Abstract
Colicin-mediated killing is an example of allelopathy, which has been found among several bacteria. Screening of 42 strains of Shigella sonnei isolated from diarrheal patients revealed that 39 (93%) S. sonnei strains were positive for colicin production against Escherichia coli DH5α. In the PCR-based detection of the colicin types, 36 (92.3%) were identified as E3, 2 (5.1%) as E3 and E8, and 1 (2.6%) as E3 and E2. Representative S. sonnei strains producing heterologous colicins exhibited antagonism against diarrheagenic Escherichia coli (DEC) groups. Although it is known that mutation in the colicin receptor renders the host resistant to colicin, there is a dearth of information on the genetic characterization of such mutants. In the fluctuation test, colicin-resistant E. coli mutants were found to occur spontaneously at the rates of 2.51 × 10−8 and 5.52 × 10−8 per generation when exposed to colicins E3 and E8 and colicins E3 and E2, respectively. Genotypic characterization of colicin-resistant E. coli (ECCr) and S. sonnei (SSCr) strains displayed mutations in the btuB gene, which encodes the receptor for vitamin B12 uptake. This gene was interrupted by various insertion sequences, such as IS1, IS2, and IS911. Complementation of ECCr and SSCr with plasmid-borne btuB (pbtuB) accomplished restoration of the colicin-susceptible phenotype. The vitamin B12 uptake assay gave an insight into the physiological relevance of the btuB mutation. Our studies provide insights into the latent influence of S. sonnei colicins in governing the existence of some of the shigellae and all of the DEC and the genetic mechanism underlying the emergence of resistance.
INTRODUCTION
Allelopathy refers to the production of toxic metabolites that suppress both the growth and survival of distinct competitors in a common niche (1, 2). Colicin is one such antimicrobial biomolecule produced by certain members of the family Enterobacteriaceae, which provides a competitive edge against microorganisms that are not immune or resistant (3–6). Colicin producers express the immunity protein constitutively, which forms a complex with the colicin protein, thereby preventing “cell suicide.” Resistance to killing is attributable to two factors: (i) a mutation in the receptor, which serves as a portal for entry into the target cell (true resistance), and (ii) an alteration in some component, which constitutes a part of the translocation machinery (tolerance) (7, 8).
In the present study, we have focused primarily on the role of colicin produced by the Shigella sonnei strains isolated from acute diarrheal patients in preventing the growth of diarrheagenic Escherichia coli (DEC) groups and spontaneous emergence of resistance to colicin due to mutation in the btuB gene, which encodes the receptor for vitamin B12 uptake. The BtuB protein also serves to localize type A/E colicins and T5-like phages (BF23 and EPS7) on the target cell surface (9, 10). The btuB gene consists of a single open reading frame, which is translated into a 614-amino-acid polypeptide. The first 20 amino acids constitute a signal peptide, which gets cleaved during secretion across the cytoplasmic membrane. The remaining 594 amino acid residues yield the mature/processed protein, which has a molecular mass of 66 kDa (11, 12). All of the BtuB molecules are capable of transporting vitamin B12 and also facilitate the killing effect of phage BF23, but only a proportion of the receptors, which are newly synthesized, can mediate colicin action (12).
There is a wealth of information on colicin production by E. coli. However, there are very few reports on the frequency and types of colicins produced by S. sonnei as well as their role in governing the population dynamics of other gut pathogens, especially in the case of polymicrobial infections. S. sonnei is the causative agent of acute bacillary dysentery, which has a low infectious dose ranging from 10 to 100 live cells. Of the four serogroups (S. sonnei, S. boydii, S. flexneri, and S. dysenteriae), S. sonnei is an exception because it is not further subdivided into subgroups, and hence, in the past colicin production was used as an additional epidemiological marker for typing S. sonnei strains (13, 14). Among the colicins produced by Shigella strains, colicin U and colicin Js are expressed by S. boydii and S. sonnei, respectively (15–17). A few studies have demonstrated that S. sonnei colicins exhibit antagonism against some of the shigellae and DEC, which are sometimes uncontrollable by any antimicrobial therapy (18). Hence, strategies for designing newer antibiotics with different modes of action are now being considered. Contrary to the indiscriminate killing approach of broad-spectrum antibiotics, colicins are highly targeted in their action, as they only cripple the target pathogen without causing collateral damage to the commensal bacteria (19). As colicins target a narrow phylogenetic range of microorganisms, the selection of mutations that confer resistance to the toxin will take place only in a small fraction of the microbial community instead of multiple species simultaneously. This study provides a unique insight into the action of S. sonnei colicins and the genetic basis of their resistance.
MATERIALS AND METHODS
Bacterial strains.
Forty-two strains of S. sonnei isolated from stool specimens of acute diarrheal patients admitted to the Infectious Diseases Hospital (IDH), Kolkata, India, were screened for colicin production using E. coli DH5α (colicin-sensitive E. coli [ECCs]) as an indicator strain. In the active surveillance, every 5th patient admitted on any two randomly selected days in a week was enrolled in the study. Three representative S. sonnei strains (IDH01791, IDH01157, and 500867) with heterologous colicin types (selected on the basis of PCR screening [20] and cross-immunity testing) were used for detecting their antagonistic activity against commensal E. coli strains that did not harbor any virulence genes, enterotoxigenic (ETEC), typical enteropathogenic (tEPEC), atypical enteropathogenic (aEPEC), enterohemorrhagic E. coli (EHEC), and enteroaggregative (EAEC) E. coli, along with S. dysenteriae type 1, S. flexneri 2a, and S. sonnei. Cloning and transformation were performed using the Mach1-T1r strain [F− Φ80lacZΔM15 ΔlacX74 hsdR(rκ− mκ+) ΔrecA1398 endA1 tonA] (Invitrogen, Carlsbad, CA).
Colicin assay.
For studying colicin production, a single colony of one of the previously confirmed strains of S. sonnei from xylose lysine desoxycholate agar (Difco, Sparks, MD, USA) was inoculated into 5 ml of Luria-Bertani (LB) broth (Difco) and incubated overnight at 37°C in a shaker. On the following day, the culture was centrifuged at 10,000 × g for 10 min. The culture supernatant from each strain was passed through a 0.22-μm filter (Millipore, Bangalore, India). About 10 μl of the crude filtrate was spotted on the Mueller-Hinton agar (MHA) (Difco) plate, previously inoculated with the log phase culture of E. coli DH5α or the other test strains. Plates were incubated at 37°C overnight and checked for the presence or absence of a zone of inhibition on the following day.
Isolation of colicin-resistant mutants.
In the colicin assay, a few colicin-resistant colonies were found to grow as suppressed colonies within the zone of inhibition on the MHA plate seeded either with E. coli DH5α or S. sonnei. Some of these colicin-resistant colonies of E. coli (ECCr) DH5α and S. sonnei (SSCr) were selected for further investigation. The colicin-resistant colonies were confirmed as E. coli and S. sonnei using biochemical tests and serological testing, respectively. In addition, pulsed-field gel electrophoresis (PFGE) was performed (21) to confirm their DNA fingerprints by comparing the colicin-susceptible progenitor and colicin-resistant strains. To detect the differences, if any, in the sugar fermentation abilities of the resistant and sensitive strains, an API 32E (bioMérieux, Marcy l'Étoile, France) test was performed according to the manufacturer's protocol.
Amplification and sequencing of the btuB gene.
The btuB gene of ECCr, SSCr, and their colicin-susceptible progenitors was amplified using the BtuB-P primer pair (Table 1). Amplicons were sized against molecular weight markers by agarose gel electrophoresis and purified using a PCR product purification kit (Qiagen, Hilden, Germany). The amplicons were sequenced with forward and reverse primers using an automated DNA sequencer (ABI 3730; Applied Biosystems, Foster City, CA). To sequence the larger amplicons of the resistant mutants, several internal primers were used (Table 1). The sequences were assembled and analyzed using DNASTAR software (DNASTAR, Inc., Madison, WI).
TABLE 1.
Primers used for sequencing the entire btuB gene
| Target | Primer(s) (5′ to 3′) | PCR conditions | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|
| BtuB-P | CGGGGTACCGATACCAGCCCGGAT and CCCAAGCTTTCAGAAGGTGTAGCT | 94°C for 1 min, 57°C for 1 min, 72°C for 1 min 30 s, 30 cycles | 1,746 | 44 |
| BtuB-D | GGATCCATGATTAAAAAAGCTTCGCTG and GGATCCTCAGAAGGTGTAGCTGCCAG | 94°C for 1 min, 57°C for 1 min, 72°C for 1 min 30 s, 30 cycles | 1,859 | This study |
| BtuB-1 | CTGCTGACGGCGTGTTCCGT and CGCCCAACAGCGTTACCCGT | 94°C for 30 s, 63°C for 45 s, 72°C for 45 s, 30 cycles | 553 | This study |
| BtuB-2 | ACGGGTAACGCTGTTGGGCG and CACTGCGTCTGGCGCCTTCA | 94°C for 30 s, 63°C for 45 s, 72°C for 45 s, 30 cycles | 587 | This study |
| BtuB-3 | TGAAGGCGCCAGACGCAGTG and CGACGCCCTTAATCCGCGCT | 94°C for 30 s, 63°C for 45 s, 72°C for 45 s, 30 cycles | 335 | This study |
| BtuB-4 | TCACGACGCGCGATGAACCC and GTTATAGCGCAGCCCGGCGT | 94°C for 30 s, 63°C for 45 s, 72°C for 45 s, 30 cycles | 383 | This study |
| BtuB3RF2 | AGTGGGAAGGCGCGTTTGAAG | Used directly for sequencing | This study | |
| BtuB3RR2 | GCAGAGGCGTTCGAGCATTAT | Used directly for sequencing | This study | |
| IS2 | GCCGCCGCAGTCGTCACAC and CCATTATCCGTCAGCCACTCCACT | 94°C for 45 s, 57°C for 45 s, 72°C for 45 s, 30 cycles | 478 | This study |
| IS911 | CCTGCTGTCCGGCGAGAACG and CAAAAGCCTCTCCGATAACACCAG | 94°C for 30 s, 70°C for 30 s, 72°C for 30 s, 25 cycles | 627 | This study |
| BtuB-5FIS1-R2 | ACGGTAGTATTGGTGCGGGTGTCG and GTGGGGCTATGTCGGGGCTAAAT | 94°C for 30 s, 70°C for 30 s, 72°C for 30 s, 25 cycles | 481 | This study |
| BtuB-6 | GATGCAATAGGCGGGGTGGTGA and CCGCTCCAGGCATCAGTAAAGTT | 94°C for 30 s, 70°C for 30 s, 72°C for 30 s, 25 cycles | 286 | This study |
Genetic complementation of ECCr and SSCr with intact btuB.
The complete btuB gene was amplified using the primer pair BtuB-D and the template DNA from an ECCs colony. The PCR product was used for Topo TA cloning (Invitrogen). One Shot Mach1-T1r competent cells (Invitrogen) were chemically transformed using the recommended protocol. The transformants were selected on LB agar plates containing 30 μg/ml of kanamycin. A PCR assay was performed using the M13 primers to confirm the btuB gene in the transformants. A single colony of the btuB-harboring clone was subjected to plasmid DNA isolation. Chemically competent ECCr and SSCr were transformed with the plasmid DNA (pbtuB). The selection was done on LB agar with kanamycin (30 μg/ml) plates. A colicin assay was performed to check for restoration of a colicin-susceptible phenotype (22).
Uptake of vitamin B12 in the colicin-susceptible, -resistant, and -complemented strains of E. coli DH5α and S. sonnei.
The uptake of vitamin B12 was quantified using the vitamin B12 assay medium (Difco). A stock concentration of vitamin B12 (400 μg/ml) (SRL, Mumbai, India) was prepared using 25% ethanol (Merck, Mumbai, India). A Lactobacillus leichmannii (ATCC 7830) strain, which is a vitamin B12-dependent auxotroph, was used as a test organism (23). After an initial standard curve using the test organism was obtained, different concentrations of vitamin B12 (80, 120, and 160 pg) were designated for the assay. Colicin-susceptible, -resistant, and -complemented strains of E. coli DH5α and S. sonnei were inoculated into normal LB broth (LBN) containing 800, 1,200, and 1,600 pg of vitamin B12, and after overnight incubation at 37°C in a shaker, the cells were centrifuged at 10,000 × g for 10 min. The culture supernatant was sterilized by passage through a 0.22-μm filter (Millipore). To 5 ml of double-strength vitamin B12 assay medium, 1 ml of the crude filtrate was added, and the volume was made up to 10 ml with sterile distilled water. In the case of controls, instead of the crude filtrate, 200, 300, and 400 μl of vitamin B12 (400 pg/ml) was added. Uninoculated and inoculated controls (without vitamin B12) were also included in the assay. After autoclaving of the above medium at 121°C for 5 min, all of the test tubes (except the uninoculated control) were inoculated with L. leichmannii and incubated overnight at 37°C in a static condition. The optical density at 530 nm (OD530) was recorded on the following day, and a graph of the OD530 versus the amount of vitamin B12 (pg) was plotted in order to compare the vitamin B12 uptake of colicin-sensitive, -resistant, and -complemented strains of E. coli and S. sonnei (24–26).
Luria-Delbrück fluctuation assay.
The Luria-Delbrück fluctuation assay (27) was adopted with a slight modification to check the average mutation rate. In brief, a series of 10 tubes containing 200 μl of LBN and a single 100-ml conical flask containing 10 ml of LBN were each inoculated with ECCs to a concentration of 500 cells/ml. Cells were incubated in a 37°C shaker until they attained an OD600 of about 1.0. One hundred-microliter aliquots were taken from each of the small cultures, and ten 100-μl aliquots were taken from the 10-ml bulk culture. These samples were spread on normal LB agar plates containing crude colicin extract from the strain IDH01157 or 500867, respectively. The plates were incubated overnight, and on the following day, the number of colonies on each plate was counted. Representative colonies were picked from each plate and subcultured onto colicin-containing plates. Further, their resistance toward colicin produced by IDH01791, IDH01157, and 500867 was confirmed. A PCR was carried out using the BtuB-P primers, and the percentage of mutants arising due to a disruption of the btuB gene by the insertion sequence (IS) element was determined. The mutation rates were estimated using the Ma-Sandri-Sarkar maximum likelihood estimator (MSS-MLE) implemented by the FALCOR webtool (http://www.keshavsingh.org/protocols/FALCOR.html) (27).
Nucleotide sequence accession numbers.
The btuB gene sequences disrupted by the IS elements in the colicin-resistant mutants were submitted to GenBank. The accession numbers for ECCrA1, ECCrA11, SSCrA2, and SSCrA11 reported in this paper are KJ494659, KC806221, KJ494660, and KC806222, respectively.
RESULTS
Detection of colicin-producing S. sonnei strains and their antagonistic activity against different enteric bacteria.
Of the 42 strains of S. sonnei screened, 39 (93%) were identified as colicin producers based on the phenotypic screening by the colicin assay using E. coli DH5α. PCR-based detection of the colicin type was carried out using the subset of 39 producers and previously published primers (20). Thirty-six strains (92.3%) were identified as colicin type E3, 2 (5.1%) as types E3 and E8, and 1 (2.6%) as types E3 and E2. Three representative S. sonnei strains, IDH01791 (colicin E3 producer), IDH01157 (producing colicins E3 and E8), and 500867 (producing colicins E3 and E2), with a heterologous colicin profile were selected for further studies. Cross-immunity tests between these colicin producers were performed to substantiate the results obtained by the PCR (Fig. 1). In addition, colicin-mediated antagonistic activity against several gut pathogens was tested. Overall, the colicin susceptibilities of the diarrheagenic E. coli (DEC) were as follows: tEPEC, 35% (n = 20); aEPEC, 55% (n = 20); EAEC, 35% (n = 20); ETEC, 55% (n = 20); and EHEC, 50% (n = 4). Among the Shigella spp., 33% of S. dysenteriae type 1 (n = 12) and 5% of S. flexneri 2a (n = 22) exhibited colicin susceptibility. The majority of the E3 type of colicin-producing S. sonnei strains (97%, n = 32) were susceptible to the IDH01157 and 500867 strains, which produce dual colicin types of E3 and E8 and E2 and E3, respectively. We found that a large proportion of the commensal E. coli (85%, n = 20) that did not harbor any of the DEC virulence genes were resistant to colicin.
FIG 1.
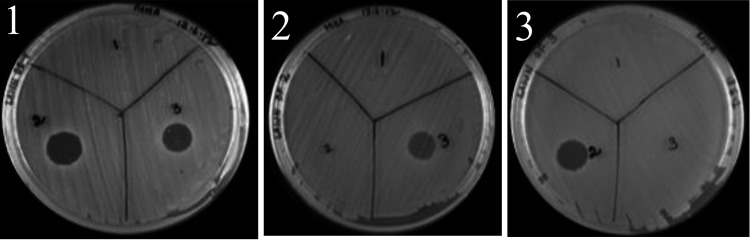
Antagonism between colicin-producing S. sonnei using the crude filter-sterilized supernatant from IDH01791 (1), IDH01157 (2), and 500867 (3) on a lawn culture of each of the aforementioned strains.
Isolation and characterization of colicin-resistant mutants based on phenotypic markers.
S. sonnei colicins exerted their lethal effect against different pathogroups of DEC. In addition, most of the S. sonnei strains, which were either heterologous or non-colicin producers, were found to be susceptible. However, in a few strains, the resistant mutants were found to grow as suppressed colonies within the zone of inhibition. Several such mutants were analyzed to gain an insight into the genetic mechanism for colicin resistance. These included colicin-resistant E. coli DH5α isolated from the zone of inhibition of the colicins produced by S. sonnei strains IDH01791 (ECCrA1 to ECCrA11), IDH01157 (ECCrB1 to ECCrB11), and 500867 (ECCrC1 to ECCrC15). Similarly, colicin-resistant S. sonnei IDH01791 isolated from the zone of inhibition of the colicin-producing conspecific S. sonnei strains IDH01157 (SSCrA1 to SSCrA11) and 500867 (SSCrB1 to SSCrB16) was also analyzed. The second category of mutants merits special attention as it marks the competition between two conspecific colicin producers. Representative mutant strains of ECCr and SSCr were confirmed as E. coli and S. sonnei, respectively, in the biochemical testing and slide agglutination assays using the specific polyclonal antisera. The API 32E test results revealed that there was no change in the biochemical fermentation profiles of the progenitor and resistant mutants (data not shown). In the PFGE, the progenitor and mutant strains were found to be identical (data not shown).
PCR-based amplification of the btuB gene.
The resistant colonies of each category of mutants were subjected to btuB PCR assays. Analysis of the PCR products by agarose gel electrophoresis showed fragments of about 3,000 bp from the majority of the ECCr colonies. In the case of SSCr, PCR products of sizes approximating 2,500 bp and 3,000 bp were obtained from most of the colonies. These unexpected large-sized amplicons indicated that the btuB gene was interrupted by the insertion of DNA fragments in the resistant mutants but not in the colicin-susceptible progenitors that gave a desired amplicon (1,746 bp).
Sequence analysis of btuB in ECCr and SSCr mutants.
DNA sequencing and BLAST analysis of representative mutants with different amplicon sizes (ECCrA1, ECCrA11, SSCrA2, and SSCrA11) showed insertion sequences (IS elements) interrupting the btuB gene. IS1 (∼770 bp) and IS2 (∼1,300 bp) were detected in ECCrA1 and ECCrA11, respectively, whereas IS911 (∼1,250 bp) and IS1 (∼770 bp) were found to disrupt the btuB gene of SSCrA2 and SSCrA11, respectively. These IS elements differed in their orientation and insertion sites within the btuB gene. The IS was incorporated in the same direction as that of btuB transcription in ECCrA11, whereas in ECCrA1, SSCrA2, and SSCrA11, the IS was oriented in the reverse direction. With the reference sequence of the E. coli btuB gene for the vitamin B12 receptor protein BtuB (GenBank accession no. M10112), the IS element was identified at positions 1392, 1689, 1176, and 1827 in ECCrA1, ECCrA11, SSCrA2, and SSCrA11, respectively. In the four mutants, the flanking region of the IS element exhibited typical direct repeats.
Complementation with intact btuB restores a colicin-susceptible phenotype.
In the colicin assay, the transformants SSCrA11btuB::IS1 and ECCrA11btuB::IS2 complemented with pbtuB exhibited susceptibility to colicin, thereby highlighting the role of intact btuB in this transfiguration. This confirms that IS elements are the exclusive determinants of colicin resistance in the aforementioned mutants.
Comparison of vitamin B12 uptake in colicin-susceptible, -resistant, and -complemented strains of E. coli and S. sonnei.
The vitamin B12 uptake assay was performed to determine the influence of the mutation in the btuB gene on the vitamin B12 uptake ability of the organism. In the cases of ECCs and SSCs, a drop in the OD530 value was recorded compared to those for ECCr and SSCr, due to normal uptake of vitamin B12 by the colicin-susceptible strains. Much less vitamin B12 remained in the spent medium that supported the growth of L. leichmannii and hence the reduction in the OD530 value. In the cases of ECCr and SSCr, there was decreased uptake of vitamin B12 by the mutant, thereby enhancing the growth of L. leichmannii, which resulted in higher OD530 values (Fig. 2). These results indicate that vitamin B12 uptake has been blocked in the resistant mutants due to defective btuB (interrupted by the IS elements). ECCr and SSCr complemented with pbtuB showed vitamin B12 uptake similar to those of the susceptible mutants, thereby highlighting the marked physiological effect of the mutation (Fig. 2).
FIG 2.
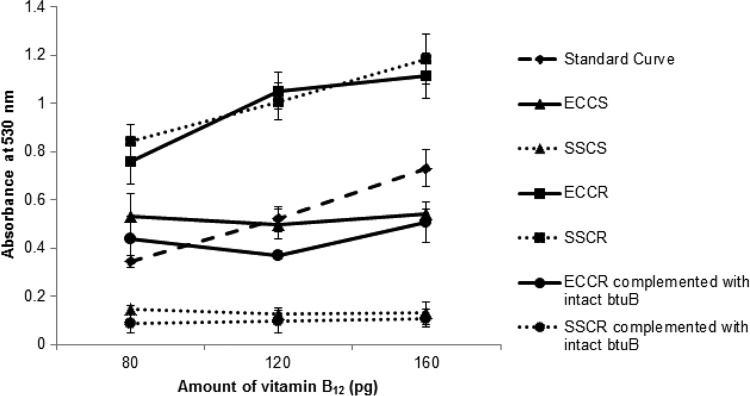
Comparison of vitamin B12 uptake in ECCs, ECCr, SSCs, and SSCr and the genetically complemented strains. The results are expressed as means and standard deviations from three independent experiments.
Estimation of the mutation rate using the fluctuation assay.
A large variation in the number of colicin-resistant colonies in a parallel series of small cultures compared with that in a series of samples taken from the bulk culture is in agreement with the fact that mutations have occurred spontaneously (see Tables S1 and S2 in the supplemental material). The rates at which ECCs became resistant to colicins E3 and E8 and colicins E3 and E2 were estimated to be 2.51 × 10−8 and 5.52 × 10−8 per generation, respectively. Further, we determined the percentage of those mutations that involved IS insertions. In the case of ECCr selected after exposure to E3 and E8 colicins produced by the strain IDH01157, 10.3% (n = 300) of the mutants exhibited a disruption of btuB by IS elements, whereas this figure was higher (18.7%, n = 300) in the case of the ECCr mutants selected on plates containing E2 and E3 colicins produced by the strain 500867.
DISCUSSION
In the past 5 years (2008 to 2012), the frequency of the occurrence of S. sonnei strains as a sole pathogen (58%, n = 83) among diarrheal patients admitted to the Infectious Diseases Hospital, Kolkata, has increased considerably. This provided us with an impetus to screen S. sonnei strains for colicin production (see Table S3 in the supplemental material). As anticipated, colicinogeny was confirmed in 92% of the strains. The majority of the S. sonnei strains isolated produced the E type of colicins (93%, 39 of 42 strains). To our knowledge, this is the first report on the occurrence of such a high frequency of E colicin-producing S. sonnei strains. The data obtained upon integration of phenotypic and genotypic methods revealed that most of them were E3 colicin producers. Among the few multicolicin producers, colicin E3 was found to co-occur either with colicin E2 or E8. Weak colicin producers seem to be very common (28). There is a possibility that the colicin types expressed by each strain are underestimated, as our screening is chiefly based on PCR and colicin assays. The multiple colicin production property confers a selective survival advantage not only against sensitive cells in a population but also against those harboring one of the colicins common to the multiproducer (3). The dynamics between colicin producers opens an interesting area of research.
There are very few reports on the role of S. sonnei colicins in governing the existence of shigellae and DEC. Considering this point, we tested the antagonistic effect of colicin-producing S. sonnei against a wide panel of DEC groups. The antagonistic spectrum suggests that most of the S. sonnei strains produced colicins that were biologically active against E. coli strains belonging to different DEC groups. Furthermore, S. sonnei strains exhibited conspecific as well as interspecific antagonism. This was evident from the hospital-based surveillance study. S. flexneri 2a and S. dysenteriae type 1 were specifically chosen for testing of their susceptibility to colicin because of an upsurge of multidrug resistance in these two strains (29), which demands a change in the antibiotic-based treatment regimen. Further, S. flexneri, particularly serotype 2a, is the serotype most endemic in Kolkata (30). However, during the past 2 decades, shigellosis caused by S. sonnei has become an emerging trend in India and other Asian countries (31–33). There are many reports on S. sonnei replacing S. flexneri as the predominant agent of bacterial dysentery in Vietnam, Thailand, Malaysia, China, and several other countries undergoing economic expansion (34). Epidemiologically, this is an important trend, as each S. sonnei strain generates its colicin(s), leading to competition that eventually reduces the frequency of the other species of bacteria belonging to the family Enterobacteriaceae (35).
The strategy employed by S. sonnei strains to direct the “microbe-kill-microbe” world in which they live provides an impetus to employ S. sonnei colicins for controlling the DEC, but paradoxically, just as colicin production is ubiquitous, so is resistance to colicin killing. However, as these toxins have a narrow spectrum of activity, the selection for resistance does not take place in several strains simultaneously. Further, colicins occur in constantly evolving combinations in nature, thereby enabling the colicin producers to keep pace with the emergent resistant ones. In addition to the emergence of resistance, the impact of the toxin on the commensal microbiota also deserves equal attention, before therapeutic applications commence. Several studies have shown that colicins leave the structure of the commensal microbiome largely undisturbed (18, 19). In our case, we found that the majority of commensal E. coli strains (85%, n = 20) were colicin resistant. Given the recent upsurge in publications highlighting the beneficial impact of commensal organisms, it is important to formulate therapeutic strategies for preserving commensal microbiota, and colicins can be exploited to that end. The limited toxicity of colicins to commensal organisms compared to that to enteric pathogens is very interesting and may be accounted for by the differences in the genetic makeups of the two groups of microorganisms. Several groups are actively pursuing research to address this long-standing question.
It has already been proven that a mutation in the btuB gene is responsible for instigating resistance to E colicins. However, there are no reports on the genetic mapping of the definitive locus within the btuB gene involved in the generation of resistance. We have focused primarily on the mutations that map btuB instead of tolQRAB, since previous studies have shown that the majority of the mutants resistant to E colicins (E2 to E8) possess mutations in the btuB gene (36). Our studies prove that the IS element-mediated disruption of the btuB gene is one of the mechanisms imparting colicin resistance. The classical Luria-Delbrück fluctuation test gave us an insight into the spontaneous nature of mutagenesis. Mutations occurring at an early stage in the small cultures gave rise to “jackpots” of resistant colonies on selective plates, thereby accounting for the high statistical variance (37). Had the mutations been induced by colicin, all mutants would appear after plating and yield a low variance in the number of resistant mutants.
Insertion sequences are mobile elements, which have a marked influence on the phenotype of an organism. These elements carry information required only for their mobilization, and they range in size from 0.7 to 2.5 kb (38–40). After exposing the ECCs and SSCs strains to a crude preparation of S. sonnei colicin, we found IS1 and IS2 disrupting the btuB gene in E. coli and IS1 and IS911 in S. sonnei. IS1 is one of the smallest “autonomous” bacterial insertion sequences isolated so far (770 bp) (41). Both IS2 (1,300 to 1,350 bp) and IS911 (1,250 bp) are members of the IS3 family, and their insertions have been reported in both the regulatory and coding regions (38). Insertional events have been reported to trigger resistance to bacteriophages (10) and to antibiotics (42, 43). However, to our knowledge, this is the first report on insertion sequences acting as one of the determinants in imparting colicin resistance by disrupting the btuB gene. We found that complementation of ECCr and SSCr with intact btuB resulted in the restoration of a colicin-susceptible phenotype and the complemented strains exhibited a degree of vitamin B12 uptake similar to those of the wild-type progenitors. Further studies are warranted to interpret the changes that the BtuB protein undergoes upon disruption of its encoding gene.
Taking into account the fact that colicins seem to play a vital role in regulating the DEC, we hypothesize that the domination by S. sonnei occurs by competitively excluding the colicin-susceptible pathogens. The propensity for resistance development against colicin may be limited if a cocktail of colicins targeting different receptors is used. In addition, understanding the mechanisms behind the emergence of resistance will enable researchers to combat this problem more efficiently.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Council of Scientific and Industrial Research (CSIR), New Delhi, India. F.C. is a recipient of an SPM award for carrying out this study under fellowship grant SPM-07/482(0087)2010-EMR-I.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04122-14.
REFERENCES
- 1.Durrett R, Levin S. 1997. Allelopathy in spatially distributed populations. J Theor Biol 185:165–171. doi: 10.1006/jtbi.1996.0292. [DOI] [PubMed] [Google Scholar]
- 2.Nakamaru M, Iwasa Y. 2000. Competition by allelopathy proceeds in traveling waves: colicin-immune strain aids colicin-sensitive strain. Theor Popul Biol 57:131–144. doi: 10.1006/tpbi.1999.1448. [DOI] [PubMed] [Google Scholar]
- 3.Gordon DM, O'Brien CL. 2006. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152:3239–3244. doi: 10.1099/mic.0.28690-0. [DOI] [PubMed] [Google Scholar]
- 4.Majeed H, Gillor O, Kerr B, Riley MA. 2011. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J 5:71–81. doi: 10.1038/ismej.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cursino L, Šmarda J, Chartone-Souza E, Nascimento AMA. 2002. Recent updated aspects of colicins of Enterobacteriaceae. Braz J Microbiol 33:185–195. doi: 10.1590/S1517-83822002000300001. [DOI] [Google Scholar]
- 6.Kleanthous C. 2010. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Rev Microbiol 8:843–848. doi: 10.1038/nrmicro2454. [DOI] [PubMed] [Google Scholar]
- 7.Feldgarden M, Riley MA. 1998. High levels of colicin resistance in Escherichia coli. Evolution 52:1270–1276. doi: 10.2307/2411296. [DOI] [PubMed] [Google Scholar]
- 8.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Masi DR, White JC, Schnaitman CA, Bradbeer C. 1973. Transport of vitamin B12 in Escherichia coli: common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J Bacteriol 115:506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl Environ Microbiol 77:2042–2050. doi: 10.1128/AEM.02504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller K, Kadner RJ. 1985. Nucleotide sequence of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol 161:904–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James R, Kleanthous C, Moore GR. 1996. The biology of E colicins: paradigms and paradoxes. Microbiology 142:1569–1580. doi: 10.1099/13500872-142-7-1569. [DOI] [PubMed] [Google Scholar]
- 13.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Wang Q, Reeves PR, Wang L. 2008. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev 32:627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 14.Vlajinac H, Krajinovic S. 1983. Colicin production as an epidemiological marker for Shigella sonnei. J Hyg (Lond) 91:273–276. doi: 10.1017/S0022172400060289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sousa MA, Farias Lde M, Oliveira PL, Moreira JS, Apolonio AC, Oliveira JS, Santoro MM, Mendes EN, Magalhaes PP. 2013. Antagonistic activity expressed by Shigella sonnei: identification of a putative new bacteriocin. Mem Inst Oswaldo Cruz 108:724–729. doi: 10.1590/0074-0276108062013008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smajs D, Pilsl H, Braun V. 1997. Colicin U, a novel colicin produced by Shigella boydii. J Bacteriol 179:4919–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smarda J, Petrzelova J, Vyskot B. 1987. Colicin Js of Shigella sonnei: classification of type colicin “7.” Zentralbl Bakteriol Mikrobiol Hyg A 263:530–540. [DOI] [PubMed] [Google Scholar]
- 18.Sousa MA, Mendes EN, Apolonio AC, Farias Lde M, Magalhaes PP. 2010. Bacteriocin production by Shigella sonnei isolated from faeces of children with acute diarrhoea. APMIS 118:125–135. doi: 10.1111/j.1600-0463.2009.02570.x. [DOI] [PubMed] [Google Scholar]
- 19.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 20.Smajs D, Micenková L, Smarda J, Vrba M, Sevčíková A, Vališová Z, Woznicová V. 2010. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol 10:288. doi: 10.1186/1471-2180-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention. 2014. Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/pulsenet/PDF/ecoli-shigella-salmonella-pfge-protocol-508c.pdf. [Google Scholar]
- 22.Sneeden JL, Loeb LA. 2003. Genetic complementation protocols. Methods Mol Biol 230:3–10. doi: 10.1385/1-59259-396-8:3. [DOI] [PubMed] [Google Scholar]
- 23.Karmi O, Zayed A, Baraghethi S, Muhammad Qadi M, Ghanem R. 2011. Measurement of vitamin B12 concentration: a review on available methods. IIOAB J 2:23–32. [Google Scholar]
- 24.Kadner RJ, Liggins GL. 1973. Transport of vitamin B12 in Escherichia coli: genetic studies. J Bacteriol 115:514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradbeer C, Woodrow ML. 1976. Transport of vitamin B12 in Escherichia coli: energy dependence. J Bacteriol 128:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadner RJ, Bassford PJ Jr. 1977. Relation of cell growth and colicin tolerance to vitamin B12 uptake in Escherichia coli. J Bacteriol 129:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall BM, Ma CX, Liang P, Singh KK. 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majeed H, Lampert A, Ghazaryan L, Gillor O. 2013. The weak shall inherit: bacteriocin-mediated interactions in bacterial populations. PLoS One 8:e63837. doi: 10.1371/journal.pone.0063837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pazhani GP, Ramamurthy T, Mitra U, Bhattacharya SK, Niyogi SK. 2005. Species diversity and antimicrobial resistance of Shigella spp. isolated between 2001 and 2004 from hospitalized children with diarrhoea in Kolkata (Calcutta), India. Epidemiol Infect 133:1089–1095. doi: 10.1017/S0950268805004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair GB, Ramamurthy T, Bhattacharya MK, Krishnan T, Ganguly S, Saha DR, Rajendran K, Manna B, Ghosh M, Okamoto K, Takeda Y. 2010. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog 2:4. doi: 10.1186/1757-4749-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S, Pazhani GP, Chowdhury G, Guin S, Dutta S, Rajendran K, Bhattacharya MK, Takeda Y, Niyogi SK, Nair GB, Ramamurthy T. 2011. Genetic characteristics and changing antimicrobial resistance among Shigella spp. isolated from hospitalized diarrhoeal patients in Kolkata, India. J Med Microbiol 60:1460–1466. doi: 10.1099/jmm.0.032920-0. [DOI] [PubMed] [Google Scholar]
- 32.Kuo CY, Su LH, Perera J, Carlos C, Tan BH, Kumarasinghe G, So T, Van PH, Chongthaleong A, Song JH, Chiu CH. 2008. Antimicrobial susceptibility of Shigella isolates in eight Asian countries, 2001-2004. J Microbiol Immunol Infect 41:107–111. [PubMed] [Google Scholar]
- 33.Vrints M, Mairiaux E, Van Meervenne E, Collard JM, Bertrand S. 2009. Surveillance of antibiotic susceptibility patterns among Shigella sonnei strains isolated in Belgium during the 18-year period 1990 to 2007. J Clin Microbiol 47:1379–1385. doi: 10.1128/JCM.02460-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt KE, Thieu Nga TV, Thanh DP, Vinh H, Kim DW, Vu Tra MP, Campbell JI, Hoang NV, Vinh NT, Minh PV, Thuy CT, Nga TT, Thompson C, Dung TT, Nhu NT, Vinh PV, Tuyet PT, Phuc HL, Lien NT, Phu BD, Ai NT, Tien NM, Dong N, Parry CM, Hien TT, Farrar JJ, Parkhill J, Dougan G, Thomson NR, Baker S. 2013. Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc Natl Acad Sci U S A 110:17522–17527. doi: 10.1073/pnas.1308632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol 7:129–133. doi: 10.1016/S0966-842X(99)01459-6. [DOI] [PubMed] [Google Scholar]
- 36.Feldgarden M, Riley MA. 1999. The phenotypic and fitness effects of colicin resistance in Escherichia coli K-12. Evolution 53:1019–1027. doi: 10.2307/2640807. [DOI] [PubMed] [Google Scholar]
- 37.Luria SE, Delbrück M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siguier P, Filee J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol 9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Touchon M, Rocha EP. 2007. Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol 24:969–981. doi: 10.1093/molbev/msm014. [DOI] [PubMed] [Google Scholar]
- 41.Johnsrud L. 1979. DNA sequence of the transposable element IS1. Mol Gen Genet 169:213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- 42.Mulvey MR, Bryce E, Boyd DA, Ofner-Agostini M, Land AM, Simor AE, Paton S. 2005. Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob Agents Chemother 49:358–365. doi: 10.1128/AAC.49.1.358-365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fournier D, Richardot C, Müller E, Robert-Nicoud M, Llanes C, Plésiat P, Jeannot K. 2013. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother 68:1772–1780. doi: 10.1093/jac/dkt098. [DOI] [PubMed] [Google Scholar]
- 44.Pan YH, Liao CC, Kuo CC, Duan KJ, Liang PH, Yuan HS, Hu ST, Chak KF. 2006. The critical roles of polyamines in regulating ColE7 production and restricting ColE7 uptake of the colicin-producing Escherichia coli. J Biol Chem 281:13083–13091. doi: 10.1074/jbc.M511365200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


