified genes that modify the severity of TDP-43-associated phenotypes in Drosophila. Overexpression of intermediate polyglutamine (polyQ) expanded versions of Ataxin 2 worsened TDP-43 toxicity and supported PolyQ Ataxin 2 expansions as an independent risk factor for ALS (43). Reducing gene dosage of Ataxin 2 (43), the endoplasmic reticulum calcium channel ITPR1 (44) or the nucleoporin Nup50 (45), reduced TDP-43 toxicity, though the mechanisms are not clear. More recently, it was shown that downregulation of the cytoplasmic polyA-binding protein or other constituents of cytoplasmic SGs, partially offsets TDP-43 neurotoxicity (46). SGs containing depots of mRNAs and temporarily stalled ribosomes are also being carefully scrutinized for their involvement in ALS caused by mutations in FUS (reviewed in 47).
A seemingly important yet relatively unexplored aspect of TDP-43 pathogenesis concerns its relationship to the cell stress response. Among the best-studied stress response pathways are those governed by the stress-activated protein kinases (SAPKs), p38 and JNK. p38 and JNK are activated in response to a wide variety of stress stimuli, including heat shock, osmotic challenge, protein synthesis inhibition, DNA damage and viral infection and orchestrate an equally varied cellular response impacting development, protein translation, apoptosis, cell cycle, as well as oxidative stress response and immune activation (reviewed in 48). Aberrant activation of JNK and p38 has been implicated in a broad range of neurodegenerative disorders (49,50), including ALS (51,52). Both JNK and p38 serve as effector kinases downstream of the MAP3K Wnd/DLK (dual leucine kinase), whose context-dependent activation regulates neuronal injury repair (53), synaptic structure remodeling (54) and axonal degeneration (55).
A hallmark of many neurodegenerative diseases is the expression of inflammatory markers within affected regions of the CNS. The potential cause and effect relationship between neuroinflammation and neurological damage in ALS and other neurodegenerative diseases has been heavily scrutinized (reviewed in 56). Frakes et al. showed that of NF-κB-mediated microglia activation contributed to motoneuron death in SOD1G93A mice (57). Additionally, extracellular mutant SOD1 was reported to trigger IL-1β release from activated microglia (58), further supporting a role for inflammatory changes in this ALS model. In contrast, the relationship between TDP-43 proteinopathy and neuroinflammation is largely unknown.
Here we report evidence that neuroinflammation is a prominent feature of TDP-43-associated neurodegeneration in Drosophila and that genetic suppression of the innate immune response conferred strong phenotypic rescue in TDP-43 transgenic flies. We also provide evidence that p38 and JNK SAPKs play important yet opposing roles in TDP-43-induced neurodegeneration in Drosophila and that p38 and JNK exert their influences, at least in part, through modulating pathologic oxidative stress and neuroinflammation. The potential relevance of these findings to human ALS is discussed.
RESULTS
Deficiency screening for TDP-43 modifier genes in Drosophila
In order to reveal genetic components potentially linked to TDP-43 neurotoxicity, we performed a genetic deficiency screen using a subset of the Bloomington deficiency kit (59). Selected deficiency stocks harboring a defined deletion of ∼50–200 genes were crossed to flies expressing full-length human UAS (upstream activating sequence)-TDP-43 transgene under the control of D42-Gal4 motoneuron-specific driver (hereafter referred to D42>TDP-43 flies) (45). Because D42>TDP-43 flies reproducibly die between 3 and 4 weeks of age, we used lifespan extension as a phenotypic readout for our primary screen, using a >10% longevity increase as an arbitrary cutoff. We randomly screened 29 non-overlapping deficiencies covering parts of the second and third chromosomes with a total coverage of ∼3000 genes. Among these, deficiency Df(3L)XS533 caught our attention by conferring at least two-fold increase in median survival (MS; Supplementary Material, Fig. S1A and B). Df(3L)XS533 did not extend the lifespan of D42-Gal4 control flies, but rather reduced the MS by half (Supplementary Material, Fig. S1C).
Df(3L)XS533 spans a large cytological segment on the third chromosome from 76B4 to 77B1 containing over 100 genes. To reduce the number of candidates, we surveyed 10 smaller deficiencies spanning 76B4 to 77B1 (Supplementary Material, Fig. S2A–E). Deficiency Df(3L)ED229, Df(3L)Exel9009 and Df(3L)BSC445 were among the strongest suppressors, increasing median longevity of D42>TDP-43 flies by 27–31% (Supplementary Material, Fig. S2B). Df(3L)Exel9009 and Df(3L)BSC445 are both contained within Df(3L)ED229 but do not overlap with each other, suggesting the possible existence of two or more modifier genes within the 76B9-76D5 cytological interval (Supplementary Material, Table S1). Intermediate suppressors included Df(3L)BSC734, Df(3L)BSC417 and Df(3L)BSC731(Supplementary Material, Fig. S2C), whereas (3L)BSC796, Df(3L)BSC830 and Df(3L)BSC839 (Supplementary Material, Fig. S2D) had little or no impact. A caveat to deficiency mapping in the presence of Gal4 is the potential for non-specific genetic interactions caused by residual UAS elements left near the breakpoint of the deletion segment (60). In fact, Df(3L)Exel6135 deficiency was lethal when crossed with D42>TDP-43 (Supplementary Material, Fig. S2A) and rendered an eyeless-phenotype when crossed to the eye-specific driver GMR-Gal4 (Supplementary Material, Fig. S2E), probably due to this mechanism. However, two pieces of evidence argue against this as a major explanation for our findings. First, multiple deficiencies covering the proximal region (Supplementary Material, Fig. S2B) showed strong phenotypic rescue. Second, within the interval of interest, Df(3L)ED229, which does not contain UAS, also conferred phenotypic rescue (Supplementary Material, Fig. S2B). Thus, the most straightforward explanation for the findings is that interval 76B4 to 77B1 contains one or more TDP-43 modifier genes.
Wnd promoted TDP-43 neurotoxicity
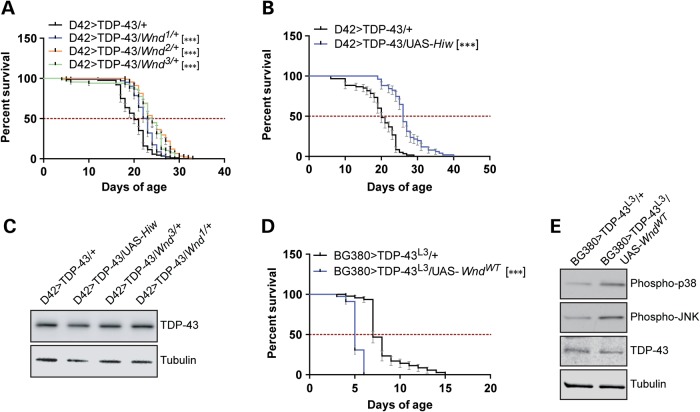
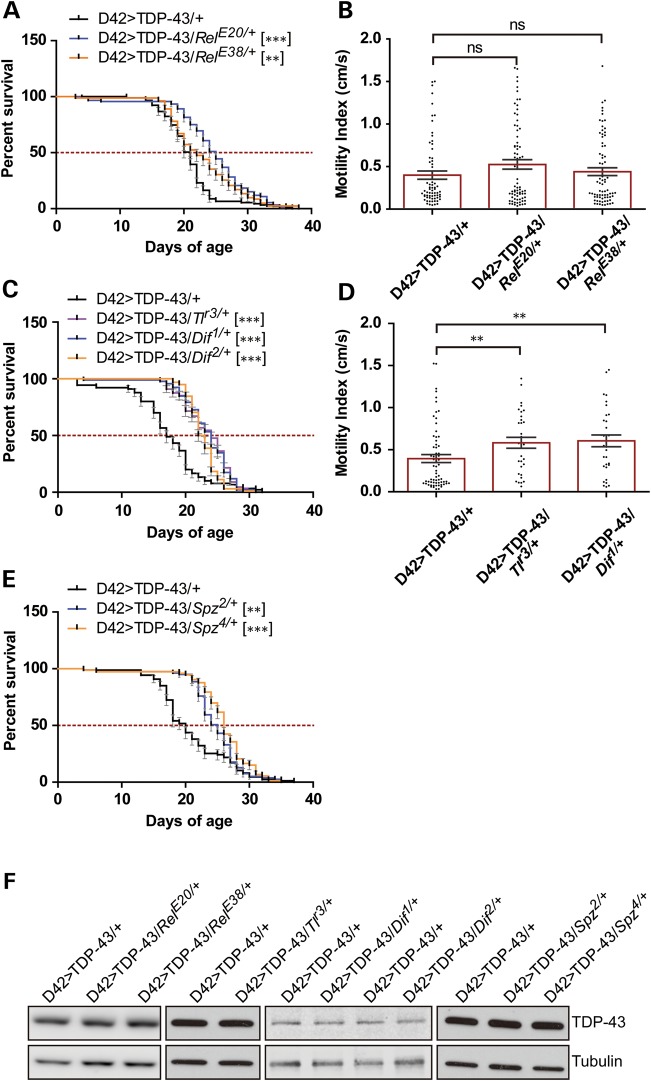
We initially focused on the 23 genes within the Df(3L)Exel9009 deficiency as candidates for TDP-43 modifiers. Among these, the MAP3K Wnd, which modulates neuromuscular junction development (61) and axonal injury repair (53,62), was of considerable interest. We therefore crossed the D42>TDP-43 flies to three different Wnd loss-of-function (LOF) mutants (61) and found that each of the Wnd alleles extended lifespan of D42>TDP-43 flies from 15 to 20% (Fig. 1A) without increasing lifespan of D42-Gal4 controls (Supplementary Material, Fig. S3A). The ubiquitin E3 ligase highwire (Hiw) regulates Wnd through proteasomal clearance (61). Overexpression of Hiw phenocopied Wnd null mutations, causing 30% increase of median longevity (Fig. 1B). Neither Wnd LOF alleles nor overexpression of Hiw altered expression of the TDP-43 transgene (Fig. 1C). A Wnd1/3compound mutant did not show any further rescue (Supplementary Material, Fig. S3B) but rather shortened longevity of D42>TDP-43 flies. This outcome may reflect critical roles for Wnd in neuron development and synaptogenesis (61).
Figure 1.
Loss-of-function mutations in Wnd reduce TDP-43 neurotoxicity. (A) Survival curve of D42>TDP-43 flies crossed to heterozygous Wnd LOF alleles. Median survival (MS) and number of animals used (N) for each genotype were: D42>TDP-43/+ (MS = 20 days, N = 88); D42>TDP-43/Wnd1/+ (MS = 23 days, N = 103); D42>TDP-43/Wnd2/+ (MS = 24 days, N = 91); D42>TDP-43/Wnd3/+ (MS = 24 days, N = 67). (B) Survival curve of D42>TDP-43 flies overexpressing wild-type Highwire (UAS-Hiw). MS and N for each genotype were: D42>TDP-43/+ (MS = 20 days, N = 60); D42>TDP-43/UAS-Hiw (MS = 26 days, N = 51). (C) Western blot analysis showing TDP-43 expression level in heads from D42>TDP-43 flies crossed to UAS-Hiw, Wnd3 or Wnd1. Tubulin was used as loading control. (D) Survival curve of BG380>TDP-43L3 female flies (containing Tub-Gal80ts) overexpressing wild-type Wnd (UAS-WndWT). The cross was performed at 18°C and newly eclosed flies shifted to 30°C at Day 0 of the survival assay. MS and N for each genotype were: BG380>TDP-43L3/+ (MS = 7 days, N = 45); BG380>TDP-43L3/UAS-WndWT (MS = 5 days, N = 78). (E) Western blot analysis showing TDP-43 expression level as well as p38 phosphorylation (equivalent to vertebrate Thr180/Tyr182) and JNK (Bsk) phosphorylation level (equivalent to vertebrate Thr183/Tyr185) in head tissues from BG380>TDP-43L3/+ and BG380>TDP-43L3/UAS-WndWT flies after incubation at 30°C for 4 days. Tubulin was used as loading control. Note: Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
In order to circumvent the potentially confounding effects of Wnd LOF during development, we crossed UAS-WndWT flies with a recombinant line that expresses temperature sensitive inhibitory Gal80ts (Tub>Gal80ts) (reviewed in 16) and a high-expressing UAS-TDP43L3 transgene. By crossing the F1 flies to the BG380-Gal4 motoneuron driver, we were able to induce concomitant expression of TDP-43 and Wnd in adult motoneurons by shifting to the permissive temperature (30°C). In keeping with the idea that Wnd promoted TDP-43 neurotoxicity, overexpression of wild-type Wnd dramatically shortened lifespan of BG380>TDP-43 flies (Fig. 1D) without impacting TDP-43 expression (Fig. 1E). Taken together, our results suggested that TDP-43 neurotoxic GOF was at least partially mediated by Wnd signaling.
Differential impacts of p38 and JNK/Bsk on TDP-43-mediated toxicity
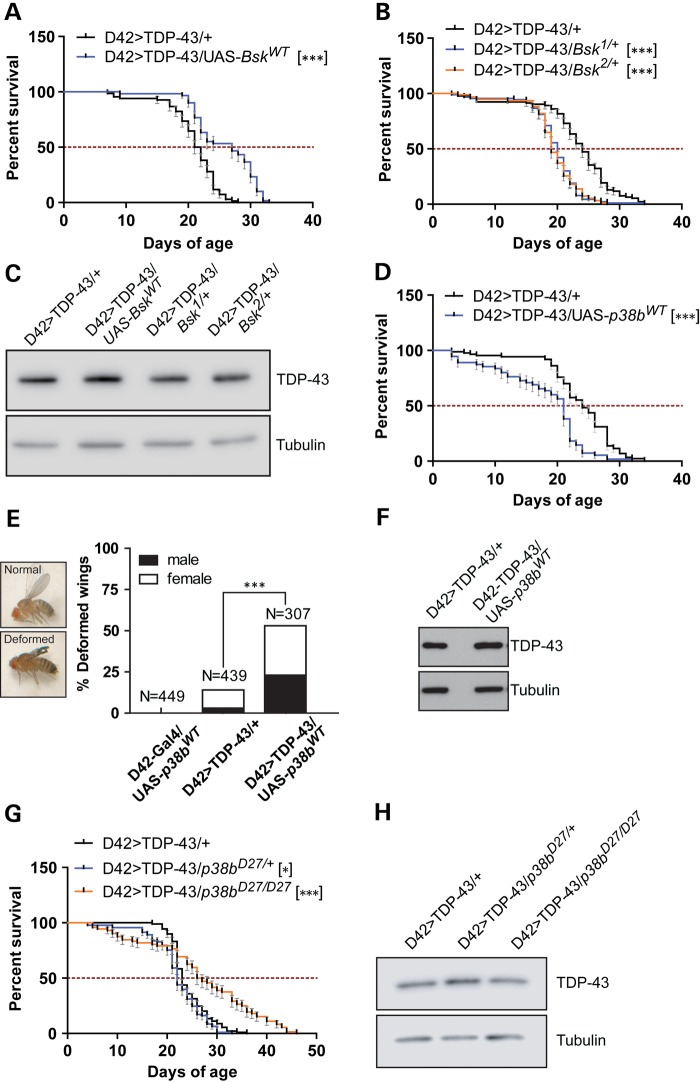
Wnd functions upstream of the SAPKs JNK and p38 (53,54). Indeed, overexpression of Wnd in fly motoneurons was sufficient to increase in JNK and p38 phosphorylation (Fig. 1E). To explore the contributions of JNK and p38 to TDP-43-induced neurodegeneration, we crossed D42>TDP-43 flies to a transgenic line that expresses the Drosophila JNK homologue Basket (Bsk) under UAS control. Expression of wild-type Bsk in motoneurons increased D42>TDP-43 fly longevity by 26% without impacting TDP-43 expression (Fig. 2A and C). On the other hand, heterozygous Bsk null alleles significantly reduced lifespan of D42>TDP-43 flies (Fig. 2B and C). Thus, JNK signaling downstream of Wnd appears neuroprotective in the D42>TDP-43 model.
Figure 2.
Differential impacts of SAPKs JNK/Bsk and p38 on TDP-43-induced neurotoxicity. (A) Survival curve of D42>TDP-43 flies overexpressing Drosophila wild-type JNK (UAS-BskWT). MS and N for each genotype were: D42>TDP-43/+ (MS = 21.5 days, N = 68); D42>TDP-43/UAS-BskWT (MS = 27 days, N = 60). (B) Survival curve of D42>TDP-43 flies crossed to heterozygous LOF Bsk alleles. MS and N for each genotype were: D42>TDP-43/+ (MS = 24 days, N = 93); D42>TDP-43/Bsk1/+ (MS = 20 days, N = 90); D42>TDP-43/Bsk2/+ (MS = 19 days, N = 94). (C) Western blot analysis for TDP-43 expression level in head tissues from D42>TDP-43 flies crossed to UAS-BskWT and Bsk mutant background. Tubulin was used as loading control. (D) Survival curve of D42>TDP-43 flies overexpressing wild-type p38b (UAS-p38bWT). MS and N for each genotype were: D42>TDP-43/+ (MS = 24 days, N = 87); D42>TDP-43/UAS-p38WT (MS = 21 days, N = 55). (E) Overexpression of p38bWT exacerbated wing deformation in D42>TDP-43 flies. Percentage of adult flies showing abnormal wing phenotype was scored and separated by gender. (F) Western blot analysis for TDP-43 expression level in head tissues from D42>TDP-43 flies crossed to UAS-p38bWT. Tubulin was used as loading control. (G) Survival curve of D42>TDP-43 flies containing heterozygous or homozygous LOF p38bD27 allele. MS and N for each genotype were: D42>TDP-43/+ (MS = 23 days, N = 89); D42>TDP-43/p38bD27/+(MS = 22 days, N = 93); D42>TDP-43/p38bD27/D27(MS = 26.5 days, N = 72). (H) Western blot analysis of TDP-43 expression level in head tissues collected from flies crossed to p38bD27 allele. Tubulin was used as loading control. Note: Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
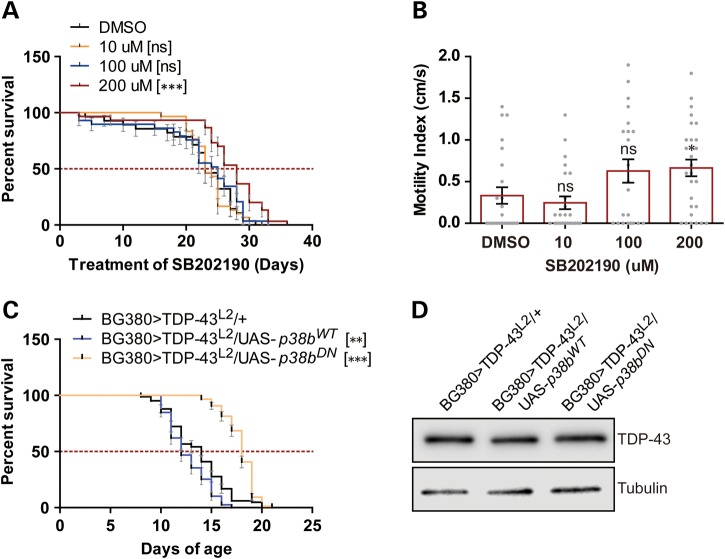
The Drosophila genome encodes three different p38 genes (p38a, p38b and p38c) with shared and unique functions (63). Interestingly, in stark contrast to the JNK/Bsk overexpression findings, overexpression of wild-type p38b in motoneurons significantly shortened lifespan of D42>TDP-43 flies (Fig. 2D) and increased the frequency of wing defects (Fig. 2E) that we and others have previously described (35,64) with no detectable increase of TDP-43 level (Fig. 2F). Homozygous deletion of p38b yielded the opposite phenotype, significantly extending the lifespan of D42>TDP-43 flies (Fig. 2G and H). The contribution of p38 signaling to TDP-43 toxicity was further investigated using small molecule p38 inhibitors. Male D42>TDP-43 flies were treated with vehicle or the p38 inhibitor SB202190 (0–200 μm) from the first day of eclosion onward. At the highest dose tested (200 μm), a significant 21% increase in MS over DMSO-treated controls was observed (Fig. 3A). The p38 inhibitor also modestly enhanced the climbing performance in aged D42>TDP-43 flies (Fig. 3B). Although less efficacious in motility improvement, a second p38 inhibitor SB203580 also extended lifespan of D42>TDP-43 flies at the highest dose tested (Supplementary Material, Fig. S4A and B).
Figure 3.
Pharmacological and genetic inhibition of p38 suppress TDP-43 toxicity during adulthood. (A) Survival curve of adult male D42>TDP-43 flies treated with the p38 inhibitor SB202190 at 10, 100 and 200 μm (concentration in food). Median survival (MS) and number of animals used (N) for each treatment condition were: DMSO (MS = 23 days, N = 28); 10 μm (MS = 23, N = 30); 100 μm (MS = 25 days, N = 29); 200 μm (MS = 28 days, N = 30). (B) Climbing performance of D42>TDP-43 flies treated with SB202190 for 20 days. MI and the number of animals used (N) for each treatment condition were: DMSO (MI = 0.3333 ± 0.09939 cm/s, N = 24); 10 μm (MI = 0.2458 ± 0.07684 cm/s, N = 24); 100 μm (MI = 0.6273 ± 0.1399 cm/s, N = 22); 200 μm (MI = 0.6643 ± 0.1004 cm/s, N = 28). Bar graph (mean ± SEM) was superimposed with scatter plot showing motility of each fly. (C) Survival curve of BG380>TDP-43L2 female flies (containing Tub>Gal80ts) overexpressing wild-type p38b (UAS-p38bWT) or dominant-negative p38b (UAS-p38bDN) flies. Cross was done in 18°C. Newly eclosed flies were then shifted to 30°C, marking the start of survival assay. MS and N for each genotype were: BG380>TDP-43L2/+ (MS = 14 days, N = 83); BG380>TDP-43L2/UAS-p38bWT(MS = 12 days, N = 79); BG380>TDP-43L2/UAS-p38bDN(MS = 18 days, N = 86). (D) Western blot analysis of TDP-43 expression level in head tissues from BG380>TDP-43L2 flies crossed to UAS-p38bWT or UAS-p38bDN. Flies were incubated at 30°C for 7 days before harvested for western blot. Tubulin was used as loading control. Note: Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
That chemical inhibition of p38 was beneficial suggested inhibiting p38 signaling during adulthood could be sufficient to repress TDP-43 mediated neurotoxicity. This idea was tested through use of a dominant-negative p38b (UAS-p38bDN) transgene under the control of temperature sensitive Gal80ts and the BG380-Gal4 motoneuron driver. After eclosion, adult flies were moved from 18 to 30°C to allow co-expression of TDP-43L2 and p38b transgenes in adult motoneurons. We found that overexpression of p38bDN increased MS, whereas overexpression of wild-type p38b decreased MS (Fig. 3C and D). Thus, in stark contrast to the JNK findings, genetic or pharmacologic interference with p38 attenuated TDP-43 toxicity in flies. In addition, these findings indicate genetic inhibition of p38 in neurons is sufficient to extend Drosophila lifespan.
TDP-43 missexpression leads to age-dependent oxidative stress that is exacerbated by Bsk LOF and p38b GOF
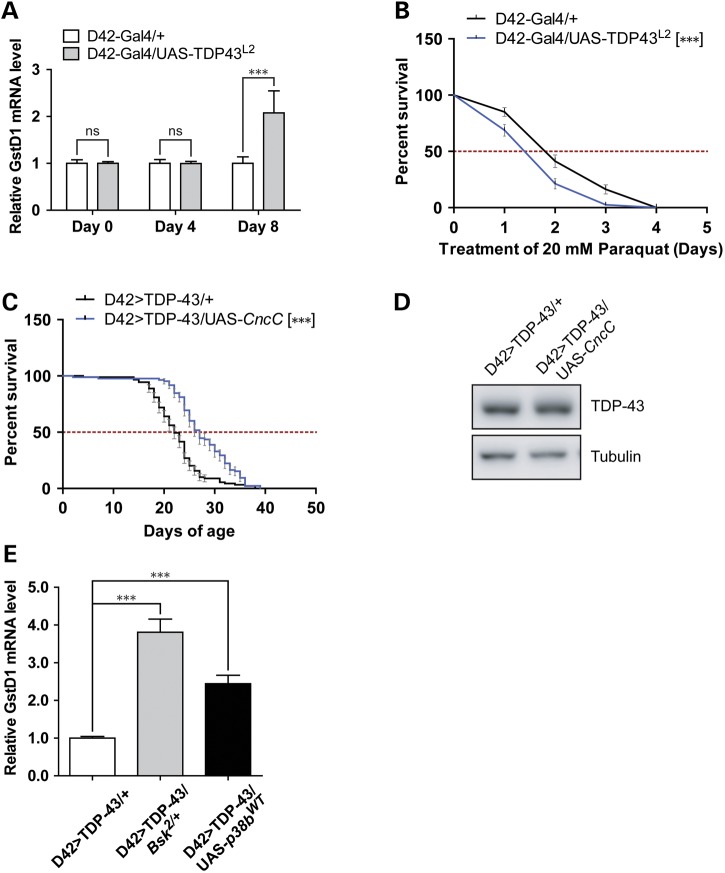
Both JNK (65) and p38 (66) kinases are required for proper oxidative stress responses in Drosophila. In light of the genetic interaction between JNK/p38 with TDP-43, we surveyed oxidative stress signaling in the TDP-43 missexpression model. Glutathione S transferase D1 (GstD1) is an oxidative responsive gene whose expression serves as a surrogate marker for reactive oxygen species (ROS) (67,68). To evaluate age-dependent oxidative stress, GstD1 mRNA levels from head extracts of newly eclosed flies (Day 0) or aged flies (Days 4 and 8) were measured by quantitative PCR (Fig. 4A). Whereas GstD1 mRNA expression was essentially unchanged in Day 0 and Day 4 D42>TDP-43 flies, GstD1 levels increased ∼2-fold in 8-day-old TDP-43 flies. Induction of GstD1 in neurons was also evidenced by higher expression of an integrated GFP reporter under control of the GstD1 promoter (67) (Supplementary Material, Fig. S5A and B). Finally, we observed increased protein carbonylation in TDP-43 expressing flies (Supplementary Material, Fig. S5C). Thus, head tissues of D42>TDP-43 flies exhibit age-dependent oxidative stress.
Figure 4.
TDP-43 overexpression in motoneurons elicits oxidative stress that is exacerbated by p38b GOF or Bsk LOF. (A) qPCR analysis measuring GstD1 mRNA level from head tissues of flies aged for 0 day, 4 days and 8 days. Levels of GstD1 in D42-Gal4/UAS-TDP-43L2 flies were compared with age-matched D42-Gal4/+ control flies (N = 4). (B) Survival curve of aged D42-Gal4/UAS-TDP-43L2 flies challenged with 20 mm paraquat. D42-Gal4/UAS-TDP-43L2 flies were aged for 10 days on normal media and then switched to paraquat containing media, marking the start of survival assay. A total of 80 flies for each genotype were used. (C) Survival curve of D42>TDP-43 flies overexpressing Cap-n-colarC (UAS-CncC). MS and N for each genotype were: D42>TDP-43/+ (MS = 22 days, N = 89); D42>TDP-43/UAS-CncC (MS = 27 days, N = 85). (D) Western blot analysis for TDP-43 expression level using head tissues from D42>TDP-43 flies crossed to UAS-CncC. Tubulin was used as loading control. (E) qPCR analysis measuring GstD1 mRNA level from head tissues of flies aged for 8 days. Levels of GstD1 in heterozygous Bsk deficient or p38b overexpressing D42>TDP-43 flies were compared with age-matched D42>TDP-43/+ control flies (N = 4). Note: Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
Increased oxidative stress levels in D42>TDP-43 flies implied these flies might have impaired defense against oxidative insult and/or show susceptibility to further oxidative insults. To test this idea, we challenged aged D42>TDP-43 flies with paraquat, a strong inducer of ROS. We observed significant decrease of survival in aged D42>TDP-43 flies (Day 10) but not newly eclosed flies (data not shown) when flies were subjected to 20 mm paraquat challenge (Fig. 4B). On the other hand, overexpression of Cap-n-colar (Cnc), a transcription factor which orchestrates transcriptional reprogramming to combat oxidative free radicals (67), increased survival (Fig. 4C and D). We also observed an even higher level of GstD1 in D42>TDP-43 flies that were Bsk haplodeficient or overexpressing UAS-p38bWT (Fig. 4E), indicating that the extent of oxidative stress correlates with reduced lifespan in these flies (Fig. 2B and D). These data suggested that, among other possible mechanisms, the impact of SAPK JNK/p38 signaling on TDP-43 neurotoxicity could be mediated through modulation of the oxidative stress response.
Expression of TDP-43 in motoneurons caused age- and dose-dependent neuroinflammation
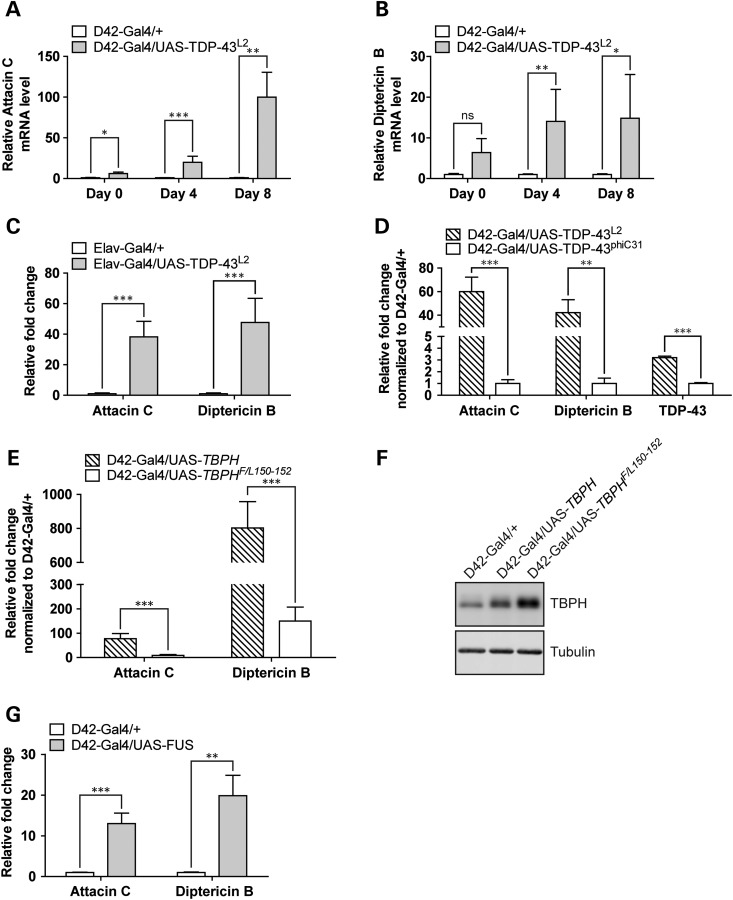
Among a myriad of other important cellular functions, JNK and p38 SAPKs are immune responsive kinases that regulate fly immunity (69–73). Given the genetic link between JNK/p38 and TDP-43, we next surveyed potential involvement of immune activation in TDP-43 pathology. Antimicrobial peptide (AMP) genes are an integral part of Drosophila humoral immunity (reviewed in 74). The AMP genes Attacin, Cecropin, Defensin, Diptericin and Drosomycin are strongly induced by infection and are frequently used as markers for neuroinflammation in Drosophila (75,76). Thus, to study neuroinflammatory response in the D42>TDP-43 flies, we measured transcriptional activation of AMP genes from fly head extracts using qPCR. Expression of TDP-43 in motoneurons strongly activated AMP genes in an age-dependent manner. For instance, relative to age-matched D42-Gal4/+ controls, D42>TDP-43/+ flies demonstrated 6-fold (Day 0), 20-fold (Day 4) and 100-fold (Day 8) increases in Attacin C expression (Fig. 5A). Although the magnitudes of induction differed, similar patterns were observed for Diptericin B (Fig. 5B), and the AMP genes Cecropin, Defensin and Drosomycin (Supplementary Material, Fig. S6A). Since the induction of Attacin C and Diptericin B was most stable among all five of the AMP genes tested, we used their expression as readout for neuroinflammation in subsequent experiments. Immune gene activation was demonstrated in independently generated TDP-43 transgenic lines (L1 and L4) and was not observed in flies overexpressing GFP (Supplementary Material, Fig. S6B), suggesting that protein overexpression in-and-of-itself is not sufficient to activate AMP genes. Finally, TDP-43-mediated neuroinflammation was not exclusive to the D42-Gal4 driver, as expression of TDP-43 under a second motoneuron driver BG380-Gal4 also caused substantial immune activation (Supplementary Material, Fig. S6B), as did expression under the Elav-Gal4 driver (Fig. 5C). By contrast, expression of TDP-43 under the glia-specific Repo-Gal4 driver weakly activated AMPs (Supplementary Material, Fig. S6D). Collectively, these results suggested that neuronal expression of TDP-43 was sufficient to cause neuroinflammation in an age-dependent manner. To further characterize TDP-43-mediated immune activation, we used a low expressing TDP-43 transgenic line that was generated through phiC31 integration (42). Compared with the UAS-TDP43L2 line that was used in our previous experiments, TDP-43 expression level was about two-thirds lower in the UAS-TDP43phiC31 line and corresponded with significant attenuation of immune responses (Fig. 5D), suggesting that TDP-43-mediated immune activation was dose dependent.
Figure 5.
Neuronal expression of TDP-43, TBPH and FUS induces antimicrobial peptide gene expression. (A) qPCR analysis measuring Attacin C mRNA level from head tissues of flies aged for 0 day, 4 days and 8 days. Expression changes in D42-Gal4/UAS-TDP-43L2 flies were compared with age-matched D42-Gal4/+ control flies with a total of four independent experiments. (B) Same as panel (A) except measuring Diptericin B mRNA level. (C) qPCR analysis measuring Attacin C and Diptericin B mRNA level from head tissues of Elav-Gal4/UAS-TDP-43L2 flies aged for 2 days. Expression changes in Elav-Gal4/UAS-TDP-43L2 flies were compared with age-matched Elav-Gal4/+ control flies with a total of four independent experiments. (D) qPCR analysis measuring Attacin C, Diptericin B and TDP-43 level from heads of D42-Gal4/UAS-TDP-43phiC31 or D42-Gal4/UAS-TDP-43L2 flies aged for 8 days. Expression changes were first normalized to age-matched D42-Gal4/+ control and induction of Attacin C and Diptericin B was compared between D42-Gal4/UAS-TDP-43phiC31 and D42-Gal4/UAS-TDP-43L2 flies in a total of four independent experiments. (E) qPCR analysis measuring Attacin C and Diptericin B level from head tissues of either D42-Gal4/UAS-TBPH or D42-Gal4/UAS-TBPHF/L150-152 flies aged for 8 days. Expression changes were first normalized to age-matched D42-Gal4/+ control. The induction of Attacin C and Diptericin B was compared between D42-Gal4/UAS-TBPH and D42-Gal4/UAS-TBPHF/L150-152 flies (N = 4). (F) Western blot analysis of TBPH protein expression level in flies used in (E). Tubulin was used as loading control. (G) qPCR analysis measuring Attacin C and Diptericin B mRNA level in heads of D42-Gal4/UAS-FUS flies aged for 4 days. Expression changes in D42-Gal4/UAS-TDP-43L2 flies were compared with age-matched D42-Gal4/+ control flies (N = 4). Note: Gene expression data were plotted with mean value. Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
Expression of TBPH and FUS in motoneurons induced AMP gene activation in aged flies
We next asked whether innate immunity was induced in other Drosophila ALS models. The Drosophila homologue of TDP-43, TBPH, is critical for appropriate synapse development and motor function in flies (77) and its missexpression caused severe motor defects, premature lethality and abnormal synapse formation (39,77). Remarkably, TBPH overexpression under the D42-Gal4 driver showed even stronger immune activation resulting 77-fold increase of Attacin C and 802-fold increase of Diptericin B in 8-day-old flies (Fig. 5E). An RNA-binding-defective mutant of TBPH, TBPHF/L150-152 (78), showed much reduced immune activation (Fig. 5E and F) in comparison to wild-type TBPH, suggesting that the hyperimmune activation mediated by TBPH overexpression was at least partially RNA-binding dependent.
Finally, to explore the generality of our findings to other ALS models, we measured Attacin C and Diptericin B levels in flies overexpressing the ALS-associated RNA-binding protein FUS/TLS (79) in motoneurons. Indeed, overexpression of wild-type FUS under the D42-Gal4 motoneuron driver caused strong immune activation (Fig. 5G). Altogether, these results demonstrated that neuronal expression of ALS disease-causing proteins TDP-43/TBPH and FUS induced strong immune activation in Drosophila, implicating neuroinflammation as a consistent marker of neurodegeneration in this model.
The immune modulatory Toll/Dif and Imd/Relish pathways contribute to TDP-43 neurotoxicity
To explore the pathologic role immune activation in D42>TDP-43 flies, we genetically disrupted the Imd/Relish pathway, which is required for AMP transcription (reviewed in 74). A heterozygous RelE20 Relish allele increased the longevity of D42>TDP-43 flies by ∼20% (Fig. 6A). A second Relish allele, RelE38, conferred a more modest lifespan extension (Fig. 6A). Neither of these alleles, however, rescued the motility defects that were usually manifested around Day 18 (Fig. 6B). Disrupting genes in the Toll/Dif (dorsal-related immunity factor) pathway, which functions parallel to the Imd/Relish pathway in AMP transcriptional activation also reduced TDP-43 toxicity. Toll receptor mutant allele Tlr3 increased MS of D42>TDP-43 flies by 41% (Fig. 6C), whereas Dif1 and Dif2 alleles extended lifespan by 41 and 44%, respectively (Fig. 6C). Genetic blockade of the Toll/Dif pathway also improved motility defects in animals aged at Day 18 (Fig. 6D).
Figure 6.
LOF mutations in the Imd/Relish and Toll/Dif pathways attenuate TDP-43-induced neurotoxicity. (A) Survival curve of D42>TDP-43 flies crossed to heterozygous Relish LOF alleles. MS and N for each genotype were: D42>TDP-43/+ (MS = 21 days, N = 91); D42>TDP-43/RelE20/+ (MS = 25 days, N = 92); D42>TDP-43/RelE38/+ (MS = 22 days, N = 82). (B) Climbing performance of D42>TDP-43 flies crossed to heterozygous Relish LOF alleles measured at Day 18. MI and the number of animals (N) used for each genotype were: D42>TDP-43/+ (MI = 0.40 ± 0.05 cm/s, N = 67); D42>TDP-43/RelE20/+ (MI = 0.52 ± 0.06 cm/s, N = 74); D42>TDP-43/RelE38/+ (MI = 0.44 ± 0.05 cm/s, N = 75). Bar graph (Mean ± SEM) was superimposed with scatter plot showing motility of each fly. (C) Survival curve of D42>TDP-43 flies crossed to heterozygous Toll and Dif LOF alleles. MS and N for each genotype were D42>TDP-43/+ (MS = 17 days, N = 90); D42>TDP-43/Tlr3/+ (MS = 24 days, N = 88); D42>TDP-43/Dif1/+ (MS = 24 days, N = 92); D42>TDP-43/Dif2/+ (MS = 22.5 days, N = 98). (D) Climbing performance of D42>TDP-43 flies crossed to heterozygous Toll and Dif LOF alleles measured at Day 18. MI and N for each genotype were: D42>TDP-43/+ (MI = 0.39 ± 0.05 cm/s, N = 66); D42>TDP-43/Tlr3/+ (MI = 0.58 ± 0.06 cm/s, N = 32); D42>TDP-43/Dif1/+ (MI = 0.60 ± 0.07 cm/s, N = 32). Bar graph (Mean ± SEM) was superimposed with scatter plot showing motility of each fly. (E) Survival curve of D42>TDP-43 flies crossed to heterozygous Spatzle LOF alleles. MS and N for each genotype were: D42>TDP-43/+ (MS = 20 days, N = 87); D42>TDP-43/Spz2/+ (MS = 25 days, N = 79); D42>TDP-43/Spz4/+ (MS = 26 days, N = 73). (F) Western blot analysis for TDP-43 expression level in head tissues from D42>TDP-43 flies crossed to different LOF alleles of the Imd/Relish and Toll/Dif pathway. Tubulin was used as loading control. Note: Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
To further evaluate the role of Toll/Dif pathway in TDP-43 neurotoxicity, we crossed D42>TDP-43 flies to harboring hypomorphic mutations in Spazle, a circulating cytokine important for activation of Toll-like receptors (80–82). Heterozygous Spatzle LOF alleles, Spz2 and Spz4, extended median longevity by 25 and 30%, respectively (Fig. 6E). Finally, none of the aforementioned alleles in either Toll/Dif or Imd/Relish pathway affect TDP-43 transgene expression (Fig. 6F). Taken together, these results strongly suggest that innate immune gene activation is an integral component of TDP-43 neurotoxicity in Drosophila.
Phenotypic rescue by TDP-43 modifier genes correlated with reduced neuroinflammation
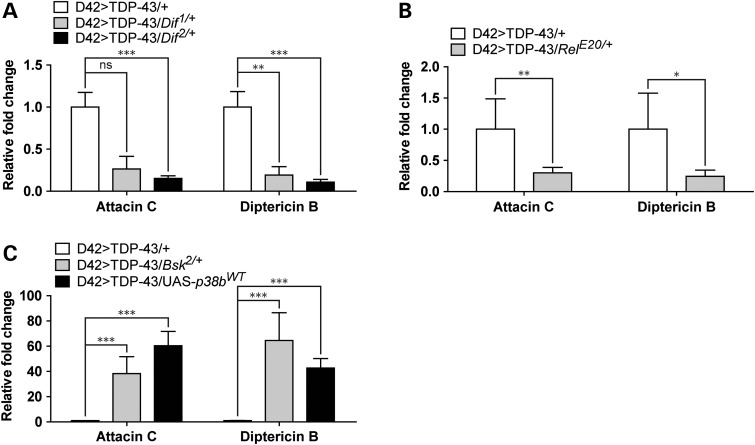
Since our data suggested that neuroinflammation was closely related to TDP-43 toxicity, we tested whether phenotypic rescue was accompanied by reduced expression of AMP genes. Consistent with this, D42>TDP-43 flies on the Dif1 background showed 3.7-fold reduction of Attacin C (P = 0.1) and 5.2-fold reduction of Diptericin B expression (Fig. 7A). Similar findings were made for the Dif2 and RelE20 alleles (Fig. 7A and B). Thus, phenotypic rescue by these alleles correlated with reduced expression of neuroinflammatory genes. On the other hand, the Bsk2 LOF mutation, which shortened lifespan of D42>TDP-43 flies, conferred a 33-fold increase of Attacin C and 37.5-fold increase of Diptericin B (Fig. 7C) versus D42>TDP-43 controls (Fig. 7C). Similarly, p38b overexpression caused a 51-fold increase of Attacin C and 32-fold increase in Diptericin B relative to D42>TDP-43 controls (Fig. 7C). These findings suggest that, in addition to influencing levels of oxidative stress, JNK and p38 kinases may differentially impact TDP-43 neurotoxicity through alterations in neuroinflammatory processes.
Figure 7.
Neuroinflammation triggered by TDP-43 missexpression requires Dif- and Rel-mediated transcription and is promoted by Bsk LOF and p38b GOF. (A) qPCR analysis measuring Attacin C and Diptericin B mRNA level from heads of 8-day-old D42>TDP-43/Dif1/+ and D42>TDP-43/Dif2/+ flies. Expression changes were compared with age-matched D42>TDP-43/+ control flies (N = 4). (B) qPCR analysis measuring Attacin C and Diptericin B mRNA levels from heads of 8-day-old D42>TDP-43/RelE20/+ flies and D42>TDP-43/+ control flies (N = 4). (C) qPCR analysis measuring Attacin C and Diptericin B mRNA level from heads of D42>TDP-43/Bsk2/+ and D42>TDP-43/UAS-p38bWT flies (N = 4). Note: Gene expression data were plotted with mean value. Error bar represented SEM. Statistical significance was summarized as asterisks shown on graph with detailed analyses listed in Supplementary Material, Table S4.
DISCUSSION
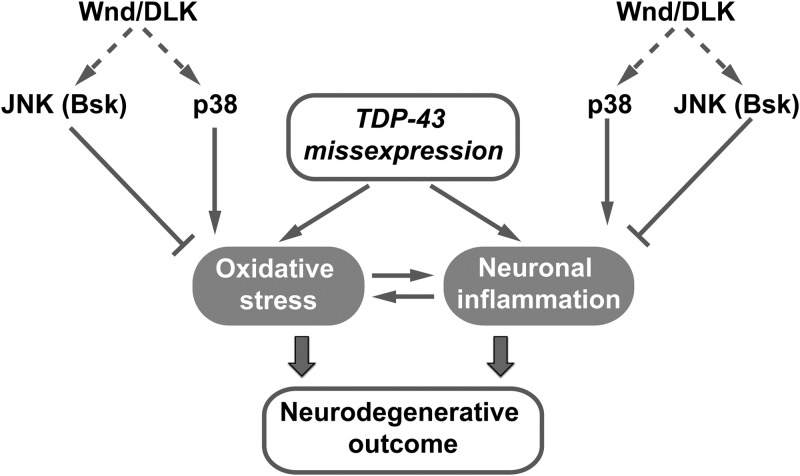
Through partial screening of a Drosophila deficiency library we identified the neuronal injury-signaling kinase Wnd/DLK and its downstream effector kinases p38 and Bsk/JNK as modifiers of TDP-43 elicited neurotoxicity. Our experiments further implicate oxidative stress and the innate immune response as key determinants of TDP-43-mediated toxicity in Drosophila motoneurons (Fig. 8). These findings support a growing literature implicating SAPKs as determinants of regenerative and degenerative responses to neuronal stress (reviewed in 83) and point toward application of anti-oxidative agents and/or immune suppressants as an avenue of therapeutic exploration in preclinical TDP-43 proteinopathy models.
Figure 8.
Working model summarizing opposing functions of JNK and p38 in TDP-43 neuropathogenesis through regulation of oxidative stress and neuroinflammation. In this model, TDP-43 missexpression in the CNS caused age-dependent oxidative stress and neuroinflammation. Wnd downstream effector kinases JNK/Bsk and p38 differentially impacted TDP-43 neurotoxicity, partially through their opposing functions maintaining neuronal homeostasis of oxidative stress responses and/or neuroinflammatory responses. SAPK p38 signaling promoted, whereas JNK/Bsk signaling antagonized oxidative stress and neuroinflammation responses that ultimately led to neurodegenerative outcome.
Drosophila Wnd was initially identified as a gene required for synaptic bouton overgrowth in Hiw mutants (61). Subsequent studies of Wnd in Drosophila and its mammalian ortholog DLK have revealed complex roles in neuronal development and axonal biology (84). DLK-dependent apoptotic signaling is strongly linked to activation of JNK and its cognate transcription factor, AP-1 (85). Against, this backdrop, a straightforward explanation for the partial rescue of TDP-43 toxicity by Wnd mutations, was that TDP-43 missexpression generates an injury signal that triggers pathologic Wnd activation. However, several findings rule out this simple model. First, we found no evidence for hyperphosphorylation of the Wnd effectors JNK and p38 in whole heads of D42>TDP-43 flies (data not shown). Second, D42>TDP-43 flies did not display an NMJ overgrowth phenotype characteristic of Wnd gain of function (data not shown). Thus, despite the fact that Wnd mutations reduce TDP-43 toxicity, we see no evidence of enhanced Wnd activation in TDP-43 transgenic flies, suggesting that constitutive Wnd signaling is sufficient to enhance TDP-43 toxicity.
Downstream of Wnd, p38 and JNK/Bsk LOF alleles consistently yielded opposing impacts on TDP-43-associated neurotoxicity, with p38 enhancing toxicity and JNK/Bsk conferring reduced toxicity. Although JNK functions as a pro-apoptotic component of the Wnd/DLK pathway (86), this is clearly not the case in context of TDP-43-induced neurodegeneration, which is caspase independent (45). Instead, our findings suggest that JNK/Bsk attenuates TDP-43-induced toxicity, at least in part, through mitigation of ROS. A pathologic role for ROS in TDP-43-mediated toxicity is supported by the findings that: (i) oxidative stress markers were elevated in aged D42>TDP-43 flies; (ii) D42>TDP-43 flies showed hypersensitivity to the oxidative stress agent paraquat; and (iii) overexpression of the anti-oxidant response transcription factor CncC extended lifespan of D42>TDP-43 flies. JNK/Bsk LOF exacerbated ROS in D42>TDP-43 flies (Fig. 4E), which is compatible with previous reports showing that Bsk deficient flies were hyper-sensitive to oxidative insults (65). A role for oxidative stress in TDP-43-induced neurotoxicity described here is consistent with studies showing that TDP-43 alters mitochondrial function (87) and deregulates protein chaperones that ensure proteostasis under conditions of stress (88). However, the precise mechanisms of ROS accumulation in TDP-43 transgenic flies are unknown and may be multifactorial.
Our findings implicate innate immunity as a second contributing factor to TDP-43-induced neurodegeneration in Drosophila. AMP genes were strongly induced when TDP-43 was expressed in neurons under several different drivers (Fig. 5). LOF mutations of the immune responsive Toll/Dif and Imd/Relish pathways conferred phenotypic rescue to TDP-43 transgenic flies, suggesting that innate immunity contributes significantly to disease in this model (Fig. 6). That AMP genes were further upregulated in D42>TDP-43 flies overexpressing p38b (Fig. 7C) suggest that p38b enhances TDP-43 toxicity, in part, by potentiating the innate immune response. Likewise, LOF mutations in JNK/Bsk potentiated AMP gene activation in TDP-43 transgenic flies (Fig. 7C), which is congruent with the deleterious impact of JNK/Bsk mutations on lifespan of TDP-43 transgenic flies (Fig. 2B). Nevertheless, how JNK/Bsk and p38 signaling contribute to TDP-43-induced AMP gene activation is not entirely clear. In flies, the relative contributions of JNK/Bsk and p38 to innate immunity are complex and context dependent. A previous study using a fat body model for fly immunity revealed that expression of AMP genes was highly JNK dependent (71), but another study concluded that JNK was dispensable for AMP induction in Drosophila S2 cell culture (89). Similarly, although the requirement of p38 in immune defense is well conserved in flies (70), members of the Drosophila p38 gene family (p38a, p38b and p38c) seem to elicit diverse, even conflicting immune functions (63,69,70). Drosophila p38a was implicated as a negative regulator of innate immunity genes (69), whereas p38b was required for full induction of AMP genes after bacterial infection (70). These findings are congruent with our observations that deletion of the p38a had minimum effects (data not shown), whereas deletion of p38b extended D42>TDP43 fly longevity (Fig. 3D). The differential contributions of p38a and p38b on TDP-43 toxicity is likely due to their limited homology and functional redundancy (63). Finally, given the far-reaching functions of p38 and JNK/Bsk in the central nervous system, it is possible that these kinases influence TDP-43 toxicity through multiple neuron autonomous and non-autonomous mechanisms. Indeed, Morfini et al. recently showed p38 activation contributed to reduced axonal transport and kinesin hyperphosphoryation in squid axons loaded with ALS-associated mutants of SOD1 (90).
Activation of the innate immune response and neuroinflammation is a recurring theme in many if not all neurodegenerative states (reviewed in 91) and our findings strongly implicate innate immunity as a contributing factor to TDP-43 toxicity in Drosophila. Immune activation could be a direct or indirect consequence of TDP-43 misexpression, though we currently favor the latter possibility. Swarup et al. showed that nuclear TDP-43 could function as co-activator to the mammalian NF-κB factor p65 which facilitated the release of pro-inflammatory cytokines from glia or macrophages (92). Recent studies have shown that innate immunity contributes to neurodegeneration in a Drosophila model of ataxia-telangiectasia and upregulation of the innate immune system itself was sufficient to trigger neurodegeneration (76,93,94). Inflammatory processes occur in human ALS and a recent study showed that inhibition of CD40 signaling conferred significant lifespan extension to SOD1 transgenic mice (95).
In conclusion, we propose that p38 and JNK, operating downstream of Wnd and possibly additional stress-activated MAP3Ks, play important and potentially opposing roles in TDP-43-induced neurodegeneration. The demonstration of oxidative stress and innate immune response activation in TDP-43 transgenic flies supports the idea that, although the pathogenetic causes of ALS are increasingly diverse, a core set of pathologic processes are ultimately engaged.
MATERIALS AND METHODS
Drosophila maintenance
Flies were maintained and aged with the standard cornmeal-yeast medium (Nutri-Fly BF, Genesee scientific) at 25°C with 12-h light–dark cycle. For Gal80ts-related temperature inducible experiments, flies were crossed at 18°C and newly eclosed progenies maintained at 30°C. For SB202190 (LC laboratories) and SB203580 (LC laboratories) treatment experiments, adult male D42>TDP-43 flies were maintained in medium containing 10, 100 or 200 μm of each inhibitor.
Drosophila stocks
The w1118 was the parental strain used for the generation of UAS-TDP-43L1-L4 transgenic lines (35) and was therefore used wild-type background control in this study. The D42>TDP-43 stock refers to a recombinant line in which UAS-TDP-43L2 and D42-Gal4 were both balanced on the third chromosome. The BG380>TDP-43L2 refers to a recombinant stock containing Tubulin>Gal-80ts and UAS-TDP-43L2 on the third chromosome and motoneuron driver BG380-Gal4 on the X chromosome (BG380-Gal4/Fm7c; Tubulin>Gal-80ts, UAS-TDP-43L2/Tm6b). BG380>TDP-43L3 is identical except used UAS-TDP-43L3 for higher TDP-43 expression (BG380-Gal4/Fm7c; Tubulin>Gal-80ts, UAS-TDP-43L3/Tm6b). The following stocks used in this study were generously provided by: Dr Collins [Wnd1, Wnd2, Wnd3 UAS-WndWT,(61) UAS-p38bWT and UAS-p38bDN (96), BG380-Gal4]; Dr Ganetzky [UAS-Hiw (97)]; Dr Downward [p38bD27 (98)]; Dr Bohmann [GstD1:GFP and UAS-CncC (67)]; Dr Voigt [UAS-TDP43phiC31 (42)]; Dr Hirth [UAS-TBPH (77)]; Dr Klima [UAS-TBPHF/L150-152 (78)]; Dr Pandey [UAS-FUS (79)]. The rest of stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University with the accession number available in Supplementary Material, Table S2.
Survival analysis
Survival analysis was performed as previously described (45). Briefly, flies were aged at 25°C and survival was recorded on a daily basis. For each individual experiment, lifespan of D42>TDP-43/+ flies were recorded every time to account for environmental impacts on longevity. For Gal80ts-related experiments, the survival assay was done in 30°C. Food was changed every 3–4 days. Greater than 10% increase in MS was used as criteria for biological significance. Log-rank (Mantel–Cox) test was used for statistical analysis.
Western blot analysis
Fly lysate was prepared by homogenizing four fly heads (two from each gender) in 80 μL of sample buffer containing 250 mm Tris (pH 6.8), 5% SDS, 10% glycerol and 5% 2-mercaptoethanol by pestle. For detecting phosphorylation, protease inhibitor cocktail (Roche) was added. Protein lysates were resolved by 10% SDS–PAGE and detected via standard western blotting as previously described (35). The following primary antibodies were used in this study: anti-TDP-43 (Proteintech, #10782-2-AP); anti-beta-Tubulin (Millipore, #05-661), anti-phospho-p38 (Cell Signaling Technology, #9211), anti-phospho-JNK (Cell Signaling Technology, #9251), anti-GFP (Santa Cruz, sc-9996), anti-DNP (Sigma D9656), anti-TBPH was a generous gift from Dr Hirth (77).
Protein carbonylation assay
The protein carbonylation assay was performed as previously described (99). Briefly, 10 fly heads were homogenized in 100 µl lysis buffer containing 200 mm sodium phosphate (pH 5.2), 1 mm EDTA, 1% SDS and protease inhibitors (Roche). The crude lysate was then cleared by centrifugation at 14,000 rpm for 10 min at room temperature. An aliquot of 20 µl supernatant was combined with 20 µl 12% SDS and 40 µl the 2,4-dintrophenylhydrazine (DNPH) buffer containing 20 mm DNPH that was pre-dissolved in 2 m HCl. The DNPH derivatization reaction was done by incubating the samples at room temperature for 1 h and then terminated by adding 30 µl of 2 m Tris (pH 7.4) buffer containing 30% glycerol. Levels of the carbonyl moiety labeled by DNP were then detected by standard western blot technique using anti-DNP antibody.
Quantitative PCR analysis
Adult flies in 1.5 ml microtube were snap-frozen in liquid nitrogen followed by vortex to separate heads. Roughly, 20 fly heads per sample were used for RNA extraction using Trizol reagent (Invitrogen). Roughly, 2 μg RNA was used for each 20 µl reverse transcription reaction (iScript cDNA synthesis Kit, Bio-Rad). For quantitative PCR using single channel detection, SYBR green supermix (iTaq Universal, Bio-Rad) was used following standard three-step PCR amplification protocol (CFX96 platform, Bio-Rad). For multiplex analysis, Taqman assay (iTaq Universal probes, Bio-Rad) was used followed by a two-step protocol (CFX96 platform, Bio-Rad). The Drosophila Mnf gene was used as housekeeping reference gene (100). Sequences for primers and Taqman probes were listed in Supplementary Material, Table S3. Gene expression data were calculated by the delta–delta Ct method. Relative fold change of gene expression was plotted as mean from four different experiments. The statistical analysis was done using the Student's t-test (CFX manager 3.1). Pair-wise comparison of each experiment can be found in Supplementary Material, Table S4. Statistical significance was summarized as n.s. P > 0.05; *0.05 < P < 0.01; **0.001 < P < 0.01; ***P < 0.001.
Climbing assay
The climbing assay was modified as previously described (44). Generally, 10–15 flies at specified age were loaded in the testing vial. To assay motility, we measured negative geotaxis behavior in flies. Flies were first tapped to the bottom of the testing vial and given 5 s to climb (Supplementary Material, Fig. S7A). Their vertical travel distance was then captured by a self-timed camera and extrapolated by ImageJ (installed with PointPicker). The motility index (MI) was calculated as speed by normalizing vertical travel distance with travel time (5 s) (Supplementary Material, Fig. S7B). A total of five trials were performed for each genotype. To avoid pseudoreplication, we chose only one trial as representative trial among all five to be included for data analysis. To choose the representative trial, average speed among all subjects per trial was calculated (Supplementary Material, Fig. S7C). The median among all five average numbers was then used to pick the representative trial (Supplementary Material, Fig. S7C). Data showing motility of each individual fly were scatter plotted with the mean value superimposed as bar graph.
Oxidative stress survival assay
Flies were incubated at 25°C on median containing 20 mm paraquat, 5% sucrose and 1% agarose. Survival recording was initiated after paraquat administration on a daily basis. Log-rank (Mantel–Cox) test was used for statistical analysis.
Statistical analysis
The statistics were done by the GraphPad Software (V5.01, San Diego, CA, USA). Log-rank (Mantel–Cox) test was performed for survival analysis. Mann–Whitney test was used for the climbing assay. Student's t-test was used for qPCR analysis (CFX manager, Bio-Rad). Chi-square test was used for analyzing deformed wing phenotype. Pair-wise comparison of each experiment can be found in Supplementary Material, Table S4. The statistical significance was summarized as n.s. P > 0.05; *0.05 < P < 0.01; **0.001 < P < 0.01; ***P < 0.001.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the NIH (R21NS067572 and R01CA180765) and ALS Association (O7LB7D).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lisa Sudmeier and Robert Kreber from the Ganezky lab at UW-Madison for her tremendous help with the NMJ analysis and deficiency screen. Our deep appreciation also goes to Dr Collins (University of Michigan-Ann Arbor), Dr Ganetzky (University of Wisconsin-Madison), Dr Downward (London Research Institute), Dr Bohmann (University of Rochester Medical Center), Dr Voigt (University Medical Center, RWTH Aachen, Germany), Dr Hirth (King's College London), Dr Klima (International Center for Genetic Engineering and Biotechnology, Trieste, Italy) and Dr Pandey (Louisiana State University Health Sciences Center) for their generous provision on fly stocks that were vital for this research.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rosen D.R. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 2.Rotunno M.S., Bosco D.A. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front Cell Neurosci. 2013;7:253. doi: 10.3389/fncel.2013.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Gendron T.F., Rademakers R., Petrucelli L. TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J. Alzheimers Dis. 2013;33(Suppl. 1):S35–345. doi: 10.3233/JAD-2012-129036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford N.J., Zhang Y.J., Baker M., Gass J.M., Finch N.A., Xu Y.F., Stewart H., Kelley B.J., Kuntz K., Crook R.J., et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabashi E., Lin L., Tradewell M.L., Dion P.A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H.D., Vande Velde C., et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 8.Van Deerlin V.M., Leverenz J.B., Bekris L.M., Bird T.D., Yuan W., Elman L.B., Clay D., Wood E.M., Chen-Plotkin A.S., Martinez-Lage M., et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitcho M.A., Bigio E.H., Mishra M., Johnson N., Weintraub S., Mesulam M., Rademakers R., Chakraverty S., Cruchaga C., Morris J.C., et al. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009;118:633–645. doi: 10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 11.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C., et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M., et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C., et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 18.Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., 3rd, Rademakers R., et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizielinska S., Gronke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O., Pietrzyk J., et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 21.Ling S.C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo W., Chen Y., Zhou X., Kar A., Ray P., Chen X., Rao E.J., Yang M., Ye H., Zhu L., et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold E.S., Ling S.C., Huelga S.C., Lagier-Tourenne C., Polymenidou M., Ditsworth D., Kordasiewicz H.B., McAlonis-Downes M., Platoshyn O., Parone P.A., et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. USA. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C., et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buratti E., Brindisi A., Pagani F., Baralle F.E. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayala Y.M., De Conti L., Avendano-Vazquez S.E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalmansingh A.S., Urekar C.J., Reddi P.P. TDP-43 is a transcriptional repressor: the testis-specific mouse acrv1 gene is a TDP-43 target in vivo. J. Biol. Chem. 2011;286:10970–10982. doi: 10.1074/jbc.M110.166587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou S.H., Wu F., Harrich D., Garcia-Martinez L.F., Gaynor R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawahara Y., Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald K.K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G.A., Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 33.Wegorzewska I., Bell S., Cairns N.J., Miller T.M., Baloh R.H. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ash P.E., Zhang Y.J., Roberts C.M., Saldi T., Hutter H., Buratti E., Petrucelli L., Link C.D. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum. Mol. Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson K.A., Kim S.H., Wassarman D.A., Tibbetts R.S. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J. Biol. Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Ray P., Rao E.J., Shi C., Guo W., Chen X., Woodruff E.A., 3rd, Fushimi K., Wu J.Y. A Drosophila model for TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y.F., Zhang Y.J., Lin W.L., Cao X., Stetler C., Dickson D.W., Lewis J., Petrucelli L. Expression of mutant TDP-43 induces neuronal dysfunction in transgenic mice. Mol. Neurodegener. 2011;6:73. doi: 10.1186/1750-1326-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romano M., Feiguin F., Buratti E. Drosophila answers to TDP-43 proteinopathies. J. Amino Acids. 2012;2012:356081. doi: 10.1155/2012/356081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin M.J., Cheng C.W., Shen C.K. Neuronal function and dysfunction of Drosophila dTDP. PLoS ONE. 2011;6:e20371. doi: 10.1371/journal.pone.0020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miguel L., Frebourg T., Campion D., Lecourtois M. Both cytoplasmic and nuclear accumulations of the protein are neurotoxic in Drosophila models of TDP-43 proteinopathies. Neurobiol. Dis. 2011;41:398–406. doi: 10.1016/j.nbd.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Ihara R., Matsukawa K., Nagata Y., Kunugi H., Tsuji S., Chihara T., Kuranaga E., Miura M., Wakabayashi T., Hashimoto T., et al. RNA binding mediates neurotoxicity in the transgenic Drosophila model of TDP-43 proteinopathy. Hum. Mol. Genet. 2013;22:4474–4484. doi: 10.1093/hmg/ddt296. [DOI] [PubMed] [Google Scholar]
- 42.Voigt A., Herholz D., Fiesel F.C., Kaur K., Muller D., Karsten P., Weber S.S., Kahle P.J., Marquardt T., Schulz J.B. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS ONE. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.H., Zhan L., Hanson K.A., Tibbetts R.S. High-content RNAi screening identifies the Type 1 inositol triphosphate receptor as a modifier of TDP-43 localization and neurotoxicity. Hum. Mol. Genet. 2012;21:4845–4856. doi: 10.1093/hmg/dds321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhan L., Hanson K.A., Kim S.H., Tare A., Tibbetts R.S. Identification of Genetic Modifiers of TDP-43 Neurotoxicity in Drosophila. PLoS ONE. 2013;8:e57214. doi: 10.1371/journal.pone.0057214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H.J., Raphael A.R., Ladow E.S., McGurk L., Weber R.A., Trojanowski J.Q., Lee V.M., Finkbeiner S., Gitler A.D., Bonini N.M. Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet. 2014;46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y.R., King O.D., Shorter J., Gitler A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tibbles L.A., Woodgett J.R. The stress-activated protein kinase pathways. Cell Mol. Life Sci. 1999;55:1230–1254. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harper S.J., Wilkie N. MAPKs: new targets for neurodegeneration. Expert Opin. Ther. Targets. 2003;7:187–200. doi: 10.1517/14728222.7.2.187. [DOI] [PubMed] [Google Scholar]
- 50.Kim E.K., Choi E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Holasek S.S., Wengenack T.M., Kandimalla K.K., Montano C., Gregor D.M., Curran G.L., Poduslo J.F. Activation of the stress-activated MAP kinase, p38, but not JNK in cortical motor neurons during early presymptomatic stages of amyotrophic lateral sclerosis in transgenic mice. Brain Res. 2005;1045:185–198. doi: 10.1016/j.brainres.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 52.Veglianese P., Lo Coco D., Bao Cutrona M., Magnoni R., Pennacchini D., Pozzi B., Gowing G., Julien J.P., Tortarolo M., Bendotti C. Activation of the p38MAPK cascade is associated with upregulation of TNF alpha receptors in the spinal motor neurons of mouse models of familial ALS. Mol. Cell Neurosci. 2006;31:218–231. doi: 10.1016/j.mcn.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Xiong X., Wang X., Ewanek R., Bhat P., Diantonio A., Collins C.A. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J. Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klinedinst S., Wang X., Xiong X., Haenfler J.M., Collins C.A. Independent pathways downstream of the Wnd/DLK MAPKKK regulate synaptic structure, axonal transport, and injury signaling. J. Neurosci. 2013;33:12764–12778. doi: 10.1523/JNEUROSCI.5160-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller B.R., Press C., Daniels R.W., Sasaki Y., Milbrandt J., DiAntonio A. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heneka M.T., Kummer M.P., Latz E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014;14:463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 57.Frakes A.E., Ferraiuolo L., Haidet-Phillips A.M., Schmelzer L., Braun L., Miranda C.J., Ladner K.J., Bevan A.K., Foust K.D., Godbout J.P., et al. Microglia induce motor neuron death via the classical NF-kappaB pathway in amyotrophic lateral sclerosis. Neuron. 2014;81:1009–1023. doi: 10.1016/j.neuron.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meissner F., Molawi K., Zychlinsky A. Mutant superoxide dismutase 1-induced IL-1beta accelerates ALS pathogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:13046–13050. doi: 10.1073/pnas.1002396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cook R.K., Christensen S.J., Deal J.A., Coburn R.A., Deal M.E., Gresens J.M., Kaufman T.C., Cook K.R. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 61.Collins C.A., Wairkar Y.P., Johnson S.L., DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 62.Xiong X., Collins C.A. A conditioning lesion protects axons from degeneration via the Wallenda/DLK MAP kinase signaling cascade. J. Neurosci. 2012;32:610–615. doi: 10.1523/JNEUROSCI.3586-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belozerov V.E., Lin Z.Y., Gingras A.C., McDermott J.C., Michael Siu K.W. High-resolution protein interaction map of the Drosophila melanogaster p38 mitogen-activated protein kinases reveals limited functional redundancy. Mol. Cell Biol. 2012;32:3695–3706. doi: 10.1128/MCB.00232-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanden Broeck L., Naval-Sanchez M., Adachi Y., Diaper D., Dourlen P., Chapuis J., Kleinberger G., Gistelinck M., Van Broeckhoven C., Lambert J.C., et al. TDP-43 loss-of-function causes neuronal loss due to defective steroid receptor-mediated gene program switching in Drosophila. Cell Rep. 2013;3:160–172. doi: 10.1016/j.celrep.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 65.Wang M.C., Bohmann D., Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- 66.Vrailas-Mortimer A., del Rivero T., Mukherjee S., Nag S., Gaitanidis A., Kadas D., Consoulas C., Duttaroy A., Sanyal S. A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and life span in Drosophila. Dev. Cell. 2011;21:783–795. doi: 10.1016/j.devcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sykiotis G.P., Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sawicki R., Singh S.P., Mondal A.K., Benes H., Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem. J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han Z.S., Enslen H., Hu X., Meng X., Wu I.H., Barrett T., Davis R.J., Ip Y.T. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol. Cell Biol. 1998;18:3527–3539. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J., Xie C., Tian L., Hong L., Wu X., Han J. Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc. Natl. Acad. Sci. USA. 2010;107:20774–20779. doi: 10.1073/pnas.1009223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delaney J.R., Stoven S., Uvell H., Anderson K.V., Engstrom Y., Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. EMBO J. 2006;25:3068–3077. doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kallio J., Leinonen A., Ulvila J., Valanne S., Ezekowitz R.A., Ramet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7:811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 73.Sluss H.K., Han Z., Barrett T., Goberdhan D.C., Wilson C., Davis R.J., Ip Y.T. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 74.Hoffmann J.A. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 75.Petersen A.J., Katzenberger R.J., Wassarman D.A. The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics. 2013;194:133–142. doi: 10.1534/genetics.113.150854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao Y., Chtarbanova S., Petersen A.J., Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. USA. 2013;110:E1752–E1760. doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diaper D.C., Adachi Y., Sutcliffe B., Humphrey D.M., Elliott C.J., Stepto A., Ludlow Z.N., Vanden Broeck L., Callaerts P., Dermaut B., et al. Loss and gain of Drosophila TDP-43 impair synaptic efficacy and motor control leading to age-related neurodegeneration by loss-of-function phenotypes. Hum. Mol. Genet. 2013;22:1539–1557. doi: 10.1093/hmg/ddt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Godena V.K., Romano G., Romano M., Appocher C., Klima R., Buratti E., Baralle F.E., Feiguin F. TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubules organization. PLoS ONE. 2011;6:e17808. doi: 10.1371/journal.pone.0017808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lanson N.A., Jr., Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., Pandey U.B. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemaitre B., Nicolas E., Michaut L., Reichhart J.M., Hoffmann J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 81.Weber A.N., Tauszig-Delamasure S., Hoffmann J.A., Lelievre E., Gascan H., Ray K.P., Morse M.A., Imler J.L., Gay N.J. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat. Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- 82.Shia A.K., Glittenberg M., Thompson G., Weber A.N., Reichhart J.M., Ligoxygakis P. Toll-dependent antimicrobial responses in Drosophila larval fat body require Spatzle secreted by haemocytes. J. Cell Sci. 2009;122:4505–4515. doi: 10.1242/jcs.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tedeschi A., Bradke F. The DLK signalling pathway – a double-edged sword in neural development and regeneration. EMBO Rep. 2013;14:605–614. doi: 10.1038/embor.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ghosh A.S., Wang B., Pozniak C.D., Chen M., Watts R.J., Lewcock J.W. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J. Cell Biol. 2011;194:751–764. doi: 10.1083/jcb.201103153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pozniak C.D., Sengupta Ghosh A., Gogineni A., Hanson J.E., Lee S.H., Larson J.L., Solanoy H., Bustos D., Li H., Ngu H., et al. Dual leucine zipper kinase is required for excitotoxicity-induced neuronal degeneration. J. Exp. Med. 2013;210:2553–2567. doi: 10.1084/jem.20122832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huntwork-Rodriguez S., Wang B., Watkins T., Ghosh A.S., Pozniak C.D., Bustos D., Newton K., Kirkpatrick D.S., Lewcock J.W. JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. J. Cell Biol. 2013;202:747–763. doi: 10.1083/jcb.201303066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang W., Li L., Lin W.L., Dickson D.W., Petrucelli L., Zhang T., Wang X. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum. Mol. Genet. 2013;22:4706–4719. doi: 10.1093/hmg/ddt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang H.Y., Hou S.C., Way T.D., Wong C.H., Wang I.F. Heat-shock protein dysregulation is associated with functional and pathological TDP-43 aggregation. Nat. Commun. 2013;4:2757. doi: 10.1038/ncomms3757. [DOI] [PubMed] [Google Scholar]
- 89.Silverman N., Zhou R., Erlich R.L., Hunter M., Bernstein E., Schneider D., Maniatis T. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 2003;278:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- 90.Morfini G.A., Bosco D.A., Brown H., Gatto R., Kaminska A., Song Y., Molla L., Baker L., Marangoni M.N., Berth S., et al. Inhibition of fast axonal transport by pathogenic SOD1 involves activation of p38 MAP kinase. PLoS ONE. 2013;8:e65235. doi: 10.1371/journal.pone.0065235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lucin K.M., Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64:110–122. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swarup V., Phaneuf D., Dupre N., Petri S., Strong M., Kriz J., Julien J.P. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J. Exp. Med. 2011;208:2429–2447. doi: 10.1084/jem.20111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maqbool A., Lattke M., Wirth T., Baumann B. Sustained, neuron-specific IKK/NF-kappaB activation generates a selective neuroinflammatory response promoting local neurodegeneration with aging. Mol. Neurodegener. 2013;8:40. doi: 10.1186/1750-1326-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petersen A.J., Rimkus S.A., Wassarman D.A. ATM kinase inhibition in glial cells activates the innate immune response and causes neurodegeneration in Drosophila. Proc. Natl. Acad. Sci. USA. 2012;109:E656–E664. doi: 10.1073/pnas.1110470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lincecum J.M., Vieira F.G., Wang M.Z., Thompson K., De Zutter G.S., Kidd J., Moreno A., Sanchez R., Carrion I.J., Levine B.A., et al. From transcriptome analysis to therapeutic anti-CD40L treatment in the SOD1 model of amyotrophic lateral sclerosis. Nat. Genet. 2010;42:392–399. doi: 10.1038/ng.557. [DOI] [PubMed] [Google Scholar]
- 96.Adachi-Yamada T., Nakamura M., Irie K., Tomoyasu Y., Sano Y., Mori E., Goto S., Ueno N., Nishida Y., Matsumoto K. p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol. Cell Biol. 1999;19:2322–2329. doi: 10.1128/mcb.19.3.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu C., Wairkar Y.P., Collins C.A., DiAntonio A. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J. Neurosci. 2005;25:9557–9566. doi: 10.1523/JNEUROSCI.2532-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cully M., Genevet A., Warne P., Treins C., Liu T., Bastien J., Baum B., Tapon N., Leevers S.J., Downward J. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell Biol. 2010;30:481–495. doi: 10.1128/MCB.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cumming R.C., Simonsen A., Finley K.D. Quantitative analysis of autophagic activity in Drosophila neural tissues by measuring the turnover rates of pathway substrates. Methods Enzymol. 2008;451:639–651. doi: 10.1016/S0076-6879(08)03235-7. [DOI] [PubMed] [Google Scholar]
- 100.Ponton F., Chapuis M.P., Pernice M., Sword G.A., Simpson S.J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011;57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.