Abstract
The inositol pyrophosphates (PP-InsPs) are a specialized group of “energetic” signaling molecules found in yeasts, plants and animals. PP-InsPs boast the most crowded three dimensional phosphate arrays found in Nature; multiple phosphates and diphosphates are crammed around the six-carbon, inositol ring. Yet, phosphate esters are also a major energy currency in cells. So the synthesis of PP-InsPs, and the maintenance of their levels in the face of a high rate of ongoing turnover, all requires significant bioenergetic input. What are the particular properties of PP-InsPs that repay this investment of cellular energy? Potential answers to that question are discussed here, against the backdrop of a recent hypothesis that signaling by PP-InsPs is evolutionarily ancient. The latter idea is extended herein, with the proposal that the primordial origins of PP-InsPs is reflected in the apparent lack of isomeric specificity of certain of their actions. Nevertheless, there are other aspects of signaling by these polyphosphates that are more selective for a particular PP-InsP isomer. Consideration of the nature of both specific and non-specific effects of PP-InsPs can help rationalize why such molecules possess so many phosphates.
Keywords: inositol pyrophosphates, structure, analogues, diphosphoinositol polyphosphates, cell-signaling, kinase, phosphorylation
Introduction and some Comments on Nomenclature
The phosphate group is a ubiquitous signaling device that establishes specificity in ligand-protein and protein-protein interactions. The phosphate’s bulk imposes geometric constraints upon these interactions. The phosphate’s negative charge at physiological pH further enhances specificity through ionic and hydrogen bonds with certain amino acid residues at physiological pH. Both proteins and small molecules can offer their phosphorylation and dephosphorylation for cell-signaling purposes. But there is one molecule in particular that belies its basic simplicity by hosting multiple phosphate recognition patterns that have extraordinary functionally versatility. This entity is a six-carbon ring structure that is systematically described as cis-1,2,3,5-trans-4,6-cyclohexanehexol. Most of us know it better as myo-inositol, or more frequently just as inositol (Figure 1). The combinatorial placement of phosphate groups around the inositol ring creates the family of signaling molecules known as the inositol phosphates. The use of inositol as this scaffold for phosphorylation is synthetically convenient for cells, as this carbohydrate is only a short offshoot from the glycolytic pathway (Sherman et al., 1977). Inositol also has the benefit of being chemically very stable - in fact, its resistance to both oxidation and high temperature has even helped it survive (in at least one of its nine possible stereoisomeric forms (Agranoff, 1985)), while being carried to Earth in meteorites (Cooper et al., 2001). Indeed, Agranoff (Agranoff, 2009) has suggested inositol could have been a constituent of prebiotic Earth.
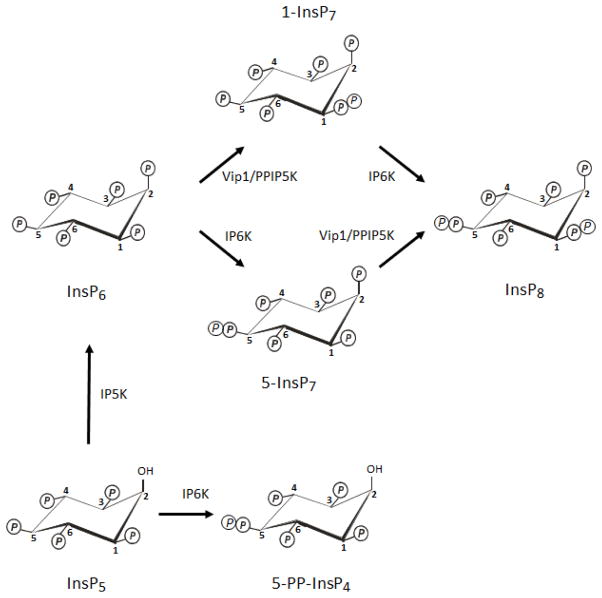
Fig. 1. Synthesis of the PP-InsPs.
The figure describes the metabolic reactions that account for the synthesis of the PP-InsPs in both yeasts and mammalian cells. The positions of the diphosphate groups were determined in the following publications: (Albert et al., 1997; Draskovic et al., 2008; Wang et al., 2012). PPIP5K (E.C. 2.7.1.158), inositol pentakisphosphate kinase; IP6K (E.C.2.7.4.21), inositol hexakisphosphate kinase, PPIP5K (E.C.2.7.4.24), diphosphoinositol pentakisphophate kinase. This figure is adapted from (Shears et al., 2013). For the brave-hearted who wish to gain insight into the unassailable logic behind the universal numbering system for the six carbon atoms of Ins, start here: (Nomenclature Committee of the International Union of Biochemistry., 1989).
In the literature, myo-inositol is abbreviated to either “Ins” or just “I”; the latter is better adapted to the spoken language so, for example, one will most often hear and see “IP8” employed in preference to “InsP8”. Either way, this represents the most densely concentrated array of phosphate groups found in Nature; “P8” indicates that eight phosphates are attached to the inositol ring. Irrespective of how you say or write it, Ins only has six hydroxyl groups, so in order to esterify eight phosphates onto this ring, there must be either one triphosphate or two diphosphates. Both kinds of derivatizations are possible, at least in vitro (Draskovic et al., 2008). It appears that the InsP8 found in metazoan cells is the form that has two diphosphates (Figure 1 and (Lin et al., 2009; Wang et al., 2012)). Hence this and other inositol phosphates with diphosphate groups are collectively most usually described as “inositol pyrophosphates”. Nevertheless, “InsP8” is a technically ambiguous term. Better would be “(PP)2-InsP4” (Shears, 2009), which explicitly describes a molecule with two diphosphates and four monophosphates, thereby distinguishing it from a different form of “InsP8” that contains one triphosphate and five monophosphates: PPP-InsP5 (Draskovic et al., 2008). However, there is increasing pressure upon today’s scientists to publish papers that access the widest possible “non-specialized audience” (Saiardi, 2012a) (read: papers that are more frequently cited). Thus, InsP8 (or IP8) are generally considered (particularly by some journal editors) to be a lesser evil because either are viewed as more user-friendly abbreviations. Presumably for similar reasons, InsP7 (or IP7) are both widely preferred in place of PP-InsP5, even though, once again, 7-phosphates can also be added to Ins in a rather different arrangement: (PP)2-InsP3 (see, for example, (Draskovic et al., 2008; Wilson et al., 2013)). This apparent conflict between Impact Factors and technical accuracy (or, depending upon your viewpoint, “foolish consistency” (Agranoff et al., 1985)), has merited a different outcome in the description of the physiologically-relevant diphosphate of Ins(1,3,4,5,6)P5. Although that particular product has six phosphates, it is invariably written as either PP-InsP4 or PP-IP4 (Wilson et al., 2013). In this case, there has been no problem accepting how inadvisable an abbreviation InsP6 would be, since of course that is how inositol hexakisphosphate is described (Figure 1). To add to this terminological miss-mash, “inositol pyrophosphates” are often abbreviated as “PP-InsPs” (or “PP-IPs”). Why “PP-InsP5” is generally avoided while “PP-InsPs” is widely used, is a point that will not be debated here. Rather, to improve the citability of this review, “InsP8”, “InsP7”, and “PP-InsPs” are the abbreviations I, too, have chosen to deploy.
Any novice approaching this field must also learn there are two naturally-occurring isomers of InsP7 in most eukaryotic cells. These are distinguished by the diphosphate group being placed at either the 1- or 5-carbon ((Albert et al., 1997; Draskovic et al., 2008; Lin et al., 2009; Wang et al., 2012); see Figure 1). InsP8 has both of these diphosphate groups ((Wang et al., 2012); see Figure 1). Two groups of enzymes synthesize inositol pyrophosphates. The 5-kinase activities of IP6K1/2/3 (E.C.2.7.4.21) (Draskovic et al., 2008; Lin et al., 2009; Saiardi et al., 1999) convert InsP6 and 1-InsP7 to 5-InsP7 and InsP8 respectively (Figure 1). Second, the 1-kinase activities of PPIP5K1/2 (E.C.2.7.4.24) (Choi et al., 2007; Fridy et al., 2007; Wang et al., 2012) phosphorylate InsP6 and 5-InsP7 to 1-InsP7 and InsP8 respectively (Figure 1). All of these reactions are reversed by specialist phosphatases: DIPPs (diphosphoinositol polyphosphate phosphohydrolases; E.C. 3.6.1.52) (Caffrey et al., 2000; Kilari et al., 2013; Safrany and Shears, 1998).
The levels of total cellular InsP7 usually lie in the 1 to 5 μM range (Barker et al., 2004; Fisher et al., 2002; Illies et al., 2007; Ingram et al., 2003), most of which appear to comprise the 5-isomer (Albert et al., 1997; Lin et al., 2009). The concentrations of InsP8 in yeast, plant and animal cells are typically each about 10–20% of those of InsP7 (Choi et al., 2005; Desai et al., 2014; Glennon and Shears, 1993; Ingram et al., 2003). Clearly, these molecules are not terribly abundant. However, there are two other factors to bear in mind. First, cellular levels of individual PP-InsPs are in roughly the same range as those of Ins(1,4,5)P3, which certainly has not impeded its own actions as an important second messenger (Streb et al., 1983). Second, there may be subcellular compartmentalization of PP-InsPs (see below).
There is one group of eukaryotes that offer a quite stunning exception to the general rule that cells maintain quite low levels of PP-InsPs. Ever since these molecule were first characterized (Europe-Finner et al., 1991; Laussmann et al., 2000; Menniti et al., 1993; Stephens et al., 1993), it has been known that their intracellular concentrations are exceptionally high in slime molds such as Dictyostelium discoideum. Their levels of InsP7 and InsP81 are known to increase further when their bacterial food supply becomes restricted and they aggregate in preparation for entering a dispersive phase of the life cycle (Laussmann et al., 2000; Pisani et al., 2014). By the time that fruiting bodies form, the concentrations of InsP7 and InsP8 are around 60 and 450 μM respectively (Pisani et al., 2014). It is hard to avoid theorizing that slime molds have uniquely exploited the properties of the PP-InsPs, perhaps as organic phosphate storage depots. Also, and unusually for eukaryotes, the pools of PP-InsPs in slime molds are supplemented with significant quantities of pyrophosphate derivatives of InsP5 (Pisani et al., 2014).
There is a significant free energy change when a diester-phosphate of a PP-InsP is hydrolyzed, due to electrostatic, solvation and resonance stabilization phenomena (Hand and Honek, 2007). Thus, such molecules are often said to be highly “energetic” (Wilson et al., 2013). Indeed, phosphate esters are a major energy currency in cells. Thus, there is a substantial bioenergetic investment in synthesizing PP-InsPs, especially as they undergo such considerable turnover (Menniti et al., 1993). It is presumed that there are specific features of the multiple phosphates in PP-InsPs that repay the cell for bankrolling their synthesis. The nature of these particular properties of PP-InsPs is a focal point for this review.
Can it be explained how PP-InsPs are multifunctional?
A number of studies into the biological actions of PP-InsPs have involved genetic experiments in which the expression of one or more isoforms of IP6K (Kcs1 in yeast) was increased or decreased/eliminated. Such an approach would be expected to modify cellular levels of all of the PP-InsPs (Figure 1), and it is not always clear which of those that are affected might be responsible for the phenotypic consequences. Nevertheless, it is still remarkable how many biological activities are perturbed by these experimentally-imposed changes in IP6K expression: maintenance of telomere length (Saiardi et al., 2005; York et al., 2005), vesicle trafficking (Saiardi et al., 2000; Saiardi et al., 2002), apoptosis (Morrison et al., 2001; Nagata et al., 2005), autophagy (Nagata et al., 2010), repair of DNA repair by homologous recombination (Jadav et al., 2013; Luo et al., 2002), transcription of glycolytic genes (Szijgyarto et al., 2011), hemostasis (Ghosh et al., 2013), phagocytic and bactericidal activities of neutrophils (Prasad et al., 2011), epigenetic modifications to chromatin (Burton et al., 2013) and exocytic insulin secretion (Illies et al., 2007). Not only do these many reports underscore the diverse actions of the PP-InsP family, they return us to an intriguing question: are PP-InsPs incredibly multifunctional, or do these alternative biological outcomes mostly lie downstream of a limited number of fundamental, over-arching processes?
Protein diphosphorylation offers a tantalizing description of how multifunctionality might arise from a single mechanism of PP-InsP action. Stephens et al (Stephens et al., 1993) were the first to suggest that PP-InsPs might be utilized as phosphate donors in phosphotransferase reactions. The first demonstration that certain proteins were phosphorylated by PP-InsPs, at least in vitro, came eleven years later (Saiardi et al., 2004). Subsequently it emerged that the β-phosphate of the diphosphate groups on the PP-InsPs are added to a pre-existing Ser-phosphate that is initially provided by a casein kinase II dependent phosphorylation event (Bhandari et al., 2007). That is, a diphosphate group is formed on the target protein. CK2 itself phosphorylates many different proteins (Ruzzene and Pinna, 2010), each of which should be considered primed for further diphosphorylation by PP-InsPs. Potentially, this mechanism might regulate many different biological activities.
An especially remarkable aspect of phosphate donation by PP-InsPs is that it is non-enzymatic (Bhandari et al., 2007; Saiardi et al., 2004). This characteristic can rationalize why PP-InsPs contain so many phosphates: in this scenario, non-enzymic protein diphosphorylation is driven by the significant free energy change that comes not just from diester phosphate hydrolysis, but also any accompanying steric and electronic rearrangements of the remaining, multiple phosphates (Bhandari et al., 2007; Hand and Honek, 2007; Saiardi et al., 2004). Nevertheless, a note of caution is warranted (see (Majerus, 2007): no-one has yet directly demonstrated that PP-InsPs can diphosphorylate proteins in vivo. This is key to advancing this hypothesis.
Snyder and colleagues (Chakraborty et al., 2010; Luo et al., 2003; Prasad et al., 2011) have separately developed another idea that could help explain why at least the 5-InsP7 produced by IP6Ks can regulate many biological processes. This proposal has developed from their observation (Luo et al., 2003) that PtdIns(3,4,5)P3-binding, pleckstrin homology (PH) domains also bind 5-InsP7. Proteins that contain these particular PH domains are normally translocated to the plasma membrane following stimulus-dependent activation of phosphoinositide 3-kinase (PI3K). This spatial rearrangement promotes assembly of multiprotein complexes and facilitates the activation of kinase cascades (Cantley, 2002). There are many cellular consequences to these signaling events, including control over protein synthesis, actin polymerization, metabolic homeostasis, cell survival, and cell cycle entry (Cantley, 2002). It is proposed that at least some of these cellular activities may be negatively regulated in those situations in which 5-InsP7, by competing with PtdIns(3,4,5)P3, inhibits signaling protein recruitment at the plasma membrane. That is, this mechanism of action offers multiple biological actions for 5-InsP7.
Snyder and colleagues reported that 5-InsP7 is a 10 to 20-fold stronger PH-domain ligand than is InsP6 (Chakraborty et al., 2010; Luo et al., 2003). That is, the 5-kinase activity of IP6K yields a molecule with an increased ability to prevent PtdIns(3,4,5)P3 from associating with PH domains (Chakraborty et al., 2010; Luo et al., 2003; Prasad et al., 2011). Our own data on this subject (Gokhale et al., 2013) agree that 5-InsP7 is more effective than InsP6, although in our hands the degree of discrimination was not more than 2-fold; we think that would not be sufficiently significant to represent the basis of a signaling phenomenon in itself. The origin of these quantitatively different data sets may in part reflect differences in PP-InsP purity. This can be impacted by the nature of the approach taken for the technically-challenging preparation of PP-InsPs (Capolicchio et al., 2013). Fortunately for future studies, some recent developments in the field of synthetic chemistry have finally made available high-quality preparations of PP-InsPs (Capolicchio et al., 2013; Capolicchio et al., 2014; Wu et al., 2013).2 Use of such standardized preparations of PP-InsPs should prove useful to all who work in this area.
In any case, we (Gokhale et al., 2013) propose that the significance of Snyder’s important work lies not in the differences of affinities of InsP6 and 5-InsP7 for PtdIns(3,4,5)P3-binding PH domains, but rather, the fact that they both bind. That is, we argue that InsP6 and 5-InsP7 both l inhibit PH domain recruitment to the plasma membrane. We (Gokhale et al., 2013) view this as a necessary process for preventing any non-sustained, stochastic increases in PtdIns(3,4,5)P3 from inappropriately recruiting and activating signaling proteins. The aberrant PtdIns(3,4,5)P3-signaling that underlies much of cancer biology is a good illustration of why careful regulation of PH-domain recruitment is so important.
Furthermore, we (Gokhale et al., 2011) have found that PPIP5K1 itself contains a cryptic PtdIns(3,4,5)P3-binding module. Thus, sustained stimulus-dependent increases in PtdIns(3,4,5)P3 cause PPIP5K1 to translocate to the plasma membrane (Gokhale et al., 2011; Gokhale et al., 2013). This, we propose, can promote a subplasmalemmal depletion of InsP6 and 5-InsP7, and their conversion to products (1-InsP7 and InsP8) that exhibit much weaker interactions with PtdIns(3,4,5)P3-binding domains (Figure 2 and (Gokhale et al., 2013)). For example, the PH domains of AKT, SIN1 and GRP1 all bind 5-InsP7 and InsP6 with higher affinities (up to 30-fold) than do 1-InsP7 and InsP8 (Gokhale et al., 2013)). How the 1-diphosphate imparts this striking specificity of action has not yet been determined. Nevertheless, these data indicate that optimum signaling by PtdIns(3,4,5)P3 in vivo may depend upon coincidence detection: the PtdIns(3,4,5)P3-mediated translocation of a PH-domain protein to the plasma membrane, plus PPIP5K1-catalyzed removal of inhibitors of that translocation (Figure 2). This is all consistent with Snyder’s (Luo et al., 2003) original demonstration that 5-InsP7 can be a multifunctional regulator (but by acting with InsP6 as an ally).
Fig. 2. The possible significance of stimulus-dependent, PtdIns(3,4,5) P3-driven compartmentalization of PPIP5K1 at the plasma membrane.
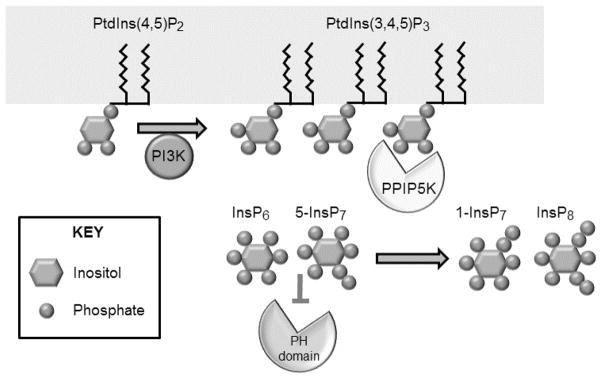
Electrostatic effects of InsP6 and 5-InsP7 are proposed to attenuate the ability of proteins with PH domains to bind to PtdIns(3,4,5)P3 in the plasma membrane (Gokhale et al., 2013). However, previous work (Gokhale et al., 2011; Gokhale et al., 2013) has demonstrated PI3K-mediated recruitment of PPIP5K1 to the plasma membrane (courtesy of its own PtdIns(3,4,5)P3-binding domain). It is hypothesized that this translocation causes a local depletion of subplasmalemmal levels of InsP6 and 5-InsP7 through their phosphorylation to 1-InsP7 and InsP8 respectively, relieving the impediment to PH domain translocation (Gokhale et al., 2013). See text for details.
The relationship between PP-InsP turnover and cellular bioenergetics
Uniquely among inositol phosphate kinases, the IP6Ks have a low affinity for ATP (Km = 1 mM (Saiardi et al., 1999; Voglmaier et al., 1996)). As a consequence, physiologically-relevant alterations in [ATP] (1–5 mM; (Soboll et al., 1978)) may impact kinase activity and hence modify the cellular levels of 5-InsP7 (Nagel et al., 2010). That is, the prevailing levels of 5-InsP7 may reflect the degree of the cell’s bioenergetic well-being. In pancreatic β-cells this may be a significant event in organismal metabolic homeostasis: increased transport of serum glucose into β-cells stimulates the rate of glycolytic ATP production, elevating levels of 5-InsP7, which promotes exocytic insulin secretion (Barker and Berggren, 2013; Illies et al., 2007; Nagel et al., 2010). It is tempting to rationalize the significance of the multiple phosphates of 5-InsP7 as their being fundamental to the polyphosphate’s “energetic” properties that closely ties its synthesis to cellular ATP availability, aided by the enzymology of IP6Ks (i.e., their low ATP affinity).
When bioenergetic health is compromised, forcing 5-InsP7 levels to fall (Nagel et al., 2010), could loss of this pyrophosphate trigger compensatory metabolic processes? There is evidence for just such an activity in a dramatic 2011 study (Szijgyarto et al., 2011). Herein, it was shown that a sustained decrease in 5-InsP7 levels may promote metabolic adjustments to restore bioenergetic health: both kcs1 yeast and IP6K1-knockout mouse embryonic fibroblasts exhibit elevated ATP levels compared to corresponding wild-type cells. Intriguingly, if this phenomenon represents a homeostatic metabolic program in the mutant cells, its mechanistic basis is counterintuitive; mitochondrial ATP synthesis is much more efficient than that produced by glycolysis, but loss of IP6K activity was associated with up-regulation of glycolytic gene expression and a profound reduction in mitochondrial function (Szijgyarto et al., 2011). Perhaps another significant metabolic event in kcs1 yeast is the conservation of ATP through a decrease in the rate of cell growth and reduction in general synthetic activity (Irvine and Denton, 2011; Szijgyarto et al., 2011).
Cells have surveillance mechanisms in place to both detect and initiate responses to relatively mild bioenergetic challenges, before ATP levels are compromised. For example, an increase in AMP concentration is a much more sensitive indicator of the onset of such metabolic stress (Hardie and Hawley, 2001). Under just such conditions, InsP8 levels also decrease (Choi et al., 2008). The mechanisms that underlie this phenomenon are unknown. The AMP-activated protein kinase would normally be one of the usual suspects to round up, but our experiments (Choi et al., 2008) indicate that particular “metabolic master switch” (Winder and Hardie, 1999) does not regulate InsP8 turnover. To add to the mystery, PPIP5Ks are apparently immune from physiological changes in [ATP]; their low Km for ATP (approx 30 μM) is more in line with that for other inositol phosphate kinases (Weaver et al., 2013).
These data described above support proposals that both 5-InsP7 and InsP8 might be “metabolic messengers”, sensing and directing homeostatic responses to cellular and organismal bioenergetic imbalance ((Burton et al., 2009; Shears, 2009; Wilson et al., 2013)). This particular role for PP-InsPs rationalizes a significant benefit that the cell could gain from the energy-intensive investment in maintaining the levels of these molecules: a raison d’être for the synthesis of a molecule with so many phosphates.
Specific effects of PP-InsPs: the possibility of receptors
While studies with IP6Ks have highlighted how 5-InsP7 can be multifunctional (see above), some experiments with the PPIP5Ks (Vip1 in Saccharomyces cerevisiae) have indicated that another PP-InsP, 1-InsP7, may have rather more specific effects. For example, in a vip1Δ strain, the Pho80-Pho85 cyclin/cyclin-dependent kinase is impaired in its response to nutrient stress (Lee et al., 2007). Normally, this cyclin kinase becomes inhibited by Pho81 when S. cerevisiae is deprived of an external source of inorganic phosphate. As a consequence, one of the substrate of Pho85, the transcription factor Pho4, becomes dephosphorylated. In this form, it stimulates expression of a phosphate transporter and a secreted acid phosphatase which work together in organic phosphate hydrolysis and assimilation (Springer et al., 2003). However, in vip1Δ cells, the Pho80-Pho85 cyclin/cyclin-dependent kinase remains active when phosphate is depleted; Pho4 is maintained in an inactive state (Lee et al., 2007).
It may be worth cautioning that the deletion of vip1 alters the degree of transcription of over 900 genes (≥2-fold change in expression (Worley et al., 2013)). Even in a strain harboring a kinase-dead allele of vip1, the transcription of more than 250 genes is affected (Worley et al., 2013). So there is a possibility that secondary genetic changes lie at the heart of this vip1Δ phenotype. That is, it may not be the loss of the vip1 kinase activity per se that is responsible for impairing the Pho4 response. For example, recent work (Worley et al., 2013) indicates Pho81 expression may be reduced in vip1Δ cells, which could provide an alternate reason for why, in this strain, Pho80-Pho85 cyclin/cyclin-dependent kinase activity is prevented from being inhibited during phosphate starvation.
O’Shea and colleagues (Lee et al., 2007; Lee et al., 2008) have presented in vitro data that offer a different explanation for the vip1Δ phenotype that is based on the catalytic activity of the kinase. They report that 1-InsP7, one of the products of Vip1, is a cofactor that augments the inhibitory activity of Pho81. Thus, absence of 1-InsP7 in vip1Δ cells could account for the Pho80-Pho85 cyclin/cyclin-dependent kinase remaining active in phosphate-depleted cells. It is especially striking that 1-InsP7 acts in a specific manner; this effect, which appears to be allosteric in nature (Lee et al., 2008), is not imitated by either InsP6 or 5-InsP7. These in vitro experiments therefore offer a candidate, receptor-based action of a single PP-InsP that is both specific and monofunctional (pending the eventual testing of InsP8).
Whether or not levels of 1-InsP7 might become elevated during phosphate depletion - a potential signaling mechanism for controlling Pho85 kinase activity - has become a controversial topic. Unfortunately, no chromatographic technique has yet been demonstrated to have the sensitivity and resolving power to both monitor the relatively low levels of 1-InsP7 in eukaryotic cells and separate these from the much higher levels of 5-InsP7 (Albert et al., 1997; Kilari et al., 2013; Lin et al., 2009; Padmanabhan et al., 2009). So total InsP7 is generally the parameter that is reported. As discussed above, levels of InsP7 fall when cellular bioenergetics is compromised (such as that which rapidly occurs following phosphate depletion (Boer et al., 2010)). Indeed, one study (Lonetti et al., 2011) has described how total InsP7 levels decrease in yeast starved of phosphate for 1 – 2 hours. In contrast, O’Shea’s group report that total InsP7 accumulates following 2 hours of phosphate deprivation (Lee et al., 2007). The reasons for these differences remain unclear.
We (Kilari et al., 2013) have attempted to investigate if 1-InsP7 might regulate the mammalian homologue of the Pho80/Pho81/Pho85 complex (p35/CDK5RAP1 (originally: C42)/CDK5 (Ching et al., 2002; Huang et al., 1999)). We (Kilari et al., 2013) found that kinase activity was not affected by either 1-InsP7, nor any other PP-InsP. In any case, unlike Pho81, recombinant CDK5RAP1 appears fully competent to inhibit cyclin kinase activity even in the absence of any ancillary factors (Ching et al., 2000).
An entirely different role for 1-InsP7 has emerged from a systems level approach involving a human genome wide RNA-interference (RNAi) screen conducted in Krishnan’s laboratory (Pulloor et al., 2014). In the latter study it was demonstrated that the kinase activities of either PPIP5K2 or PPIP5K2 up-regulate IFN-β expression during viral invasion (Pulloor et al., 2014). IFN-β initiates anti-viral protein synthesis, and can also stimulate the ability of natural killer cells and cytotoxic T-cells to destroy the virus-infected cells (Sin et al., 2012). An illustration of the biological value of this pro-inflammatory pathway comes from the observation that the over-expression of either of the two PPIP5K isoforms in HEK cells affords them protection against infection by the influenza A virus (Pulloor et al., 2014).
It appears that it is the 1-InsP7 synthesized by the PPIP5Ks that promotes the inflammatory response to viral attack. Assays performed in vitro showed that as little as 0.5 μM 1-InsP7 enhance the degree of phosphorylation and activation of IRF3, a transcription factor for IFN-β expression (Pulloor et al., 2014). Although InsP8 also elicites this effect, it is much less potent. Moreover, 5-InsP7 is ineffective (Pulloor et al., 2014). These data strongly point to a specific, receptor-based mechanism of action. It might now be productive to investigate if levels of 1-InsP7 increase following cell infiltration by certain viruses, since such a response would speak further to the possibility that 1-InsP7 might act in a signaling capacity.
It is intriguing that IRF3 phosphophorylation is not supported by a synthetic analogue in which the diphosphate of 1-InsP7 was replaced with a phosphonoacetic acid ester (Pulloor et al., 2014). This reagent, synthesized by Potter’s group (Riley et al., 2012), cannot diphosphorylate proteins due to the chemical stability of its P-C bond. Thus, it is possible that phosphate transfer from the natural 1-InsP7 ligand contributes to its regulation of IRF3 phosphorylation. On the other hand, the diphosphate group of 1-InsP7 also has a higher degree of negative charge than does a phosphonoacetate (Wang et al., 2014b). Thus, the analogue may have been ineffective in this assay because it could not fully recapitulate electrostatic interactions of 1-InsP7 with a target protein. Moreover, it has been demonstrated that protein diphosphorylation by PP-InsPs is not specific to a particular member of this family; they all do it (Bhandari et al., 2007). That does not fit with the isomeric specificity with which 1-InsP7 regulates IRF3 (see above).
The metabolic and immune systems are closely integrated. Thus, nutrient excess can induce an inflammatory response by activating some of the same pathways that react to viral attack (Bieghs and Trautwein, 2013). For example, IFN-β transcription is stimulated by the addition of palmitate to cell cultures, a phenomenon that is exploited as a model for nutrient overload (McCall et al., 2010). Could 1-InsP7 participate in that response too? If it did, the kinase activities of the PPIP5Ks might then be considered a pharmacological target for the treatment of metabolic inflammation, such as that which occurs in obesity and diabetes (Bieghs and Trautwein, 2013). It might initially seem overly optimistic to suggest that the catalytic site of one member of such a large family of inositol phosphate kinases could be specifically druggable. However, PPIP5Ks have been found to host what may yet turn out to be a rare target for pharmacological intervention. On the surface of the kinase domain, there is a second substrate-binding site that is proposed to help commandeer substrate from the bulk phase, prior to its delivery into the catalytic pocket; this two-stage, “catch-and-pass” reaction mechanism has no known precedent among other small molecule kinases (Wang et al., 2014b). Moreover, compared to the catalytic pocket, this second, “substrate capture” site appears more tolerant to ligand modification, even to the extent that bulky hydrophobic groups can be added (Wang et al., 2014b).
Compartmentalization of PP-InsP signaling?
Early visualizations of inositol phosphates and other small-molecule second messengers depicted them as moving virtually unhindered throughout the cell (e.g. (Allbritton et al., 1992)). These ideas have now yielded to models of subcellular microdomains in which there is restricted diffusion of intracellular signals, allowing them to accumulate and act locally (Alekseev et al., 2012; Zaccolo et al., 2006). This case for compartmentalization is strengthened by a body of evidence describing reduced rates of diffusion in the narrow, submembranous space (Alekseev et al., 2012). Here for example, another so-called “diffusible” signal, cAMP, has been reported to exhibit a diffusion coefficient some seven orders of magnitude less than that observed in the bulk of the cytoplasm (Alekseev et al., 2012). Mobility may be restricted by physical barriers (e.g. protein aggregates) and electrostatic interactions with membranes (Alekseev et al., 2012).
For some time, compartmentalization of the turnover of both InsP6 and PP-InsPs has been felt likely to be important for mediating at least some of the cell-signaling activities of these molecules (Burton et al., 2009; Saiardi et al., 2004). This situation reflects the fact that InsP6 is the molecule that superficially is most similar to the PP-InsPs, and of course, InsP6 is also considerably more abundant. InsP6 can, for example, inhibit protein diphosphorylation by PP-InsPs (Saiardi et al., 2004). The thinking has been that to prevent InsP6 competing with PP-InsPs for a protein’s ligand-binding site, maybe there are spatially separated pools of these two groups of molecules (Saiardi et al., 2004).
There is evidence that compartmentalization of InsP6 may be brought about by Mg2+-enhanced binding of the polyphosphate to membranes (Poyner et al., 1993); in the latter study it was hypothesized that the role of the metal ion was to form a bridge between the polyphosphate and the positively charged phospholipids. InsP6 bound more tightly to membranes than did InsP5 isomers suggesting that it was the number of phosphates that is more important than the nature of their arrangement around the inositol ring (Poyner et al., 1993). The participation of Mg2+ in this binding phenomenon may be faciliated by InsP6 predominantly existing in a penta-Mg2+ salt inside cells (Torres et al., 2005). Perhaps Mg2+ also facilitates both InsP6 and the PP-InsPs being compartmentalized by their association with membranes. In fact, in preliminary experiments using surface plasmon resonance, we (Gokhale and Shears, unpublished) have obtained small but reproducible signals indicating InsP6 and PP-InsPs can associate with phospholipid vesicles in a Mg2+-dependent manner.
Further research is required in order to understand the extent to which InsP6 and PP-InsPs might be compartmentalized in vivo (see below), and the mechanisms involved. Should InsP6 and PP-InsPs exhibit restricted mobility in submembranous regions of the cell, it would strengthen the viability of some hypotheses concerning mechanisms of action of these polyphosphates. One proposed mechanism of action of PP-InsPs - protein diphosphorylation - could be enhanced by localized synthesis of these polyphosphates in close proximity to cellular membranes. Another example of compartmentalization is receptor-dependent translocation of PPIP5K1 to the plasma membrane, which has been hypothesized to locally reduce the levels of InsP6 and 5-InsP7 in the subplasmalemmal zone (Gokhale et al., 2013). The lower the rate of InsP6 and 5-InsP7 diffusion from the bulk phase and across this narrow, unstirred layer underneath the membrane (Alekseev et al., 2012), the longer it will take to replete this zone with those polyphosphates. As discussed above (and in Figure 2), such a phenomenon may assist PPIP5K1 in regulating PtdIns(3,4,5)P3-signaling. It is also worth mentioning that competition between 5-InsP7 and PtdIns(3,4,5)P3 for PH domains is subject to an order-of-addition phenomenon in vitro: 5-InsP7 is a 50-fold more potent inhibitor when it is added before as opposed to after PtdIns(3,4,5)P3 is bound to the AKT PH domain (Chakraborty et al., 2010). If this is also the case in vivo, then once the PH domains are occupied by PtdIns(3,4,5)P3, the proteins would be somewhat protected from being displaced from the plasma membrane by the InsP6 and 5-InsP7 that diffuses into the subplasmalemmal zone from the bulk phase.
The examination of ideas on compartmentalization of InsP6 and PP-InsPs in vivo will probably have to await the development of probes that can spatially resolve each of these individual polyphosphates. This is a research direction that is very much in its infancy and fraught with profound technical problems. For example, even though InsP6 is relatively abundant, fluorescent probes would have to be designed that can distinguish this polyphosphate from InsP5 isomers and even inorganic polyphosphates (Kolozsvari et al., 2014). A tetranaphthoimidazolium-based fluorescent probe for InsP6 was recently described (Lee et al., 2014). However, this reagent did not yield suitable signals with live cells until after they were incubated with exogenous InsP6 (Lee et al., 2014). InsP6 that is added to cells in this manner appears to be endocytosed into intracellular vesicles (Windhorst et al., 2013), raising the possibility that was also the fate of the tetranaphthoimidazolium complex (rather than it accessing the cytosol). Nevertheless, it is hoped that future work will build upon this initial step towards the goal of imaging the spatial dynamics of cellular InsP6 and, ultimately, pools of PP-InsP too.
General Conclusions
Phylogenetic analysis (Bennett et al., 2006), as well as other considerations (Saiardi, 2012b), have together argued that PP-InsP signaling is evolutionarily more ancient than is Ins(1,4,5)P3-mediated Ca2+ mobilization. Indeed, recent solving of the crystal structure of an Entamoeba histolytica IP6K - a proposed “living fossil” - has rationalized how an IP3K might have evolved from a primarily InsP6-phosphorylating kinase (Wang et al., 2014a). If signaling by PP-InsPs really was primeval, maybe some of their initial mechanisms of action might have exhibited less ligand specificity and subtlety than we are now used to characterizing. Could some versions of these putative non-specific regulatory processes have survived to this day? Perhaps one answer to that question is provided by 5-InsP7-mediated activation of insulin secretion (Illies et al., 2007): the dose/response relationship indicates 10 μM is a maximally-effective concentration. At this dosage, the 1-, 3-, 4- and 5-isomers of InsP7 were equally effective (Illies et al., 2007). Is this ligand promiscuity3 a confirmatory example of the non-specific nature of an evolutionarily ancient PP-InsP signaling response? In fact, insulin-like proteins themselves, as well as the process of protein secretion via exocytosis, are also long in the tooth, evolutionarily-speaking (Adelson, 1971; Burgoyne and Morgan, 2003; Chan and Steiner, 2000). Protein diphosphorylation is also not specific to any particular PP-InsP (Bhandari et al., 2007). (It might also be noted that protein diphosphorylation depends upon the action of CK2 which is itself one of the most evolutionarily conserved protein kinases (Pinna, 2002)).
If indeed signaling by PP-InsPs has such a long history, then there must have been considerable selection pressure to maintain the considerable investment of cellular energy in the synthesis and maintenance of such molecules. It is easy to appreciate that these bioenergetic demands could be repaid by the process of protein diphosphorylation by the PP-InsPs (Bhandari et al., 2007; Saiardi et al., 2004). The large free energy change that drives protein diphosphorylation relies not just upon the hydrolysis of a diphosphate bond per se, but also the accompanying steric and electronic rearrangements of the remaining multiple phosphates (Hand and Honek, 2007). That is, the requirement for many phosphates can be rationalized. However, as mentioned above (and see (Majerus, 2007)), the field still awaits a direct demonstration that this signaling mechanism operates in vivo. That being said, perhaps PP-InsPs donate phosphate to other targets that remain to be identified.
There is another property of PP-InsPs that may rationalize the cell investment in such highly-phosphorylated molecules; their overall, highly negative character may facilitate delocalized electrostatic interactions with positively charged protein domains. That is, non-specific (“brute-force”) electrostatics, dependent largely upon phosphate number, may overcome the requirement that a protein be presented with a particular three dimensional phosphate array (Lemmon et al., 2002). Such interactions may in part explain how binding of PtdIns(3,4,5)P3 to PH domains is inhibited by both InsP6 and 5-InsP7 (Shears et al., 2011). Perhaps these non-specific electrostatic interactions occur nearer to the surface of the protein rather than inside the ligand-binding pocket, thereby modulating the electrostatically polarized nature of the PH domain which is important for it being “steered” towards PtdIns(3,4,5)P3 in the plasma membrane (Lumb and Sansom, 2012). Such a relatively weak and rather non-specific mechanism might also be viewed as a process that is evolutionarily ancient. Perhaps some stereospecific interactions of PP-InsPs with PH domains (Gokhale et al., 2013) evolved more recently, following the appearance of a kinase activity that adds the 1-diphosphate to PP-InsPs.
Building on the original idea that the signaling roles of PP-InsPs are coordinated with the overall cellular phosphate balance (Bennett et al., 2006; Saiardi, 2012b), there is now much evidence that PP-InsPs are proactive metabolic messengers (Shears, 2009; Wilson et al., 2013). As discussed above, short-term bioenergetic crises in cells appear to correlate with decreases in levels of 5-InsP7 and/or InsP8 (Choi et al., 2008; Lonetti et al., 2011; Nagel et al., 2010). A sustained drop in cellular level of 5-InsP7 appears to promote compensatory metabolic reprogramming to assist a bioenergetic recovery (Szijgyarto et al., 2011). Conversely, an elevation in InsP7 levels may direct cellular responses to elevated serum glucose. For example, in pancreatic β-cells, glucose-mediated increases in ATP might increase InsP7 levels to promote insulin secretion (Illies et al., 2007; Nagel et al., 2010). Furthermore, reasoning presented above argues that it might be profitable to investigate if an elevation in 1-InsP7 synthesis might occur in response to pro-inflammatory nutrient excess. Such direct, functionally-significant relationships between cellular bioenergetic status and “high-energy” PP-InsP synthesis speak directly to a regulatory interface between the cell signaling and bioenergetic roles of phosphate esters. Arguably this is, at present, the most enticing answer to: “why so many phosphates?”
Acknowledgments
Work in the authors’ laboratory was supported by the Intramural Research Program of the NIH/National Institute of Environmental Health Sciences. I thank Drs. Chris Barker and Adolfo Saiardi for their insightful comments.
Footnotes
Not only the concentrations but also the nature of the PP-InsP isomers in slime molds differs from those in other eukaryotes. Dictyostelium and Polysphondylium species contain both 5-InsP6, 6-InsP6 and 5,6-InsP8 (Laussmann et al., 1998) (c.f. the structures in Figure 1). Polysphondylium also synthesize 1/3,5-InsP8 (Laussmann et al., 1998). Recent work with Arabidopsis suggests plants synthesize yet another InsP8 isomer by a PPIP5K/Vip1 dependent pathway that may not involve an IP6K (Desai et al., 2014).
These accomplishments prompt me also to acknowledge the important contributions made by other colleagues who battled with the chemical synthesis of PP-InsP synthesis in years past (Albert et al., 1997; Falck et al., 1995; Reddy et al., 1997; Zhang et al., 2009). The significance of this work should not be overlooked, particularly as these were pioneering syntheses at a time when the field garnered little attention. Nevertheless, I hope it will be considered fair to suggest that the more recently produced materials should prove to be more widely used research tools because their purity and durability are improved (see (Capolicchio et al., 2013; Capolicchio et al., 2014) for more discussion).
This apparent lack of specificity has little physiological consequence, as the 3- and 4-isomers do not occur naturally, and (as yet) there is no evidence the 1-isomer reaches levels that would have an effect upon exocytosis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adelson JW. Enterosecretory proteins. Nature. 1971;229:321–325. doi: 10.1038/229321a0. [DOI] [PubMed] [Google Scholar]
- Agranoff BW. Phosphorylated derivatives of myo-inositol. Fed Proc. 1985;45:2629–2633. [PubMed] [Google Scholar]
- Agranoff BW. Turtles All the Way: Reflections on myo-Inositol. J Biol Chem. 2009;284:21121–21126. doi: 10.1074/jbc.X109.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranoff BW, Eisenberg F, Jr, Hauser G, Hawthorn JN, Michell RH. Comment on Abbreviations. In: Bleasdale JE, Eichberg J, Hauser G, editors. Inositol and Phosphoinositides. Metabolism and Regulation. Clifton: Humna Press; 1985. p. xxi. [Google Scholar]
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy KK, Falck JR, Bröker M, Shears SB, Mayr GW. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseev AE, Reyes S, Selivanov VA, Dzeja PP, Terzic A. Compartmentation of membrane processes and nucleotide dynamics in diffusion-restricted cardiac cell microenvironment. J Mol Cell Cardiol. 2012;52:401–409. doi: 10.1016/j.yjmcc.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allbritton NL, Meyer T, Stryer L. Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science. 1992;258:1812–1815. doi: 10.1126/science.1465619. [DOI] [PubMed] [Google Scholar]
- Barker CJ, Berggren PO. New Horizons in Cellular Regulation by Inositol Polyphosphates: Insights from the Pancreatic beta-Cell. Pharmacol Rev. 2013;65:641–669. doi: 10.1124/pr.112.006775. [DOI] [PubMed] [Google Scholar]
- Barker CJ, Wright J, Hughes PJ, Kirk CJ, Michell RH. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell Mol Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, Molina H, Pandey A, Werner JK, Jr, Juluri KR, Xu Y, Prestwich GD, Parang K, Snyder SH. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci U S A. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieghs V, Trautwein C. The innate immune response during liver inflammation and metabolic disease. Trends Immunol. 2013;34:446–452. doi: 10.1016/j.it.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD, Morgan A. Secretory granule exocytosis. Physiol Rev. 2003;83:581–632. doi: 10.1152/physrev.00031.2002. [DOI] [PubMed] [Google Scholar]
- Burton A, Azevedo C, Andreassi C, Riccio A, Saiardi A. Inositol pyrophosphates regulate JMJD2C-dependent histone demethylation. Proc Natl Acad Sci U S A. 2013;110:18970–18975. doi: 10.1073/pnas.1309699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Hu X, Saiardi A. Are Inositol Pyrophosphates Signalling Molecules? J Cell Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- Caffrey JJ, Safrany ST, Yang X, Shears SB. Discovery of Molecular and Catalytic Diversity Among Human Diphosphoinositol Polyphosphate Phosphohydrolases: An Expanding NUDT Family. J Biol Chem. 2000;275:12730–12736. doi: 10.1074/jbc.275.17.12730. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Capolicchio S, Thakor DT, Linden A, Jessen HJ. Synthesis of Unsymmetric Diphospho-Inositol Polyphosphates. Angew Chem Int Ed Engl. 2013;52:6912–5916. doi: 10.1002/anie.201301092. [DOI] [PubMed] [Google Scholar]
- Capolicchio S, Wang H, Thakor DT, Shears SB, Jessen HJ. Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation. Angew Chem Int Ed Engl. 2014 doi: 10.1002/anie.201404398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, Saleh M, Snowman AM, Moran TH, Mezey E, Snyder SH. Inositol pyrophosphates inhibit akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SJ, Steiner DF. Insulin Through the Ages: Phylogeny of a Growth Promoting and Metabolic Regulatory Hormone. Amer Zool. 2000;40:213–222. [Google Scholar]
- Ching YP, Pang AS, Lam WH, Qi RZ, Wang JH. Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. J Biol Chem. 2002;277:15237–15240. doi: 10.1074/jbc.C200032200. [DOI] [PubMed] [Google Scholar]
- Ching YP, Qi Z, Wang JH. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene. 2000;242:285–294. doi: 10.1016/s0378-1119(99)00499-0. [DOI] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Choi JH, Shears SB. Cellular Energetic Status Supervises the Synthesis of Bis-Diphosphoinositol Tetrakisphosphate Independently of AMP-Activated Protein Kinase. Molecular Pharmacology. 2008;74:527–536. doi: 10.1124/mol.107.044628. Ref Type: Journal (Full) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Cooper G, Kimmich N, Belisle W, Sarinana J, Brabham K, Garrel L. Carbonaceous meteorites as a source of sugar-related organic compounds for the early Earth. Nature. 2001;414:879–883. doi: 10.1038/414879a. [DOI] [PubMed] [Google Scholar]
- Desai M, Rangarajan P, Donahue JL, Williams SP, Land ES, Mandal MK, Phillippy BQ, Perera IY, Raboy V, Gillaspy GE. Two Inositol Hexakisphosphate Kinases Drive Inositol Pyrophosphate Synthesis in Plants. Plant J. 2014 doi: 10.1111/tpj.12669. [DOI] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, Snyder SH, Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Europe-Finner GN, Gammon B, Newell PC. Accumulation of [3H]-inositol into inositol polyphosphates during development of dictyostelium. Biochem Biophys Res Commun. 1991;181:191–196. doi: 10.1016/s0006-291x(05)81400-7. [DOI] [PubMed] [Google Scholar]
- Falck JR, Reddy KK, Saady M, Mioskowski C, Shears SB, Tan Z, Safrany ST. Synthesis and structure of cellular mediators: inositol polyphosphate diphosphates. J A C S. 1995;117:12172–12175. [Google Scholar]
- Fisher DI, Safrany ST, McLennan AG, Cartwright JL. Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J Biol Chem. 2002;277:47313–47317. doi: 10.1074/jbc.M209795200. [DOI] [PubMed] [Google Scholar]
- Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Shukla D, Suman K, Lakshmi BJ, Manorama R, Kumar S, Bhandari R. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood. 2013;122:1478–1486. doi: 10.1182/blood-2013-01-481549. [DOI] [PubMed] [Google Scholar]
- Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J. 1993;293:583–590. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, Shears SB. PPIP5K1 Modulates Ligand Competition Between Diphosphoinositol Polyphosphates and PtdIns(3,4,5)P3 for Polyphosphoinositide-Binding Domains. Biochem J. 2013;453:413–426. doi: 10.1042/BJ20121528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Shears SB. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem J. 2011;434:415–426. doi: 10.1042/BJ20101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand CE, Honek JF. Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: a semi-empirical investigation. Bioorg Med Chem Lett. 2007;17:183–188. doi: 10.1016/j.bmcl.2006.09.066. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Hawley SA. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays. 2001;23:1112–1119. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- Huang D, Patrick G, Moffat J, Tsai LH, Andrews B. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc Natl Acad Sci U S A. 1999;96:14445–14450. doi: 10.1073/pnas.96.25.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, Yang S-N, Barma DK, Falck JR, Saiardi A, Barker CJ, Berggren P-O. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Ingram SW, Safrany ST, Barnes LD. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene and the effects on growth rate, morphology, and intracellular diadenosine 5′, 5‴-P1, P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem J. 2003;369:519–528. doi: 10.1042/BJ20020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF, Denton RM. Cell biology. Pyro-technic control of metabolism. Science. 2011;334:770–771. doi: 10.1126/science.1214727. [DOI] [PubMed] [Google Scholar]
- Jadav RS, Chanduri MV, Sengupta S, Bhandari R. Inositol Pyrophosphate Synthesis by Inositol Hexakisphosphate Kinase 1 is Required for Homologous Recombination Repair. J Biol Chem. 2013;288:3312–3321. doi: 10.1074/jbc.M112.396556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilari RS, Weaver JD, Shears SB, Safrany ST. Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett. 2013;587:3464–3470. doi: 10.1016/j.febslet.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolozsvari B, Parisi F, Saiardi A. Inositol phosphates induce DAPI fluorescence shift. Biochem J. 2014;460:377–385. doi: 10.1042/BJ20140237. [DOI] [PubMed] [Google Scholar]
- Laussmann T, Hansen A, Reddy KM, Reddy KK, Falck JR, Vogel G. Diphospho-myo-inositol phosphates in Dictyostelium and Polysphondylium: identification of a new bisdiphospho-myo-inositol tetrakisphosphate. FEBS Lett. 1998;426:145–150. doi: 10.1016/s0014-5793(98)00329-9. [DOI] [PubMed] [Google Scholar]
- Laussmann T, Pikzack C, Thiel U, Mayr GW, Vogel G. Diphospho-myo-inositol phosphates during the life cycle of Dictyostelium and Polysphondylium. Eur J Biochem. 2000;267:2447–2451. doi: 10.1046/j.1432-1327.2000.01264.x. [DOI] [PubMed] [Google Scholar]
- Lee M, Moon JH, Jun EJ, Kim G, Kwon YU, Lee JY, Yoon J. A tetranaphthoimidazolium receptor as a fluorescent chemosensor for phytate. Chem Commun (Camb) 2014;50:5851–5853. doi: 10.1039/c4cc02036g. [DOI] [PubMed] [Google Scholar]
- Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 2002;513:71–76. doi: 10.1016/s0014-5793(01)03243-4. [DOI] [PubMed] [Google Scholar]
- Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, Falck JR, Shears SB, York JD, Mayr GW. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J Biol Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetti A, Szijgyarto Z, Bosch D, Loss O, Azevedo C, Saiardi A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J Biol Chem. 2011;286:31966–31974. doi: 10.1074/jbc.M111.266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb CN, Sansom MS. Finding a needle in a haystack: the role of electrostatics in target lipid recognition by PH domains. PLoS Comput Biol. 2012;8:e1002617. doi: 10.1371/journal.pcbi.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase C1 mutant yeast. Biochemistry. 2002;41:2509–2515. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- Majerus PW. A Discrete Signaling Function for an Inositol Pyrophosphate. Sci STKE. 2007;416:pe72. doi: 10.1126/stke.4162007pe72. [DOI] [PubMed] [Google Scholar]
- McCall KD, Holliday D, Dickerson E, Wallace B, Schwartz AL, Schwartz C, Lewis CJ, Kohn LD, Schwartz FL. Phenylmethimazole blocks palmitate-mediated induction of inflammatory cytokine pathways in 3T3L1 adipocytes and RAW 264. 7 macrophages. J Endocrinol. 2010;207:343–353. doi: 10.1677/JOE-09-0370. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-b in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- Nagata E, Saiardi A, Tsukamoto H, Satoh T, Itoh Y, Itoh J, Shibata M, Takizawa S, Takagi S. Inositol hexakisphosphate kinases promote autophagy. Int J Biochem Cell Biol. 2010;42:2065–2071. doi: 10.1016/j.biocel.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Nagel A, Barker CJ, Berggren PO, Illies C. Diphosphosinositol polyphosphates and energy metabolism: assay for ATP/ADP ratio. Methods Mol Biol. 2010;645:123–131. doi: 10.1007/978-1-60327-175-2_8. [DOI] [PubMed] [Google Scholar]
- Nomenclature Committee of the International Union of Biochemistry. Numbering of atoms in myo-inositol. Recommendations 1988. Biochem J. 1989;258:1–2. [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna LA. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- Pisani F, Livermore T, Rose G, Chubb JR, Gaspari M, Saiardi A. Analysis of Dictyostelium discoideum inositol pyrophosphate metabolism by gel electrophoresis. PLoS ONE. 2014;9:e85533. doi: 10.1371/journal.pone.0085533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyner DR, Cooke F, Hanley MR, Reynolds DJM, Hawkins PT. Characterization of metal ion-induced 3hinositol hexakisphosphate binding to rat cerebellar membarnes. J Biol Chem. 1993;268:1032–1038. [PubMed] [Google Scholar]
- Prasad A, Jia Y, Chakraborty A, Li Y, Jain SK, Zhong J, Roy SG, Loison F, Mondal S, Sakai J, Blanchard C, Snyder SH, Luo HR. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat Immunol. 2011;12:752–760. doi: 10.1038/ni.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulloor NK, Nair S, Kostic AD, Bist P, Weaver JD, Tyagi R, Uchil PD, York JD, Snyder SH, Garcia-Sastre A, Lin R, Shears SB, Xavier RJ, Krishnan MN. Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response. PLoS Pathog. 2014;10:e1003981. doi: 10.1371/journal.ppat.1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KM, Reddy KK, Falck JR. Synthesis of 2- and 5-Diphospho-myo-inositol pentakisphosphate (2- and 5-PP-InsP5), intracellular mediators. Tetrahedron Lett. 1997;38:4951–4952. [Google Scholar]
- Riley AM, Wang H, Weaver JD, Shears SB, Potter BVL. First synthetic analogues of diphosphoinositol polyphosphates: interaction with PPIP5 kinase. Chem Commun. 2012;48:11292–11294. doi: 10.1039/c2cc36044f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Safrany ST, Shears SB. Turnover of bis-diphosphoinositol tetrakisphosphate in a smooth muscle cell line is regulated by b2- adrenergic receptors through a cAMP-mediated, A-kinase-independent mechanism. EMBO J. 1998;17:1710–1716. doi: 10.1093/emboj/17.6.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A. Cell signalling by inositol pyrophosphates. Subcell Biochem. 2012a;59:413–443. doi: 10.1007/978-94-007-3015-1_14. [DOI] [PubMed] [Google Scholar]
- Saiardi A. How inositol pyrophosphates control cellular phosphate homeostasis? Adv Biol Regul. 2012b;52:351–359. doi: 10.1016/j.jbior.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Bhandari A, Resnick R, Cain A, Snowman AM, Snyder SH. Inositol Pyrophosphate: Physiologic Phosphorylation of Proteins. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The Inositol Hexakisphosphate Kinase Family: Catalytic Flexibility, and Function in Yeast Vacuole Biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman A, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length via PI3K-related protein kinases. Proc Nat Acad Sci USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Nat Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Gokhale NA, Wang H, Zaremba A. Diphosphoinositol polyphosphates: what are the mechanisms? Adv Enzyme Regul. 2011;51:13–25. doi: 10.1016/j.advenzreg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Weaver JD, Wang H. Structural insight into inositol pyrophosphate turnover. Adv Biol Regul. 2013;53:19–27. doi: 10.1016/j.jbior.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman WR, Rasheed A, Mauck LA, Wiecko J. Incubations of testis myo-inositol-1-phosphate synthase with D-(5-18O)glucose 6-phosphate and with H218O show no evidence of Schiff base formation. J Biol Chem. 1977;252:5672–5676. [PubMed] [Google Scholar]
- Sin WX, Li P, Yeong JP, Chin KC. Activation and regulation of interferon-beta in immune responses. Immunol Res. 2012;53:25–40. doi: 10.1007/s12026-012-8293-7. [DOI] [PubMed] [Google Scholar]
- Soboll S, Scholz R, Heldt HW. Subcellular metabolite concentrations. Dependence of mitochondrial and cytosolic ATP systems on the metabolic state of perfused rat liver. Eur J Biochem. 1978;87:377–390. doi: 10.1111/j.1432-1033.1978.tb12387.x. [DOI] [PubMed] [Google Scholar]
- Springer M, Wykoff DD, Miller N, O’Shea EK. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 2003;1:E28. doi: 10.1371/journal.pbio.0000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LR, Radenberg T, Thiel U, Vogel G, Khoo K-H, Dell A, Jackson TR, Hawkins PT, Mayr GW. The detection, purification, structural characterization and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial store in pancreatic cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–68. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Torres J, Domínguez S, Cerdá FM, Obal G, Mederos A, Irvine RF, Dìaz A, Kremer C. Solution behaviour of myo-inositol hexakisphosphate in the presence of multivalent cations. Prediction of a neutral pentamagnesium species under cytosolic/nuclear conditions. Journal of Inorganic Biochemistry. 2005;99:828–840. doi: 10.1016/j.jinorgbio.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Voglmaier SM, Bembenek ME, Kaplin AI, Dormán G, Olszewski JD, Prestwich GD, Snyder SH. Purified inositol hexakisphosphate kinase is an ATP synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc Nat Acad Sci USA. 1996;93:4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, DeRose EF, London RE, Shears SB. IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family. Nature Communications. 2014a;5:4178. doi: 10.1038/ncomms5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Falck JR, Hall TM, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol. 2012;8:111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Godage HY, Riley AM, Weaver JD, Shears SB, Potter BVL. Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery. Chemistry & Biology. 2014b;21:689–699. doi: 10.1016/j.chembiol.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JD, Wang H, Shears SB. The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci Rep. 2013;33:228–241. doi: 10.1042/BSR20120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MS, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem J. 2013;452:369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- Winder WW, Hardie DG. AMP-activated protein kinase, a metabolic master switch: possible roles in type 2 diabetes. Am J Physiol. 1999;277:E1–10. doi: 10.1152/ajpendo.1999.277.1.E1. [DOI] [PubMed] [Google Scholar]
- Windhorst S, Lin H, Blechner C, Fanick W, Brandt L, Brehm MA, Mayr GW. Tumour cells can employ extracellular Ins(1,2,3,4,5,6)P6 and multiple inositol-polyphosphate phosphatase 1 (MINPP1) dephosphorylation to improve their proliferation. Biochem J. 2013;450:115–125. doi: 10.1042/BJ20121524. [DOI] [PubMed] [Google Scholar]
- Worley J, Luo X, Capaldi AP. Inositol Pyrophosphates Regulate Cell Growth and the Environmental Stress Response by Activating the HDAC Rpd3L. Cell Rep. 2013;3:1476–1482. doi: 10.1016/j.celrep.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Dul BE, Trevisan AJ, Fiedler D. Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate second messengers. Chem Sci. 2013;4:405–410. doi: 10.1039/C2SC21553E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- Zaccolo M, Di BG, Lissandron V, Mancuso L, Terrin A, Zamparo I. Restricted diffusion of a freely diffusible second messenger: mechanisms underlying compartmentalized cAMP signalling. Biochem Soc Trans. 2006;34:495–497. doi: 10.1042/BST0340495. [DOI] [PubMed] [Google Scholar]
- Zhang H, Thompson J, Prestwich GD. A scalable synthesis of the IP7 isomer, 5-PP-Ins(1,2,3,4,6)P5. Org Lett. 2009;11:1551–1554. doi: 10.1021/ol900149x. [DOI] [PMC free article] [PubMed] [Google Scholar]