Abstract
Malaria patients are frequently coinfected with HIV and mycobacteria causing tuberculosis, which increases the use of coadministered drugs and thereby enhances the risk of pharmacokinetic drug-drug interactions. Activation of the pregnane X receptor (PXR) by xenobiotics, which include many drugs, induces drug metabolism and transport, thereby resulting in possible attenuation or loss of the therapeutic responses to the drugs being coadministered. While several artemisinin-type antimalarial drugs have been shown to activate PXR, data on nonartemisinin-type antimalarials are still missing. Therefore, this study aimed to elucidate the potential of nonartemisinin antimalarial drugs and drug metabolites to activate PXR. We screened 16 clinically used antimalarial drugs and six major drug metabolites for binding to PXR using the two-hybrid PXR ligand binding domain assembly assay; this identified carboxymefloquine, the major and pharmacologically inactive metabolite of the antimalarial drug mefloquine, as a potential PXR ligand. Two-hybrid PXR-coactivator and -corepressor interaction assays and PXR-dependent promoter reporter gene assays confirmed carboxymefloquine to be a novel PXR agonist which specifically activated the human receptor. In the PXR-expressing intestinal LS174T cells and in primary human hepatocytes, carboxymefloquine induced the expression of drug-metabolizing enzymes and transporters on the mRNA and protein levels. The crucial role of PXR for the carboxymefloquine-dependent induction of gene expression was confirmed by small interfering RNA (siRNA)-mediated knockdown of the receptor. Thus, the clinical use of mefloquine may result in pharmacokinetic drug-drug interactions by means of its metabolite carboxymefloquine. Whether these in vitro findings are of in vivo relevance has to be addressed in future clinical drug-drug interaction studies.
INTRODUCTION
Malaria, which is caused by infection with parasitic protozoans of the genus Plasmodium, is still a major global health burden, with an estimated 207 million cases and 627,000 deaths worldwide in 2012 (1). The emerging resistance of the parasite to artemisinins (2) and the need to treat malaria patients coinfected with HIV and/or mycobacteria causing tuberculosis (3) increasingly necessitates the use of combination drug therapies and coadministration of drugs, respectively, which also may be accompanied by a higher risk for drug-drug interactions. Mechanistically, these may arise from the inhibition or induction of metabolism and/or transport of coadministered drugs. Competitive or noncompetitive enzyme inhibition may result in adverse toxic effects due to higher-than-expected drug concentrations, whereas clinically relevant induction may result in therapeutic failure due to insufficient drug levels. The interaction potential of antimalarial drugs due to the inhibition of cytochrome P450 (CYP) drug-metabolizing enzymes has been analyzed quite extensively in vitro, and it has been shown to result in some clinically relevant drug-drug interactions, as exemplified by the interaction of quinine/quinidine with drugs metabolized by CYP2D6 (4). In contrast, the induction of drug metabolism and transport by antimalarials has not been addressed equally well. With respect to clinically relevant induction, it is known only that artemisinin-type antimalarial drugs, especially artemisinin itself, autoinduce their elimination (5) by increasing cytochrome P450-dependent metabolism (6), thereby suggesting a significant drug-drug interaction potential of the compounds. Drug-drug interactions by artemisinin-induced CYP2B6 and CYP3A4 expression and activities have been further predicted by quantitative modeling (7).
The induction of drug metabolism and transport by xenobiotics, which include many drugs, is mediated by xenosensing nuclear receptors, among which pregnane X receptor (PXR) (NR1I2) and the closely related constitutive androstane receptor (CAR) (NR1I3) are of outstanding importance in humans, as they are activated by a wide array of structurally highly diverse chemicals (8). Xenobiotics activate these receptors by inducing their transcriptional activity, which in the case of PXR typically requires binding of the compound to the receptor as a ligand (8). Upon activation, both receptors induce the expression of a battery of genes involved in absorption, distribution, metabolism, and excretion (ADME), comprising phase I cytochrome P450 and phase II conjugating drug-metabolizing enzymes, as well as phase 0/III drug transporters in the intestines and liver (9, 10). In previous studies, we and others showed that the artemisinin-type antimalarial drugs artemisinin, artemether, and arteether, as well as the pharmacologically inactive artemisinin metabolite deoxyartemisinin, are ligands of human PXR and human and mouse CAR, and by that, induce ADME gene expression (11–13). Other artemisinin-type drugs, such as artesunate and dihydroartemisinin, showed weak human CAR inverse agonism, with differential effects on CAR isoforms (13). When screening 21 antimalarial drugs and 11 major drug metabolites, we did not identify any drug outside the artemisinin class as binding to and thereby activating CAR (13). However, the potential of nonartemisinin antimalarials in activating PXR has not yet been analyzed.
This study aimed to elucidate the activation of PXR by nonartemisinin antimalarial drugs and major drug metabolites. By using the cellular mammalian two-hybrid PXR ligand binding domain assembly assay to screen ligand binding, as well as further cellular assays to detect ligand-dependent cofactor interactions and PXR-dependent reporter gene activity, we identified carboxymefloquine, the main metabolite of mefloquine, which is a major drug used in antimalarial prophylaxis, as a novel agonist and activator of PXR. These results were further substantiated by the observation of carboxymefloquine-driven ADME gene expression in intestinal and hepatic cells. The data presented here indicate that the clinical use of mefloquine may result in drug-drug interactions by means of its metabolite.
MATERIALS AND METHODS
Chemicals.
Rifampin and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (Taufkirchen, Germany). The supply sources of the antimalarial drugs and drug metabolites have been noted previously (13).
Plasmids.
Expression plasmids carrying genes encoding fusion proteins of the GAL4-DNA binding domain (DBD) and helix 1 of the human PXR ligand binding domain (LBD) (amino acids 132 to 188) or the receptor interaction domain (RID) of human corepressor silencing mediator of retinoid acid and thyroid hormone receptor (SMRT) have been published (11). The respective fusion of the RID of human coactivator SRC1 was previously described (14). Expression plasmids carrying genes encoding fusion proteins of the VP16-activation domain (AD) and human PXR LBD (amino acids 108 to 434) or part of it (amino acids 189 to 434) have been described (11).
The expression plasmids carrying genes encoding human farnesoid X receptor (FXR), liver X receptor alpha (LXRα), and LXRβ were constructed by cloning the respective open reading frames, amplified by PCR from the cDNA of the human intestinal cell lines Caco-2 and LS174T, using the appropriate primers, into expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA). The respective 5′ upstream primers introduced optimized Kozak consensus sequences. The human PPARγ1 expression plasmid was described previously (15) and kindly provided by T. Tanaka (University of Tokyo, Japan). All further expression plasmids carrying genes encoding human and mouse nuclear receptors have been described (11).
Annealed complementary oligonucleotides, designed using the BLOCK-iT RNAi Designer (Invitrogen) to target human PXR (5′-GCT GAC ATG TCA ACC TAC ATG-3′ [sh-PXR#1, positions 2569 to 2589 of GenBank accession no. NM_003889] and 5′-GAC ACT ACC TTC TCC CAT TTC-3′ [sh-PXR#3, positions 2326 to 2346] or nonmammalian sequence 5′-CAA CAA GAT GAA GAG CAC CAA-3′ [sh-CTR]), were cloned into pENTR/U6 vector (Invitrogen), and the inserts were sequenced. The two sh-PXR plasmids were kindly provided by B. A. Kandel (Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, Germany).
The following firefly luciferase reporter gene plasmids have been published: GAL4-dependent pGL3-G5 (14), CYP3A4 enhancer/promoter reporter gene plasmid pGL3-CYP3A4(−7830/Δ7208 to 364) and reporter gene plasmid with a trimer of CYP3A23 direct repeat (DR) 3 motif (16), reporter gene plasmid with a dimer of the MDR1-DR4(I) (17), CYP2B6 enhancer/promoter reporter gene plasmid pB-1.6k/PB/XREM (18), reporter gene plasmids containing a dimer of the hepatic nuclear factor alpha (HNF4α)-response element (RE) of the human PXR promoter or tetramer of the glucocorticoid receptor (GR)-RE of the tyrosine aminotransferase promoter, respectively (11), and p4xACO-Luc, Addgene plasmid 16533, kindly provided by B. Vogelstein (19). The reporter gene plasmid, containing a dimer of the consensus nuclear receptor inverted repeat (IR) 1 motif, was constructed by self-ligation of the appropriate double-stranded oligonucleotides via added BamHI/BglII sites and cloning into the BglII site of pGL3-Tk(−105) (17).
Cell culture, transient transfections, and reporter gene assays.
The COS-1 cells were cultivated as described previously (14). The culture of HepG2 cells, purchased from ATCC (Manassas, VA), and the generation and culture of HepG2-PXR cells stably transfected with a human PXR expression plasmid have been described (20). One day before transfection, the COS-1 cells were plated at 3 × 104 cells per well and the HepG2 and HepG2-PXR cells at 1.5 × 105 cells per well in 24-well plates. The transfections were performed in triplicate using plasmid DNAs totaling 0.2 μg and Effectene transfection reagent (Qiagen, Hilden, Germany), according to the manufacturer's protocol. Mammalian two-hybrid PXR assembly, coactivator, and corepressor interaction assays were performed as described previously (11), using the plasmids specified in the figure legends. CYP3A4 and CYP2B6 enhancer/promoter reporter gene assays were performed as described previously (16). The following firefly luciferase reporter gene plasmids were used for the analysis of nuclear receptor activation by carboxymefloquine: CYP3A4 (human PXR [hPXR] and mouse PXR [mPXR]), CYP2B6 (hCAR and mCAR), DR3 (vitamin D receptor [VDR]), DR4 (thyroid hormone receptor alpha [TRα], TRβ, LXRα, and LXRβ), HNF4α-RE (HNF4α), GR-RE (GRα), IR1 (FXR), and p4xACO-Luc (PPARγ1). Normalization plasmids pRL-CMV (Renilla luciferase; Promega, Madison, WI) or pCMVβ (β-galactosidase; Clontech, Mountain View, CA) were cotransfected to adjust for variable transfection efficiencies. Subsequently, the cells were treated with chemicals for 6 h or 24 h, as specified in the figure legends. The cells were lysed with passive lysis buffer (Promega), and the firefly luciferase and β-galactosidase activities were analyzed as described previously (21). To determine Renilla luciferase activities, 100 μl of an assay solution consisting of 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM CaCl2, and 1 μM coelenterazine was injected automatically into 20 μl of cell lysate, and luminescence was measured immediately for 10 s with an AutoLumat Plus (Berthold, Bad Wildbad, Germany).
Primary human hepatocytes.
Human hepatocytes were isolated from the tissue samples of human liver resections, which were obtained from patients who underwent partial hepatectomy because of primary or secondary liver tumors, as described previously (22). The experimental procedures were performed according to the institutional guidelines for liver resections of tumor patients, which included each patient's written informed consent, and these were approved by the local ethics committee of the Eberhard-Karls University of Tübingen, Germany. For the induction experiments, the isolated cells were seeded at 1.5 × 106 cells per well into collagen type I-coated 6-well plates and treated with chemicals, as described previously (20).
siRNA-mediated gene knockdown.
Primary human hepatocytes were seeded at 9.6 × 105 cells/well into collagen type I-coated 6-well plates. After 8 to 10 h, the cells were transfected with nontargeting negative-control siRNA (Silencer select negative-control 1) or predesigned Silencer select siRNA targeting 5′-GGC TAT CAC TTC AAT GTC A-3′ (positions 1987 to 2005 of GenBank accession no. NM_003889) of PXR (s16911), which were provided by Life Technologies (Darmstadt, Germany), at a final concentration of 20 nM using Lipofectamine RNAiMAX (Life Technologies). The culture medium was renewed daily, and the cells were harvested for RNA analysis 72 h after transfection. During the last 24 h, the cells were treated with chemicals.
Quantitative real-time reverse transcription-PCR analysis.
Total RNA and first-strand cDNA were prepared by established procedures (21). The integrity of the RNA samples was confirmed by formaldehyde agarose gel electrophoresis.
The absolute quantification of ABCB1, CYP2B6, CYP3A4, and 18S rRNA gene expression levels in LS174T cells (ATCC) was performed with cDNA corresponding to 25 ng or 25 pg (18S rRNA) total RNA using the 7500 real-time PCR system (Life Technologies), as described previously (20). The respective TaqMan assays are indicated below.
For the relative quantification analyses of ADME genes in primary human hepatocytes, cDNA samples (25 ng each) were preamplified for 14 cycles using a mix of primer-probe sets of the respective genes (omitting the set of 18S rRNA) and TaqMan PreAmp master mix (Life Technologies), according to the Fluidigm specific target amplification protocol (Fluidigm, South San Francisco, CA); the mixture was finally diluted 1:5 with nuclease-free H2O. A 48.48 Dynamic Array or FLEXsix gene expression integrated fluidic circuit (Fluidigm) was loaded with these diluted preamplified samples and the primer-probe sets of the respective genes, the latter of which were added before into TaqMan gene expression master mix (Life Technologies). TaqMan real-time quantitative PCR was performed using the BioMark HD system (Fluidigm), according to the manufacturer's protocol. The assays were done in triplicate. The following TaqMan assays have been reported: ABCB1 and CYP2B6 (11), UGT1A3 (23), and EPHX1 and 18S rRNA (20). TaqMan gene expression assays, consisting of predesigned commercial primer-probe sets (Life Technologies), were used to quantify the other genes: Hs00166123_m1 (ABCC2), Hs00868409_s1 (CYP2A6), Hs00946140_g1 (CYP2C8), Hs00604506_m1 (CYP3A4), Hs01114267_m1 (PXR), Hs99999902_m1 (RPLP0), and Hs00272374_m1 (SLCO1B1). The data were analyzed using the BioMark real-time PCR analysis software and further processed by applying the ΔΔCT method. The 18S rRNA levels were used to normalize the gene expression levels.
Protein analysis.
Total protein homogenates were prepared by lysing cells for 30 min on ice in 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 1% (vol/vol) Nonidet P-40, supplemented with Halt protease inhibitor cocktail (Thermo Scientific, Rockford, IL) and quantified using the bicinchoninic acid method. Thirty micrograms of each protein sample was analyzed by SDS-polyacrylamide gel electrophoresis and subsequent Western blotting (20), using the antibodies WB-3A4 and INHIBITION MAB-2B6 (BD Biosciences, Heidelberg, Germany) specific for CYP3A4 and CYP2B6, respectively, and β-actin antibody (clone AC-15) from Sigma-Aldrich. The protein bands were quantified with the digital charge-coupled-device camera Stella and AIDA software (Raytest, Straubenhardt, Germany).
Data analysis.
The mean or median values from at least three independent experiments were used for statistical analysis, unless indicated otherwise. Multiple comparisons were performed using one-way or two-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test, as described in the figure legends. The comparisons with a hypothetical value were performed using a one-sample t test or Wilcoxon signed-rank test. The Wilcoxon signed-rank test was used exclusively for the hepatocyte experiments, in which large interindividual variability was observed. All calculations were done with GraphPad Prism 6.03 (GraphPad Software, La Jolla, CA).
RESULTS
Screening of antimalarial drugs and drug metabolites for binding to PXR.
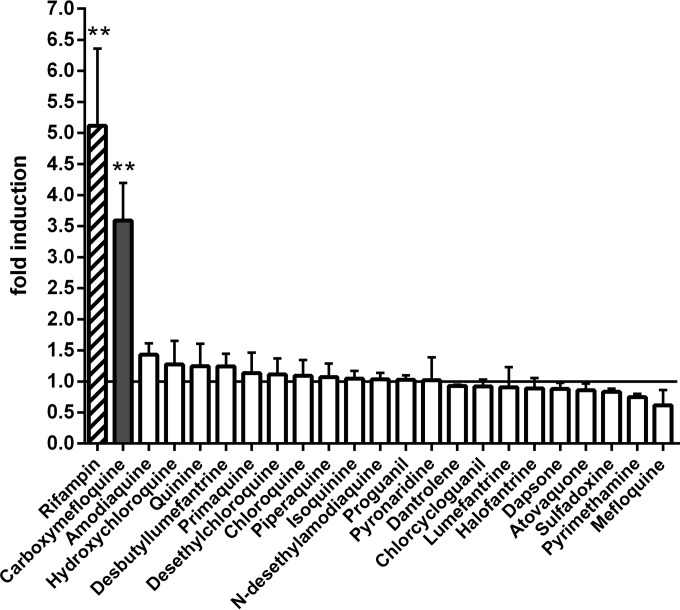
The mammalian two-hybrid assembly assay of human PXR, which relies on the ligand-dependent interaction of helix 1 with the remainder of the ligand binding domain (11), was applied to screen the PXR binding characteristics of 16 antimalarial drugs and six major drug metabolites. Artemisinin-type compounds, which have been analyzed for their activation of the xenobiotic receptors PXR and CAR (11, 13), were excluded. As several of the compounds proved to be highly cytotoxic to the HepG2 cells used in this assay (data not shown), treatment duration had to be limited to 6 h, which was still sufficient to generate a significant induction with the prototypical PXR ligand rifampin at a concentration of 10 μM (Fig. 1). The short treatment time further allowed for using compounds at concentrations up to 30 μM without being cytotoxic, with the single exception of desbutyl-lumefantrine, which could be used only at 10 μM. Among the tested compounds, only carboxymefloquine [Ro 21-5104; 2,8-bis(trifluoromethyl)-4-quinoline carboxylic acid], the major metabolite of mefloquine (24), significantly induced the assembly of the human PXR ligand binding domain (Fig. 1). These data indicate that carboxymefloquine may be a ligand of PXR.
FIG 1.
Carboxymefloquine induces the assembly of the human PXR ligand binding domain. HepG2 cells, transiently transfected with firefly luciferase reporter gene plasmid pGL3-G5, normalization plasmid pRL-CMV, and expression plasmids carrying genes encoding GAL4-DBD/PXR-LBD (positions 132 to 188) and VP16-AD/PXR-LBD (positions 189 to 434) fusion proteins were treated with the indicated antimalarials or vehicle DMSO for 6 h. Rifampin was used at 10 μM and all antimalarials were used at 30 μM, except desbutyl-lumefantrine, which was used at 10 μM. The columns show the mean fold induction ± standard deviation (SD) of normalized firefly luciferase activities by chemical treatment, with the activities of cells treated only with DMSO designated 1. Statistically significant differences to this value were calculated by one-sample t test and P values were corrected for multiple testing by Bonferroni's method. **, P < 0.01.
Carboxymefloquine is an agonist of PXR.
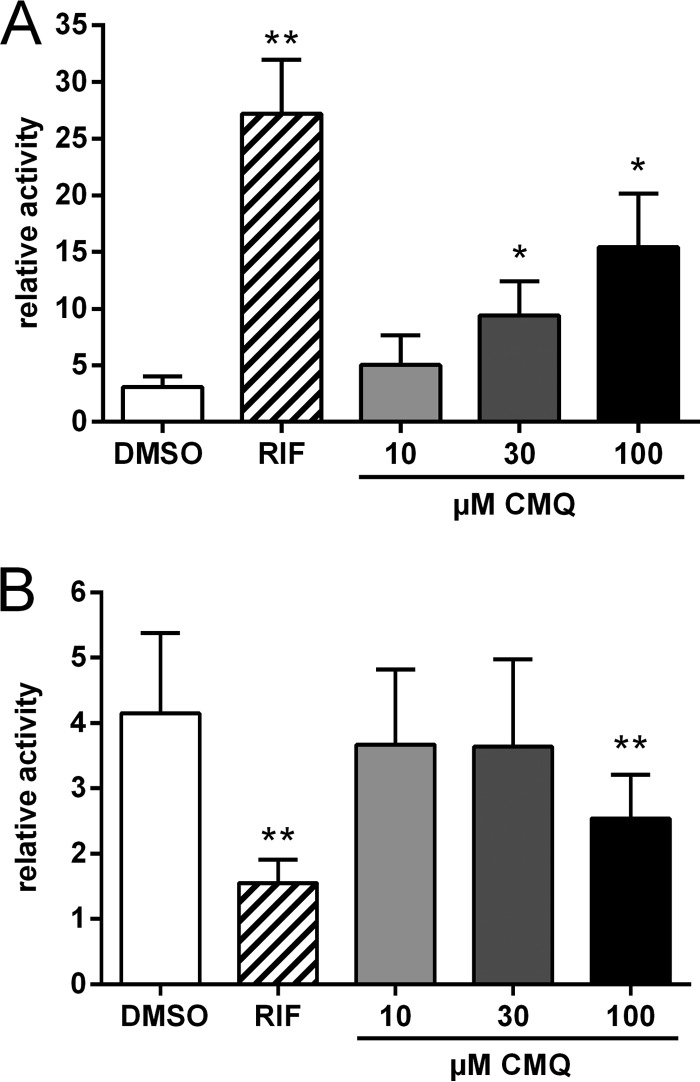
Nuclear receptor assembly assays cannot discriminate between agonist and antagonist ligands (25). Thus, we next analyzed whether carboxymefloquine shows an induction of coactivator recruitment and release of corepressor interaction, both of which are characteristics of PXR agonists (20). Figure 2A shows that the compound induced the interaction of PXR with the coactivator SRC1 in a dose-dependent manner. A significant release of corepressor SMRT from PXR was observed at the highest dose of 100 μM (Fig. 2B). In summary, these data suggest that carboxymefloquine acts as a PXR agonist.
FIG 2.
Carboxymefloquine is an agonist of PXR. HepG2-PXR cells, transiently transfected with firefly luciferase reporter gene plasmid pGL3-G5, normalization plasmid pCMVβ, and expression plasmids carrying genes encoding VP16-AD/PXR-LBD (positions 108 to 434) and GAL4-DBD/SRC1-RID (A) or GAL4-DBD/SMRT-RID (B) were treated with 10 μM rifampin (RIF), the indicated doses of carboxymefloquine (CMQ), or 0.1% DMSO for 24 h. The columns show the mean ± SD of relative normalized firefly luciferase activities compared to the activities of cells cotransfected with empty vector pVP16-AD and respective GAL4-DBD fusion protein expression plasmid and treated with DMSO only, which was designated 1 and is indicated by the broken line. The data were analyzed by repeated-measures one-way ANOVA with Dunnett's multiple-comparison test. *, P < 0.05; **, P < 0.01.
Carboxymefloquine specifically activates human PXR.
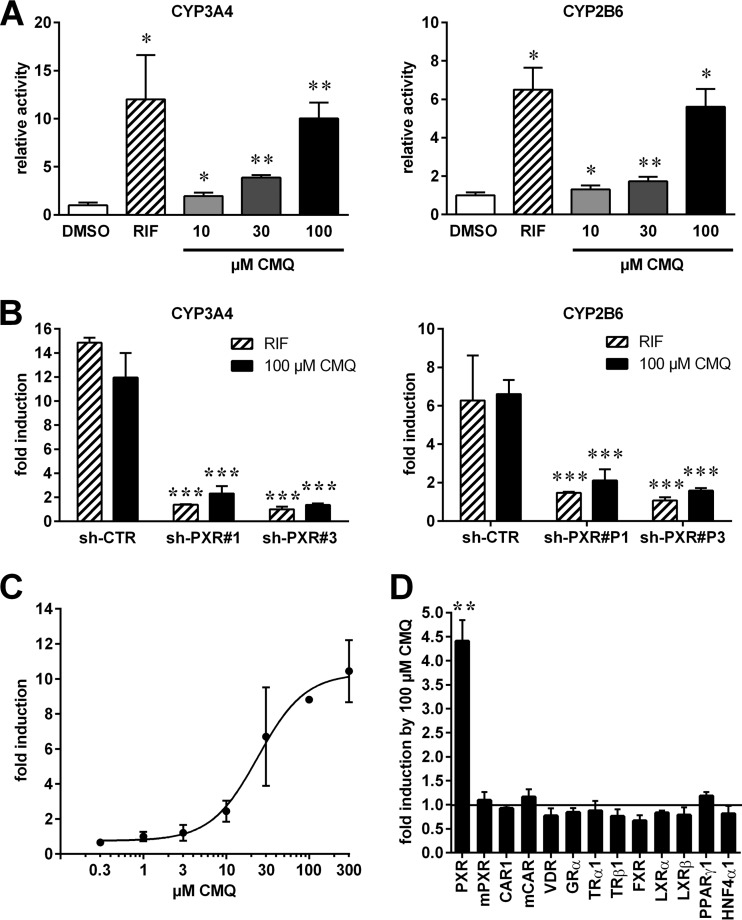
As an agonist, carboxymefloquine should induce transcriptional activation by PXR. Thus, HepG2-PXR cells, which express high levels of PXR due to stable transfection of a respective expression plasmid (20), were transiently transfected with PXR-dependent CYP3A4 and CYP2B6 enhancer/promoter reporter genes and treated with carboxymefloquine. Figure 3A shows that the compound significantly induced the transcriptional activation of CYP3A4 and CYP2B6 reporters in a dose-dependent manner. PXR knockdown by transient cotransfection of short hairpin RNA (shRNA) expression plasmids, which carry PXR-specific siRNA sequences, further confirmed that the induction of both reporters by carboxymefloquine was indeed mediated by PXR (Fig. 3B). A dose-response analysis revealed that carboxymefloquine activated human PXR, with a 50% effective concentration (EC50) of 24 μM (95% confidence interval [CI], 13.4 to 43.3 μM) (Fig. 3C). Next, we analyzed the specificity of activation by carboxymefloquine. COS-1 cells were cotransfected with nuclear receptor expression plasmids and the appropriate respective reporter genes. Figure 3D shows that among the 13 nuclear receptors that were analyzed, only human PXR was activated by carboxymefloquine. Notably, carboxymefloquine activated neither mouse PXR nor CAR or VDR, the two closest relatives of PXR. In summary, these data suggest that carboxymefloquine is a species-specific activator of PXR, strongly activating the human receptor but not the mouse homologue.
FIG 3.
Carboxymefloquine specifically activates human PXR. (A) HepG2-PXR cells were transiently transfected with CYP3A4 or CYP2B6 enhancer/promoter reporter gene plasmids, as indicated, and normalization plasmid pCMVβ. The transfected cells were treated with 0.1% DMSO, 10 μM rifampin (RIF), or the indicated doses of carboxymefloquine (CMQ) for 24 h. The columns show the mean ± SD of relative normalized luciferase activities compared to the mean activities of cells treated with DMSO only, which was designated 1. The data were analyzed by repeated-measures one-way ANOVA with Dunnett's multiple-comparison test. (B) HepG2-PXR cells were transfected as in panel A, except that the shRNA expression plasmids carrying negative-control siRNA (sh-CTR) or PXR-specific siRNA (sh-PXR#1 and sh-PXR#3) sequences were cotransfected. The cells were treated for 24 h, as indicated. The columns show the mean fold induction ± SD of normalized luciferase activities by chemical treatment, with the activities of appropriately transfected cells treated with DMSO designated 1. The data were analyzed by two-way ANOVA with Dunnett's multiple-comparison test. (C) HepG2-PXR cells were transfected with CYP3A4 enhancer/promoter reporter gene plasmid and normalization plasmid pCMVβ. The graph shows the dose response of induction by carboxymefloquine. (D) COS-1 cells were transiently cotransfected with combinations of the indicated mouse (m) or human nuclear receptor expression plasmids and the appropriate respective promoter reporter gene plasmids (as specified in Materials and Methods) and normalization plasmid pCMVβ. The transfected cells were treated with 0.1% DMSO or 100 μM carboxymefloquine for 24 h. The columns show the mean fold induction ± SD of normalized luciferase activities by carboxymefloquine, with the activities of the appropriately transfected cells treated with DMSO designated 1. The data were analyzed by one-sample t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Carboxymefloquine induces the expression of drug-metabolizing enzyme and transporter genes in human intestinal and hepatic cells.
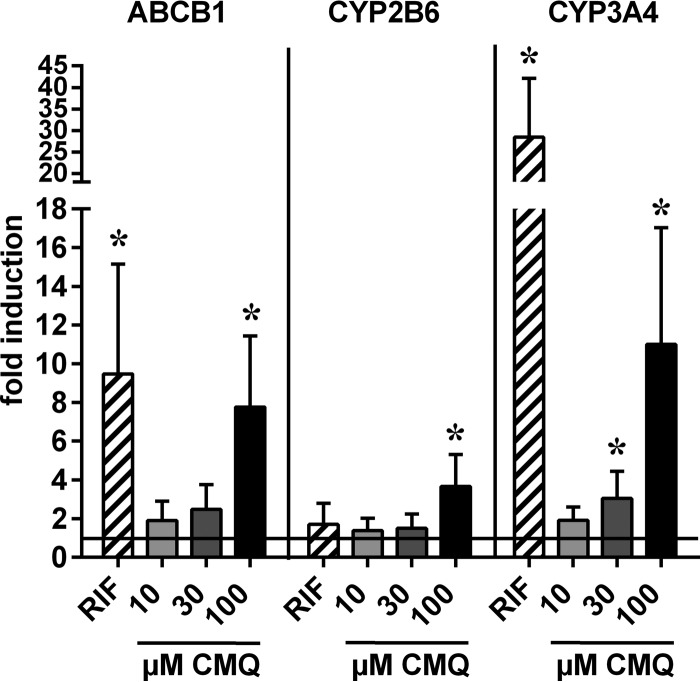
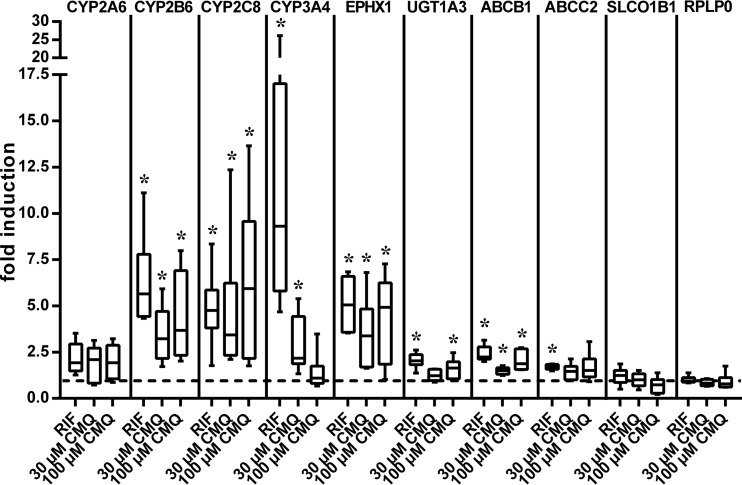
To analyze whether carboxymefloquine induces endogenous PXR target genes, human intestinal LS174T cells and primary human hepatocytes, both proven models of PXR activation due to strong expression of the receptor (11, 17), were treated with the compound, and the expression of selected PXR target genes was analyzed by quantitative real-time reverse transcription-PCR (RT-PCR). Figure 4 shows that carboxymefloquine induced the expression of ABCB1, which encodes the drug transporter multidrug resistance protein 1 (MDR1)/P-glycoprotein (Pgp), and of cytochrome P450 genes CYP2B6 and CYP3A4 in LS174T cells in a dose-dependent manner. In primary human hepatocytes, a more comprehensive set of PXR target genes was analyzed. Carboxymefloquine significantly induced the median mRNA expression of CYP2B6, CYP2C8, EPHX1, UGT1A3, and ABCB1. The expression of CYP2A6 and ABCC2 appeared to be induced, but it did not reach statistical significance, whereas the expression of SLCO1B1 was not altered (Fig. 5). Interestingly, CYP3A4 was induced only by 30 μM carboxymefloquine, whereas increasing the dose to 100 μM did not result in further induction. Consistently, RPLP0, which is not a PXR target gene, was induced neither by rifampin nor carboxymefloquine. Together, these data clearly demonstrate the capacity of carboxymefloquine to induce the expression of ADME genes.
FIG 4.
Carboxymefloquine induces the expression of cytochrome P450 genes and ABCB1 in intestinal LS174T cells. The cells were treated for 48 h with 0.1% DMSO, 10 μM rifampin (RIF), or the indicated doses of carboxymefloquine (CMQ). mRNA expression of the indicated genes was quantified by TaqMan real-time RT-PCR and normalized to the expression levels of 18S rRNA. The columns show the mean fold induction ± SD of mRNA expression by chemical treatment, with the expression in the cells treated with DMSO designated 1. The data were analyzed by one-sample t test. *, P < 0.05.
FIG 5.
Carboxymefloquine coordinately induces the expression of ADME genes in primary human hepatocytes. Primary human hepatocyte cultures from 6 donors were each treated with 0.1% DMSO, 30 μM rifampin (RIF), or the indicated doses of carboxymefloquine (CMQ) for 48 h. mRNA expression of the indicated genes was quantified by TaqMan real-time RT-PCR and normalized to the expression levels of 18S rRNA. The data are shown as the fold induction of mRNA expression by chemical treatment, with the expression in the cells treated with only DMSO designated 1; these data are presented as box and whisker plots, with boxes representing the 25th to 75th percentiles, medians indicated by horizontal lines, and whiskers showing the minimum and maximum values. The data were analyzed by Wilcoxon signed-rank test. *, P < 0.05.
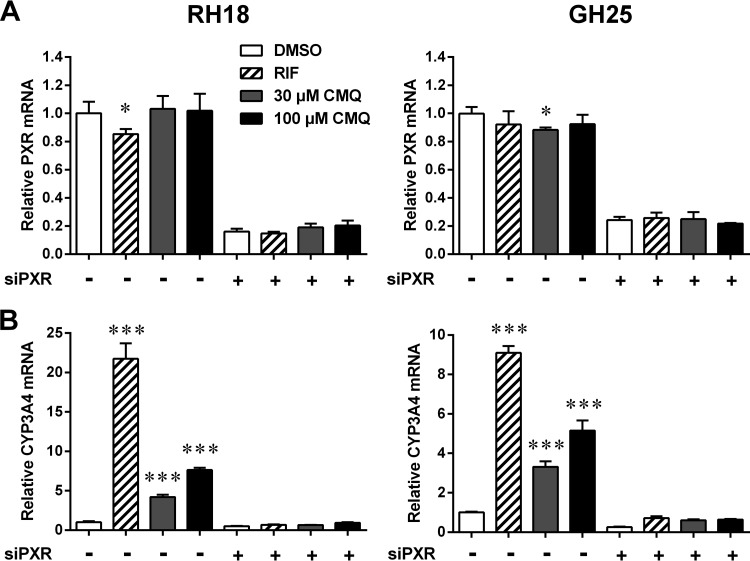
PXR mediates carboxymefloquine-dependent induction of gene expression in primary human hepatocytes.
In contrast to LS174T cells, which do not express detectable levels of CAR (11), primary human hepatocytes show prominent CAR expression (26). Even if we previously showed (13) and confirmed again here that carboxymefloquine does not act as a human CAR agonist, it is still possible that the compound indirectly activates this receptor and by doing so induces the expression of ADME genes in primary human hepatocytes. Therefore, siRNA-mediated knockdown of PXR expression was performed in primary human hepatocytes of two different donors to unequivocally demonstrate that PXR mediates the induction of gene expression by carboxymefloquine. The transfection of PXR-specific siRNA resulted in 75 to 80% knockdown of PXR mRNA expression (Fig. 6A), which equally eliminated the induction of CYP3A4 expression by rifampin and carboxymefloquine (Fig. 6B). In contrast to treatment for 48 h (see Fig. 5), carboxymefloquine further showed dose-dependent induction of CYP3A4 expression in the hepatocytes of both donors if the cells were treated for 24 h only.
FIG 6.
PXR mediates induction of CYP3A4 by carboxymefloquine in primary human hepatocytes. Primary human hepatocytes of two donors (RH18 and GH25) were transfected with PXR-specific siRNA (+) or negative-control siRNA (−). The cells were treated for 24 h with 0.1% DMSO, 10 μM rifampin (RIF), or carboxymefloquine (CMQ), as indicated. mRNA expression of PXR (A) and CYP3A4 (B) was quantified by TaqMan real-time RT-PCR and normalized to the expression of 18S rRNA levels. The columns show the means ± SD from triplicate measurements. The expression data are presented relative to the mean expression of cells transfected with control siRNA and treated with DMSO, which was designated 1. The data were analyzed by two-way ANOVA with Dunnett's multiple-comparison test. Significant differences to the respective cells treated with DMSO are indicated. *, P < 0.05; ***, P < 0.001.
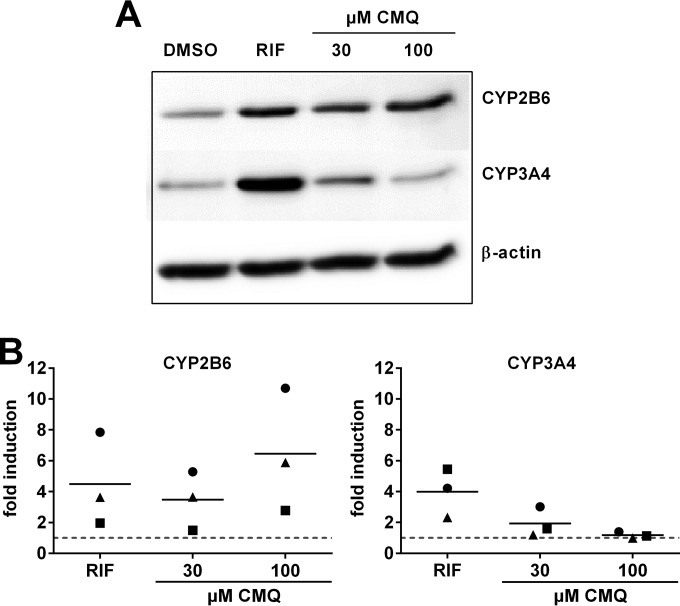
Induction of cytochrome P450 genes by carboxymefloquine results in elevated protein levels in primary human hepatocytes.
The induction of gene expression is only functionally relevant if it results in equally elevated protein levels. Thus, we exemplarily analyzed cytochrome P450 protein expression in primary human hepatocytes that were treated with carboxymefloquine. Figure 7 shows that the CYP2B6 protein was induced by the compound in a dose-dependent manner. In contrast, only the treatment with 30 μM carboxymefloquine resulted in somewhat elevated CYP3A4 protein levels, whereas 100 μM did not exert any effect. Thus, regarding CYP2B6 and CYP3A4, induction on the protein level proved to reflect the mRNA data. In conclusion, these data indicate that induction by carboxymefloquine, at least of the expression of CYP2B6, may be of functional relevance.
FIG 7.
Carboxymefloquine induces the protein expression of CYP2B6. Primary human hepatocyte cultures from 3 donors were each treated with 30 μM rifampin (RIF) or the indicated doses of carboxymefloquine (CMQ) for 48 h. Cytochrome P450 and β-actin protein expression were analyzed in total protein homogenates by immunoblotting using specific antibodies. (A) Representative Western blot analysis of a single culture. (B) Fold induction of protein expression by chemical treatment was calculated compared to the expression levels in the respective cells treated with DMSO only, which was designated 1 and is indicated by the broken line. The symbols denote cultures from individual donors.
DISCUSSION
Screening a range of clinically relevant antimalarial drugs and drug metabolites revealed most of these compounds to be ineffective in terms of inducing the assembly of the PXR ligand binding domain, which was used in this study as a surrogate of ligand binding to PXR. The notable exception was carboxymefloquine, the main metabolite of mefloquine. It is worthy to note that since the pronounced cytotoxicities of most compounds limited the applicable doses and treatment period, it formally cannot be ruled out that PXR assembly may be induced weakly by some other antimalarial at higher doses and/or with longer treatment.
In this study, carboxymefloquine proved to be an agonist and specific activator of human PXR without apparent cross talk with other nuclear receptors. The EC50 of 24 μM for PXR activation in vitro is close to the upper limit of the observed blood drug levels of 5 to 18 μM, which are achieved by multiple prophylactic doses of the parent drug mefloquine (27). This may indicate that the activation of PXR by the metabolite is possibly of relevance in vivo, even if intrahepatic metabolite levels, which are critical for PXR activation, are currently unknown. The intracellular concentrations of carboxymefloquine depend on the concentration of the parent drug mefloquine and the rate of its metabolism. A mean plasma half-life of 3 weeks (27) and biliary excretion (28) suggest slow metabolism and enterohepatic circulation of the drug, thereby allowing repeated cycles of hepatic metabolism. Accordingly, carboxymefloquine blood levels rise constantly over a span of weeks, especially after repeated dosing, as in prophylaxis (27), and they exceed mefloquine levels after the first day (29).
The knockdown of PXR completely eliminated the induction of ADME gene expression by carboxymefloquine in primary human hepatocytes, as was exemplarily shown for the prototypical PXR target gene CYP3A4. Thus, phenobarbital-type indirect activation of CAR (8), the other xenosensing nuclear receptor in humans, was ruled out as the underlying mechanism, and PXR was unequivocally shown to be involved in carboxymefloquine-dependent induction.
Carboxymefloquine further proved to dose-dependently induce the expression of ADME genes, which in the case of CYP3A4 was observed only if the treatment lasted for 24 h. Treatments lasting for 48 h seemed to reduce the extent of CYP3A4 induction by 30 μM and to inhibit CYP3A4 induction by 100 μM carboxymefloquine. Thus, increasing doses and long-lasting treatments impaired the PXR-mediated induction of CYP3A4 by carboxymefloquine. Although a toxic effect or general inhibition of PXR activation by high doses of the compound itself is ruled out, because the effect was specific for CYP3A4, we can only speculate about the mechanism underlying this phenomenon. Theoretically, increasing the dose and time of incubation with carboxymefloquine may inhibit a specific signaling pathway or factor that is required for hepatic CYP3A4 expression. Alternatively, a signaling pathway or factor that specifically inhibits CYP3A4 expression may be activated. Either way, such a mechanism has to counteract PXR-dependent induction by the compound. The elucidation of this mechanism requires further studies.
Mefloquine is an effective therapeutic and prophylactic antimalarial. Despite an ongoing debate on its safety because of controversial reports on the frequency and severity of neuropsychiatric adverse effects, mefloquine is still widely used and even regarded as indispensable for specific patient groups, such as pregnant women, infants, small children, and long-term travelers (30). Furthermore, it is used in WHO-recommended artemisinin combination therapy with artesunate (31). Mefloquine is mainly metabolized by CYP3A4, as was shown by coincubation/coadministration with the isozyme-specific inhibitor ketoconazole and the CYP3A4 inducer rifampin, both in human hepatocytes in vitro and human volunteers in vivo (24, 32, 33). The pharmacologically inactive carboxymefloquine (34) is regarded as the major metabolite (35). As carboxymefloquine proved in this study to induce the expression of drug-metabolizing enzymes and transporters by activating PXR, mefloquine may cause pharmacokinetic drug-drug interactions by means of its metabolite, thereby potentially reducing plasma drug levels and thus the efficacy of the drugs being coadministered. This may be especially relevant in prophylactic use, for which mefloquine is taken for at least several weeks. The absence of autoinduction of elimination during multiple dosing of mefloquine (27) and the lack of an effect on the erythromycin breath test (36), which is a measure of hepatic CYP3A4 activity, indicate that mefloquine is not inducing CYP3A4 activity in vivo. At first sight, this is contradictory to the induction of CYP3A4 mRNA in primary human hepatocytes by short-term 24-h treatment with carboxymefloquine. However, it may be related to the inhibitory effect of high doses and longer treatment, which we also observed. Furthermore, mefloquine was reported to inhibit the metabolism of primaquine (37), as did the established CYP3A inhibitor ketoconazole, thereby suggesting the inhibition of CYP3A4 enzymatic activity. However, the lack of an effect of mefloquine on the erythromycin breath test (36) also argues against that assumption. The published in vivo data and our in vitro results suggest that drugs susceptible to a pharmacokinetic interaction with mefloquine will preferentially be metabolized by CYP2B6 and CYP2C8 or transported by MDR1/Pgp, all of which were demonstrated here to be consistently induced in vitro by carboxymefloquine, rather than metabolized by CYP3A4.
The data from two of the drug interaction studies with mefloquine support the hypothesis that the drug, by means of its metabolite carboxymefloquine, may result in a clinically relevant induction of drug metabolism and transport. First, in patients receiving repeated doses of artemisinin together with a single dose of mefloquine on day 1, the area under the plasma concentration-time curve (AUC) of artemisinin on day 2 was significantly reduced compared to that at day 1, which was not observed in the patients treated with artemisinin alone (38). As artemisinin is metabolized mainly by CYP2B6 (39), the reduction of its AUC may be caused by carboxymefloquine-dependent induction of the enzyme. Second, the ritonavir steady-state AUC was significantly decreased by mefloquine (36). The induction of the CYP3A4-dependent metabolism of ritonavir did not occur, as the half-life of the drug was not changed. Because ritonavir is also transported by MDR1/Pgp, the authors speculated that mefloquine or its metabolite carboxymefloquine may induce MDR1/Pgp in the gut wall and, by that, reduce bioavailability (36). Having shown that treatment with carboxymefloquine induces the expression of the ABCB1 gene, which encodes MDR1/Pgp, in intestinal and hepatic cells, here we provide evidence for the above notion. Additional evidence for mefloquine-dependent induction of drug metabolism in vivo comes from a case report showing that mefloquine increased the number of epileptic seizures in a patient successfully treated with valproic acid and carbamazepine (40). The physician reported a reduced half-life of valproic acid, whereas the half-life of carbamazepine did not change, thereby indicating that mefloquine may have specifically accelerated valproic acid metabolism. While carbamazepine is metabolized mainly by CYP3A4 (41), whose activity is not induced in vivo by mefloquine (27, 36), valproic acid has been shown to be metabolized by CYP2A6, CYP2B6, and CYP2C9 (42). Thus, the induction of hepatic CYP2B6 by carboxymefloquine may explain the observation.
Our study further shows that it is important to include major drug metabolites in addition to the parental drugs when analyzing xenobiotic induction by activating nuclear receptors, as they may also be receptor ligands. To the best of our knowledge, carboxymefloquine is the first example of a drug metabolite that activates PXR while its parental drug is inactive in this respect.
In conclusion, we have shown here in cellular systems that carboxymefloquine induces ADME gene expression by PXR activation in vitro, which indicates that the parental drug mefloquine may cause pharmacokinetic drug-drug interactions due to the induction of drug metabolism and transport by means of its metabolite. Even if pharmacokinetic data from a few clinical studies seem to support this hypothesis, final proof of carboxymefloquine-mediated induction of ADME genes by mefloquine in vivo still requires a controlled clinical study. The increasing necessity of the coadministration of antimalarial drugs with antiretroviral and/or antituberculosis medications in treating malaria patients coinfected with HIV and/or tuberculosis strongly indicates the need to evaluate the pharmacokinetic interaction potentials of antimalarials, including their major metabolites; this is the case even if this and our previous study (13) indicate that the induction of drug metabolism and transport by activating the human xenosensors PXR and CAR is not a concern for most nonartemisinin antimalarials.
ACKNOWLEDGMENTS
We thank K. Abuazi Rincones for expert technical assistance. We acknowledge B. Burkhardt for the preparation of primary human hepatocytes.
B. A. Kandel, B. Vogelstein, and T. Tanaka kindly provided plasmids. The nonprofit foundation Human Tissue and Cell Research (Regensburg, Germany) kindly provided hepatocytes for the siRNA experiments.
This work was partially supported by the German Federal Ministry of Education and Research (BMBF) HepatoSys network grants 0313081 (to A.K.N.) and 0313080I (to O.B.), by the Deutsche Forschungsgemeinschaft (Germany) grant KE 1629/1-1 (to M.S.), and by the Robert Bosch Foundation, Stuttgart, Germany.
This work contains parts of the doctoral thesis of R.P. and bachelor thesis of S.T.
REFERENCES
- 1.World Health Organization. 2013. World malaria report 2013. World Health Organization, Geneva, Switzerland: http://www.who.int/malaria/publications/world_malaria_report_2013/wmr2013_no_profiles.pdf?ua=1. [Google Scholar]
- 2.Na-Bangchang K, Karbwang J. 2013. Emerging artemisinin resistance in the border areas of Thailand. Expert Rev Clin Pharmacol 6:307–322. doi: 10.1586/ecp.13.17. [DOI] [PubMed] [Google Scholar]
- 3.Valadas E, Gomes A, Sutre A, Brilha S, Wete A, Hänscheid T, Antunes F. 2013. Tuberculosis with malaria or HIV co-infection in a large hospital in Luanda, Angola. J Infect Dev Ctries 7:269–272. doi: 10.3855/jidc.2703. [DOI] [PubMed] [Google Scholar]
- 4.Giao PT, de Vries PJ. 2001. Pharmacokinetic interactions of antimalarial agents. Clin Pharmacokinet 40:343–373. doi: 10.2165/00003088-200140050-00003. [DOI] [PubMed] [Google Scholar]
- 5.Simonsson US, Jansson B, Hai TN, Huong DX, Tybring G, Ashton M. 2003. Artemisinin autoinduction is caused by involvement of cytochrome P450 2B6 but not 2C9. Clin Pharmacol Ther 74:32–43. doi: 10.1016/S0009-9236(03)00092-4. [DOI] [PubMed] [Google Scholar]
- 6.Asimus S, Elsherbiny D, Hai TN, Jansson B, Huong NV, Petzold MG, Simonsson US, Ashton M. 2007. Artemisinin antimalarials moderately affect cytochrome P450 enzyme activity in healthy subjects. Fundam Clin Pharmacol 21:307–316. doi: 10.1111/j.1472-8206.2007.00471.x. [DOI] [PubMed] [Google Scholar]
- 7.Xing J, Kirby BJ, Whittington D, Wan Y, Goodlett DR. 2012. Evaluation of P450 inhibition and induction by artemisinin antimalarials in human liver microsomes and primary human hepatocytes. Drug Metab Dispos 40:1757–1764. doi: 10.1124/dmd.112.045765. [DOI] [PubMed] [Google Scholar]
- 8.di Masi A, De Marinis E, Ascenzi P, Marino M. 2009. Nuclear receptors CAR and PXR: molecular, functional, and biomedical aspects. Mol Aspects Med 30:297–343. doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. 2002. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 10.Wang YM, Ong SS, Chai SC, Chen T. 2012. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol 8:803–817. doi: 10.1517/17425255.2012.685237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burk O, Arnold KA, Nussler AK, Schaeffeler E, Efimova E, Avery BA, Avery MA, Fromm MF, Eichelbaum M. 2005. Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol 67:1954–1965. doi: 10.1124/mol.104.009019. [DOI] [PubMed] [Google Scholar]
- 12.Simonsson US, Lindell M, Raffalli-Mathieu F, Lannerbro A, Honkakoski P, Lang MA. 2006. In vivo and mechanistic evidence of nuclear receptor CAR induction by artemisinin. Eur J Clin Invest 36:647–653. doi: 10.1111/j.1365-2362.2006.01700.x. [DOI] [PubMed] [Google Scholar]
- 13.Burk O, Piedade R, Ghebreghiorghis L, Fait JT, Nussler AK, Gil JP, Windshügel B, Schwab M. 2012. Differential effects of clinically used derivatives and metabolites of artemisinin in the activation of constitutive androstane receptor isoforms. Br J Pharmacol 167:666–681. doi: 10.1111/j.1476-5381.2012.02033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold KA, Eichelbaum M, Burk O. 2004. Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept 2:1. doi: 10.1186/1478-1336-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tachibana K, Kobayashi Y, Tanaka T, Tagami M, Sugiyama A, Katayama T, Ueda C, Yamasaki D, Ishimoto K, Sumitomo M, Uchiyama Y, Kohro T, Sakai J, Hamakubo T, Kodama T, Doi T. 2005. Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing different PPAR isoforms. Nucl Recept 3:3. doi: 10.1186/1478-1336-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hustert E, Zibat A, Presecan-Siedel E, Eiselt R, Mueller R, Fuss C, Brehm I, Brinkmann U, Eichelbaum M, Wojnowski L, Burk O. 2001. Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab Dispos 29:1454–1459. [PubMed] [Google Scholar]
- 17.Geick A, Eichelbaum M, Burk O. 2001. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem 276:14581–14587. doi: 10.1074/jbc.M010173200. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. 2003. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- 19.He TC, Chan TA, Vogelstein B, Kinzler KW. 1999. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell 99:335–345. doi: 10.1016/S0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffart E, Ghebreghiorghis L, Nussler AK, Thasler WE, Weiss TS, Schwab M, Burk O. 2012. Effects of atorvastatin metabolites on induction of drug-metabolizing enzymes and membrane transporters through human pregnane X receptor. Br J Pharmacol 165:1595–1608. doi: 10.1111/j.1476-5381.2011.01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burk O, Tegude H, Koch I, Hustert E, Wolbold R, Glaeser H, Klein K, Fromm MF, Nuessler AK, Neuhaus P, Zanger UM, Eichelbaum M, Wojnowski L. 2002. Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine. J Biol Chem 277:24280–24288. doi: 10.1074/jbc.M202345200. [DOI] [PubMed] [Google Scholar]
- 22.Knobeloch D, Ehnert S, Schyschka L, Büchler P, Schoenberg M, Kleeff J, Thasler WE, Nussler NC, Godoy P, Hengstler J, Nussler AK. 2012. Human hepatocytes: isolation, culture, and quality procedures. Methods Mol Biol 806:99–120. doi: 10.1007/978-1-61779-367-7_8. [DOI] [PubMed] [Google Scholar]
- 23.Riedmaier S, Klein K, Hofmann U, Keskitalo JE, Neuvonen PJ, Schwab M, Niemi M, Zanger UM. 2010. UDP-glucuronosyltransferase (UGT) polymorphisms affect atorvastatin lactonization in vitro and in vivo. Clin Pharmacol Ther 87:65–73. doi: 10.1038/clpt.2009.181. [DOI] [PubMed] [Google Scholar]
- 24.Fontaine F, de Sousa G, Burcham PC, Duchêne P, Rahmani R. 2000. Role of cytochrome P450 3A in the metabolism of mefloquine in human and animal hepatocytes. Life Sci 66:2193–2212. doi: 10.1016/S0024-3205(00)00546-4. [DOI] [PubMed] [Google Scholar]
- 25.Pissios P, Tzameli I, Kushner P, Moore DD. 2000. Dynamic stabilization of nuclear receptor ligand binding domains by hormone or corepressor binding. Mol Cell 6:245–253. doi: 10.1016/S1097-2765(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 26.Pascussi JM, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. 2000. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: consequences on cytochrome P450 gene regulation. Mol Pharmacol 58:1441–1450. doi: 10.1124/mol.58.6.1441. [DOI] [PubMed] [Google Scholar]
- 27.Mimica I, Fry W, Eckert G, Schwartz DE. 1983. Multiple-dose kinetic study of mefloquine in healthy male volunteers. Chemotherapy 29:184–187. doi: 10.1159/000238195. [DOI] [PubMed] [Google Scholar]
- 28.Mu JY, Israili ZH, Dayton PG. 1975. Studies of the disposition and metabolism of mefloquine HCl (WR 142,490), a quinolinemethanol antimalarial, in the rat. Limited studies with an analog, WR 30,090. Drug Metab Dispos 3:198–210. [PubMed] [Google Scholar]
- 29.Schwartz DE, Eckert G, Hartmann D, Weber B, Richard-Lenoble D, Ekue JM, Gentilini M. 1982. Single dose kinetics of mefloquine in man. Plasma levels of the unchanged drug and of one of its metabolites. Chemotherapy 28:70–84. [DOI] [PubMed] [Google Scholar]
- 30.Schlagenhauf P, Adamcova M, Regep L, Schaerer MT, Rhein HG. 2010. The position of mefloquine as a 21st century malaria chemoprophylaxis. Malar J 9:357. doi: 10.1186/1475-2875-9-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. 2010. Guidelines for the treatment of malaria, 2nd ed. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2010/9789241547925_eng.pdf?ua=1. [Google Scholar]
- 32.Ridtitid W, Wongnawa M, Mahatthanatrakul W, Chaipol P, Sunbhanich M. 2000. Effect of rifampin on plasma concentrations of mefloquine in healthy volunteers. J Pharm Pharmacol 52:1265–1269. doi: 10.1211/0022357001777243. [DOI] [PubMed] [Google Scholar]
- 33.Ridtitid W, Wongnawa M, Mahatthanatrakul W, Raungsri N, Sunbhanich M. 2005. Ketoconazole increases plasma concentrations of antimalarial mefloquine in healthy human volunteers. J Clin Pharm Ther 30:285–290. doi: 10.1111/j.1365-2710.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 34.Basco LK, Gillotin C, Gimenez F, Farinotti R, Le Bras J. 1991. Absence of antimalarial activity or interaction with mefloquine enantiomers in vitro of the main human metabolite of mefloquine. Trans R Soc Trop Med Hyg 85:208–209. doi: 10.1016/0035-9203(91)90022-Q. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz DE, Weber W, Richard-Lenoble D, Gentilini M. 1980. Kinetic studies of mefloquine and of one of its metabolites, Ro 21-5104, in the dog and in man. Acta Trop 37:238–242. [PubMed] [Google Scholar]
- 36.Khaliq Y, Gallicano K, Tisdale C, Carignan G, Cooper C, McCarthy A. 2001. Pharmacokinetic interaction between mefloquine and ritonavir in healthy volunteers. Br J Clin Pharmacol 51:591–600. doi: 10.1046/j.1365-2125.2001.01393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bangchang KN, Karbwang J, Back DJ. 1992. Primaquine metabolism by human liver microsomes: effect of other antimalarial drugs. Biochem Pharmacol 44:587–590. doi: 10.1016/0006-2952(92)90453-P. [DOI] [PubMed] [Google Scholar]
- 38.Svensson US, Alin H, Karlsson MO, Bergqvist Y, Ashton M. 2002. Population pharmacokinetic and pharmacodynamic modelling of artemisinin and mefloquine enantiomers in patients with falciparum malaria. Eur J Clin Pharmacol 58:339–351. doi: 10.1007/s00228-002-0485-y. [DOI] [PubMed] [Google Scholar]
- 39.Svensson US, Ashton M. 1999. Identification of the human cytochrome P450 enzymes involved in the in vitro metabolism of artemisinin. Br J Clin Pharmacol 48:528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jallon P. 1988. Use of mefloquine in epileptic patients. J Neurol Neurosurg Psychiatry 51:732. doi: 10.1136/jnnp.51.5.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerr BM, Thummel KE, Wurden CJ, Klein SM, Kroetz DL, Gonzalez FJ, Levy RH. 1994. Human liver carbamazepine metabolism. Role of CYP3A4 and CYP2C8 in 10,11-epoxide formation. Biochem Pharmacol 47:1969–1979. [DOI] [PubMed] [Google Scholar]
- 42.Kiang TK, Ho PC, Anari MR, Tong V, Abbott FS, Chang TK. 2006. Contribution of CYP2C9, CYP2A6, and CYP2B6 to valproic acid metabolism in hepatic microsomes from individuals with the CYP2C9*1/*1 genotype. Toxicol Sci 94:261–271. doi: 10.1093/toxsci/kfl096. [DOI] [PubMed] [Google Scholar]