Abstract
In order to understand the genetic background and dissemination mechanism of carbapenem resistance and fosfomycin resistance in Enterobacteriaceae isolates, we studied a clinical Escherichia coli strain HS102707 isolate and an Enterobacter aerogenes strain HS112625 isolate, both of which were resistant to carbapenem and fosfomycin and positive for the blaKPC-2 and fosA3 genes. In addition, a clinical Klebsiella pneumoniae strain HS092839 isolate which was resistant to carbapenem was also studied. A 70-kb plasmid was successfully transferred to recipient E. coli J53 by a conjugation test. PCR and Southern blot analysis showed that blaKPC-2 was located on this plasmid. The complete sequence of pHS102707 showed that this plasmid belongs to the P11 subfamily (IncP1) and has a replication gene, several plasmid-stable genes, an intact type IV secretion system gene cluster, and a composite transposon Tn1721-Tn3 that harbored blaKPC-2. Interestingly, a composite IS26 transposon carrying fosA3 was inserted in the Tn1721-tnpA gene in pHS102707 and pHS112625, leading to the disruption of Tn1721-tnpA and the deletion of Tn1721-tnpR. However, only IS26 with a truncated Tn21-tnpR was inserted in pHS092839 at the same position. To our knowledge, this is the first report of fosA3 and blaKPC-2 colocated in the same Tn1721-Tn3–like composite transposon on a novel IncP group plasmid.
INTRODUCTION
The increasing incidences of carbapenem-resistant bacteria, which are frequently resistant to most antibiotics, have renewed interest in revisiting the clinical use of old antibiotics, such as fosfomycin, for treating infections (1–3). Fosfomycin remains active against most Enterobacteriaceae isolates, with low-level resistance mediated most often by mutations in chromosomal loci, including glpT (4), murA (3), and so on. Recently, two novel plasmid-encoded fosfomycin-inactivating enzymes, FosA3 and FosC2, were found among CTX-M-producing Escherichia coli isolates in Japan, China, and South Korea (5–7). In the majority of isolates expressing these enzymes, the corresponding genes are located on an IS26-flanked composite transposon inserted into the vicinity of blaCTX-M.
The most prevalent class A carbapenemases in clinical Enterobacteriaceae isolates are the Klebsiella pneumoniae carbapenemase (KPC) enzymes, and the KPC-2 variant is the most common (8). In Europe and the United States, blaKPC-2 is frequently located within a Tn3-based transposon, Tn4401 (8, 9). Similarly, Shen et al. (10) showed that the majority of carbapenem-resistant isolates, obtained from six eastern cities in China, carried blaKPC-2 within related but more complex chimeric elements. They comprise Tn1721, Tn3, Tn4401, and ISKpn8 fragments and a segment similar to the plasmid RA3. This complex composite transposon possesses Tn1721-derived termini that match the intact transposase gene and an internal blaKPC-2-bearing 2-kb fragment showing significant sequence similarity to Tn4401 (10). In this study, we report the coexistence of fosA3 and blaKPC-2 on a single transposon found within a conjugative IncP plasmid, which was identified in two temporally and spatially related clinical isolates belonging to distinct Enterobacteriaceae species. To our knowledge, this is the first report of a transposon carrying the fosfomycin resistance gene fosA3 and the carbapenemase gene blaKPC-2.
MATERIALS AND METHODS
Bacterial strains and antibiotic susceptibility testing.
Two clinical isolates, of E. coli strain HS102707 and Enterobacter aerogenes strain HS112625, were collected after they were found to not be sensitive to imipenem and fosfomycin in routine antibiotic susceptibility testing. Among 78 blaKPC-positive clinical K. pneumoniae isolates for which we sequenced the genetic environment of blaKPC, strain HS092839 has a blaKPC-2 environment similar to that of E. coli strain HS102707 or E. aerogenes strain HS112625; that is, a similar blaKPC-2-carrying transposon was inserted into a similar 70-kb plasmid. Therefore, the K. pneumoniae HS092839 isolate was also included in this study.
The MICs of amikacin, ceftazidime, cefotaxime, imipenem, ertapenem, meropenem, piperacillin-tazobactam, and fosfomycin were determined by the Etest technique (AB Biodisk, Sweden) for the three isolates mentioned above, their transconjugants, and recipient E. coli J53. The results of the susceptibility tests were interpreted by Clinical and Laboratory Standards Institute (CLSI) criteria for E. coli and even for species other than E. coli (11).
MLST.
The sequence types (STs) of E. coli strain HS102707 were were determined by analyzing housekeeping genes in two schemes: (i) eight genes used in the Pasteur project, including dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA, and (ii) seven genes used in the Achtman project, including adk, fumC, gyrB, icd, mdh, purA, and recA. The ST of K. pneumoniae strain HS092839 was determined with seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB). The results were compared with information in the multilocus sequence typing (MLST) databases (available at http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html, http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html, and http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
Bacterial conjugation.
Conjugation was performed using donor and recipient E. coli J53 cells mixed at a ratio of 1:1 in broth culture as described previously (12). Transconjugants were selected on MacConkey agar containing ampicillin (100 mg/liter) and sodium azide (100 mg/liter). Putative transconjugant colonies were selected and identified by the Vitek system and further confirmed by a blaKPC-2 PCR assay.
Plasmid analysis.
Plasmids were extracted from 100-ml overnight cultures with the Qiagen plasmid midi kit (Qiagen, Germany) and examined by agarose gel electrophoresis. Plasmid sizes were estimated by comparison with E. coli V517 plasmid bands (54.2, 7.3, 5.6, 5.2, 3.9, 3.1, 2.7, and 2.1 kb) (13). The primers targeting blaKPC genes were described previously (14).
Sequencing of pHS102707.
One microgram of pHS102707 was used to generate a fragment library using the Ion Plus fragment library kit (Life Technologies). DNA sequencing and sequence assembly were performed using the Ion Torrent platform (Life Technologies). Gaps were closed by sequencing amplicons generated by primer-walking PCR performed on linking clones. Open reading frames were identified and annotated by searching against the NCBI nonredundant protein database.
Southern hybridization.
Purified plasmids were electrophoresed in a 1.0% agarose gel, transferred to a positively charged nylon membrane (Roche Applied Science), and probed with a PCR-amplified blaKPC-2 probe according to the protocol specified by the DIG-High Prime DNA labeling and detection kit (Roche Applied Science).
Analysis of the genetic environment of blaKPC-2.
As preliminary screening suggested that pHS102707, pHS112625, and pHS092839 shared a blaKPC-2 genetic context similar to that of pKP048 (10), a PCR mapping approach was used to examine the relevant regions of the former plasmids. A series of primers with a common annealing temperature of ∼60°C (see Table S1 and Fig. S1 in the supplemental material) was designed to allow for the generation of overlapping PCR fragments spanning the region of interest. When a standard primer pair failed to yield a product, alternative outer primers from the same PCR mapping set were used to span the region of variation. All relevant amplicons obtained were sequenced.
Nucleotide sequence accession numbers.
The complete sequences of the plasmid pHS102707, a 15,464-bp region of pHS112625, and a 15,499-bp region of pHS092839 were deposited in GenBank under the accession numbers KF701335, KF724506, and KJ210592, respectively. The sequences of the relaxase genes of pHS092839 and pHS112625 were deposited under accession numbers KJ210594 and KJ210593, respectively.
RESULTS AND DISCUSSION
Fosfomycin resistance is uncommon among Enterobacteriaceae clinical isolates.
As part of a broader study focused on carbapenem-resistant Enterobacteriaceae isolates obtained at Huashan Hospital, Shanghai, from August 2006 to December 2011, two isolates, from E. coli strain HS102707 (Ec-07) and E. aerogenes strain HS112625 (Ea-25), which exhibited resistance to fosfomycin and intermediate resistance to imipenem, were chosen for further characterization. A third multidrug-resistant, fosfomycin-sensitive Enterobacteriaceae isolate from this same collection, K. pneumoniae strain HS092839 (Kp-39), carrying a similar plasmid and Tn1721 transposon structure, was also analyzed for comparison. The MICs of selected antimicrobial agents for Ec-07, Ea-25, and Kp-39 were determined (Table 1), which confirms that all three were resistant or intermediately resistant to imipenem. The MICs of fosfomycin for the Kp-39, Ec-07, and Ea-25 transconjugants were 0.38 mg/liter, 48 mg/liter, and >1,024 mg/liter, respectively. All three isolates tested positive for blaKPC-2 by PCR, and Ec-07 and Ea-25 were also positive for fosA3.
TABLE 1.
MICs of the studied strains
| Strain | MIC (mg/liter) ofa: |
|||||||
|---|---|---|---|---|---|---|---|---|
| AK | IMP | CAZ | ETP | CTX | MEM | TZP | FOS | |
| E. coli J53 | 2 | 0.25 | 0.25 | 0.06 | 0.064 | 0.015 | 4 | 1 |
| K. pneumoniae HS092839 (Kp-39) | >256 | >32 | >256 | >32 | >256 | 32 | >256 | 4 |
| E. coli J53 HS092839 transconjugant | 2 | 4 | 8 | 1.5 | 4 | 1 | >256 | 0.38 |
| Enterobacter aerogenes HS112625 (Ea-25) | 8 | 8 | 32 | 24 | 32 | 8 | >256 | 1024 |
| E. coli J53 HS112625 transconjugant | 2 | 4 | 8 | 1 | 6 | 0.5 | >256 | 48 |
| E. coli HS102707 (Ec-07) | 8 | 6 | 6 | 1 | 4 | 0.5 | >256 | >1,024 |
| E. coli J53 HS102707 transconjugant | 1.5 | 4 | 6 | 1 | 4 | 0.25 | >256 | >1,024 |
AK, amikacin; IMP, imipenem; CAZ, ceftazidime; ETP, ertapenem; CTX, cefotaxime; MEM, meropenem; TZP, piperacillin-tazobactam; FOS, fosfomycin.
The E. coli (Ec-07) and K. pneumoniae (Kp-39) isolates were also characterized by MLST and found to belong to ST46 and ST11, respectively, according to corresponding species-specific MLST schemes of Pasteur. Consistent with previous reports, ST11 has been shown to be a predominant K. pneumoniae sequence type identified among clinical isolates from China (15, 16). In contrast, E. coli ST46 (or ST5 in Achtman MLST schemes) has not been reported to be a dominant clone in China or elsewhere (17–19).
fosA3 and blaKPC-2 are carried on the same conjugative plasmid.
In order to further explore the genetic basis of resistance, we performed conjugation experiments with each of the three primary isolates serving as a donor and the azide-resistant E. coli J53 as the recipient. Transconjugants were selected on medium containing ampicillin and azide. Next, electrophoretic plasmid profiles of the parental isolates were compared with those of matching transconjugants. Based on these data, it was evident that the parent Ea-25 and Kp-39 isolates carried multiple plasmids. By contrast, Ec-07 appeared to carry only a single plasmid. The three transconjugants derived from each of the primary clinical isolates appeared to each harbor a single plasmid of approximately 70 kb in size (Fig. 1a). As expected, plasmid bands of the transconjugants matched corresponding bands of each of the parent isolates. blaKPC-2-directed PCR and Southern blot analysis of the plasmid DNA extracted from both parent donor isolates and the transconjugants showed that blaKPC-2 was located on these ∼70-kb plasmids (Fig. 1b). Electrophoresis of EcoRI- and HindIII-digested plasmids purified from the three transconjugants showed that the restriction profiles of Ec-07-derived pHS102707, Kp-39-derived pHS092839, and Ea-25-derived pHS112625 were similar, but some differences were still seen among these three plasmids (Fig. 1c). The different MICs for fosfomycin between the Ec-07 and Ea-25 transconjugants may be due to the differences between plasmid pHS102707 and pHS112625.
FIG 1.

Electrophoretic profiles of plasmids (a), hybridization with a blaKPC-2-specific probe (b), and plasmids digested with EcoRI and HindIII (c). (a and b) In lanes 1, 3, and 5, plasmids were extracted from parental isolate K. pneumoniae HS092839, E. coli HS102707, and E. aerogenes HS112625, respectively; in lanes 2, 4, and 6, plasmids were extracted from E. coli J53 HS092839 transconjugant, E. coli J53 HS102707 transconjugant, and E. coli J53 HS112625 transconjugant, respectively. The asterisk in panel b indicates a faint blaKPC-2-positive band. (c) In lanes 1, 2, and 3, plasmids derived from isolates K. pneumoniae HS092839, E. coli HS102707, and E. aerogenes HS112625 were digested with EcoRI; in lanes 4, 5, and 6, plasmids derived from isolates K. pneumoniae HS092839, E. coli HS102707, and E. aerogenes HS112625 were digested with HindIII.
Complete sequence of the E. coli-derived pHS102707 plasmid.
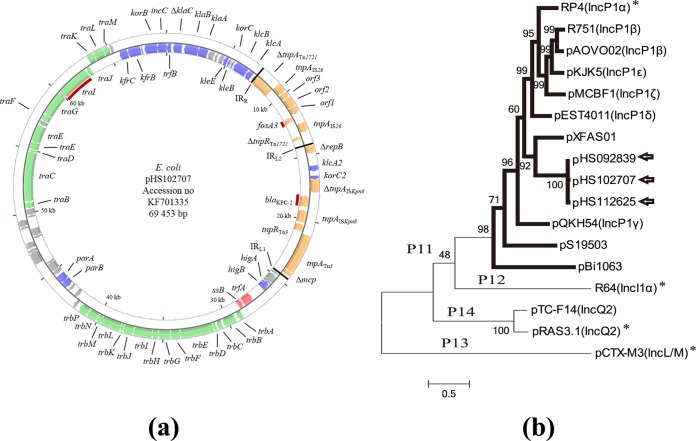
In order to further define the genetic context of blaKPC-2 and fosA3 in E. coli strain HS102707, we determined the entire sequence of the plasmid pHS102707, which was 69,453 bp in length with a G+C content of 48.9%. Using the approach by Norman et al. (20), we determined that pHS102707 carries genes involved in replication, stability, propagation, and adaptation (Fig. 2). It has one replication gene, trfA, in a replication module, and several plasmid stability genes, including parAB, higA, and higB, etc. blaKPC-2 was located within a complex chimeric element derived from Tn1721, Tn3, and other mobile elements. This complex element was inserted in the stability module of pHS102707. Critically, consistent with its ready mobilization by conjugation, pHS102707 encodes a full complement of conjugation machinery, including a type IV secretion system (T4SS), a relaxase, and a cognate origin of transfer (oriT) sequence. The pHS102707 T4SS is encoded by the trbA-trbP and traA-traM gene clusters.
FIG 2.
(a) Schematic map of pHS102707, an IncP plasmid found in the carbapenem-resistant E. coli isolate investigated in this study. Genes shown in red, blue, green, and orange are involved in replication, stability, propagation, and adaptation, respectively. Genes encoding unknown functions or that are not directly related to the above-mentioned roles are indicated in gray and shown unlabeled. Red bars highlight blaKPC-2, fosA3, and the relaxase gene (traI). Tn1721-specific inverted repeats that define the boundaries of the associated mosaic transposon are labeled IRL1 and IRR; a third matching internal repeat sequence is labeled IRL2. (b) Inferred phylogenetic relationships of the plasmid-coding relaxase homologs. Seventeen protein sequences were aligned and the tree was generated with MEGA5 (21, 22) using the maximum-likelihood method. The relaxase sequences obtained in this study are indicated by blank arrows, while others were taken from Alvarado et al. (25). The sequences marked by an asterisk denote the ones taken from the prototype plasmid of each subfamily.
Approximately 50% of Gammaproteobacteria plasmids are potentially transmissible (conjugative and mobilizable) (23). Relaxase is the only component common to all transmissible plasmids (24, 25). Based upon putative protein sequence similarities, a total of 741 sequences of relaxase encoded by 673 plasmids have been taken from 1,730 plasmid sequences deposited in GenBank (25). All the plasmids with relaxase can be classified into one of six families (MOBP, MOBF, MOBV, MOBQ, MOBH, and MOBC) and 31 subfamilies (23–25). MOBP was the most represented (273 plasmids) by a degenerate primer MOB typing (DPMT) method (26). Within each family, subfamilies corresponding to phylogenetic clades contain more members (e.g., MOBF12, which harbors relaxases encoded by IncF plasmids, and MOBP11, which groups relaxases encoded by IncP plasmids) (23). Phylogenetic analysis shows that the three relaxases encoded by pHS102707, pHS092839, and pHS112625 were grouped as the P11 relaxase subfamily (Fig. 2b). Relaxase protein sequence comparison of pHS102707 revealed 30.5% identity with RP4 (Inc P1α), the prototype plasmid for the P11 subfamily. The relaxase most similar to that of pHS102707, pXFAS01, exhibited 47% identity; no identity was found with any other non-P11 subfamily relaxases by NCBI blast.
The blaKPC-2 and fosA3 genes are borne on a novel mosaic transposon.
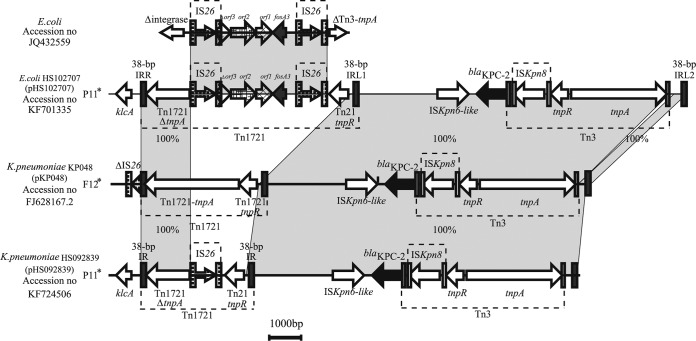
The fosA3 gene in pHS102707 is embedded in a variant of the recently described blaKPC-2-bearing transposon found on pKP048 (Fig. 3). blaKPC-2 is located in a Tn1721-Tn3 chimeric element flanked by two 38-bp inverted repeat sequences, IRR and IRL2. In pHS102707 and pHS092839, Tn1721-Tn3 was inserted close to the gene klcA, but in pKP048, it was located next to a truncated IS26-tnpA. pKP048 belongs to the F family (IncF), while pHS102707, pHS092839, and pHS112625 belong to the P11 subfamily (IncP1). This suggests that a block mobilization of the complex transposon may happen among IncP1 plasmids. Interestingly, EcoRI and HindIII digestion profiles (Fig. 1c) show that the three P11 subfamily plasmids are not identical, and some differences exist.
FIG 3.
Comparative analysis of the blaKPC-2 and fosA3 bearing mosaic Tn1721-Tn3-derived transposons present on pHS102707. The genes are depicted as arrows according to the direction of transcription. blaKPC-2 and fosA3 are shown in black and dark gray, respectively. The inverted repeats are indicated by the variable, vertical gray bars. Regions with similar sequences are indicated in gray between the different plasmids. The asterisk indicates that the plasmid type was determined by the degenerate primer MOB typing (DPMT) method (25).
The blaKPC-2-flanking regions among pHS102707, pHS092839, and pKP048 on pHS102707 showed a high degree of synteny. The similar regions contained Tn3, blaKPC-2, and the ISKpn6-like transposase gene. However, several insertion and/or deletion events had taken place within the Tn1721-like region. Tn1721 was intact in pKP048, carrying intact termini and transposase genes. In pHS092839, IS26 and a truncated Tn21-tnpR had been inserted into Tn1721, resulting in the truncated Tn1721-tnpA and deletion of Tn1721-tnpR. Remarkably, in pHS102707, a DNA fragment newly found in this study, IS26-orf3-orf2-orf1-fosA3-IS26-Tn21-tnpR, had been inserted into Tn1721-tnpA. The IS26 composite transposon in pHS102707 is the same as that found in E. coli isolated from livestock (GenBank accession no. JQ432559) (26).
The plasmid-borne fosA3 fosfomycin resistance gene was first reported in E. coli isolates collected between 2002 and 2007 in Japan (7) and has since been reported in China and South Korea. Where characterized, fosA3 has been found in an IS26-associated context on different plasmids from E. coli and K. pneumoniae (5, 26) and has frequently been linked physically to one of several blaCTX-M variants and, occasionally, to the aminoglycoside resistance-encoding rmtB gene. Our findings, combined with those of the present study, suggest that the IS26 composite transposon is highly mobile, appearing in the plasmid harboring blaCTX-M, as well as the transposon with blaKPC-2. Additionally, it is interesting to note that the two IS26 insertions in pHS102707 and pHS092839 occurred in the exactly the same position as in pKP048. pHS102707 has an IS26-mediated composite transposon in comparison to pHS092839, highlighted here by the insertion of fosA3. Since IS26 is often present in the vicinity of a long list of resistance genes, this insertion sequence is likely to contribute to the accelerated emergence of other elements carrying fosA3 alongside various repertoires of preexisting resistance determinants.
In conclusion, we report from this study an IS26-flanked composite transposon which has mobilized fosA3 onto a blaKPC-2-bearing Tn1721-Tn3-derived mosaic transposon in an ST11-type K. pneumoniae isolate. This brings together genes coding for resistance to two classes of last-line antimicrobial agents on a single mobilizable element that itself resides on a larger conjugative plasmid.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Stephen H. Gregory (Department of Medicine, Rhode Island Hospital and the Warren Albert Medical School of Brown University) for his contributions to writing and editing this manuscript.
This work was supported by the National Natural Science Foundation of China (grants 81071396 and 31170082), the National Clinical Key Subject and the Science (to X.J.), the Technology Commission of Shanghai Municipality (grant 114119a0500), the Shanghai Municipal Commission of Health and Family Planning (grant JWKJ-RCYQ-201204, awarded to G.L.), and the 973 program, Ministry of Science and Technology, China (grant 2015CB554202). D.B. was partially funded by the British Council, and K.R. and H.-Y.O. received a grant from the China Scholarship Council Sino-UK Higher Education Research Partnership for Ph.D. Studies.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03061-14.
REFERENCES
- 1.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. 2010. Fosfomycin for the treatment of multidrug-resistant, including extended spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis 10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez L, Ocampo L, Rosario C, Sumano H. 2010. Pharmacokinetics of disodium fosfomycin in broilers and dose strategies to comply with its pharmacodynamics versus Escherichia coli. Poult Sci 89:2106–2115. doi: 10.3382/ps.2010-00892. [DOI] [PubMed] [Google Scholar]
- 3.Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, Terada S, Muratani T, Matsumoto T, Nakahama C, Tomono K. 2010. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents 35:333–337. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Couce A, Briales A, Rodríguez-Rojas A, Costas C, Pascual A, Blázquez J. 2012. Genomewide overexpression screen for fosfomycin resistance in Escherichia coli: MurA confers clinical resistance at low fitness cost. Antimicrob Agents Chemother 56:2767–2769. doi: 10.1128/AAC.06122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, Arakawa Y. 2012. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and K. pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother 67:2843–2847. doi: 10.1093/jac/dks319. [DOI] [PubMed] [Google Scholar]
- 6.Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, Chen Z, Liu JH. 2012. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother 56:2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin modifying enzymes. Antimicrob Agents Chemother 54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI. 2014. Performance standards for antimicrobial susceptibility testing, vol 34, 24th informational supplement. Approved standard M100-s24. CLSI, Wayne, PA. [Google Scholar]
- 12.Wu Q, Zhang YB, Han LZ, Sun J, Ni Y. 2009. Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob Agents Chemother 53:271–272. doi: 10.1128/AAC.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendes RE, Bell JM, Turnidge JD, Yang Q, Yu Y, Sun Z, Jones RN. 2008. Carbapenem-resistant isolates of Klebsiella pneumoniae in China and detection of a conjugative plasmid (blaKPC-2 plus qnrB4) and a blaIMP-4 gene. Antimicrob Agents Chemother 52:798–799. doi: 10.1128/AAC.01185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodford N, Tierno PM Jr, Young K, Tysall L, Palepou MF, Ward E, Painter RE, Suber DF, Shungu D, Silver LL, Inglima K, Kornblum J, Livermore DM. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob Agents Chemother 48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Jiang X, Wang Y, Li G, Tian Y, Liu H, Ai F, Ma Y, Wang B, Ruan F, Rajakumar K. 2014. Contribution of β-lactamases and the porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 58:1214–1217. doi: 10.1128/AAC.02045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, Chen Y, Tian S, Zhao J, Shen D, Han L. 2013. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect 19:E509–E515. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 17.Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. 2011. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect 17:1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Siu LK, Lin JC, Wu TL, Fung CP, Wang JT, Lu PL, Chuang YC. 2013. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect Dis 13:599. doi: 10.1186/1471-2334-13-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 beta-lactamase producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18(31):pii=20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549. [DOI] [PubMed] [Google Scholar]
- 20.Norman A, Hansen LH, Sørensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci 364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcillan-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 24.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarado A, Garcillán-Barcia MP, de la Cruz F. 2012. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7:e40438. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho PL, Chan J, Lo WU, Law PY, Li Z, Lai EL, Chow KH. 2013. Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J Appl Microbiol 114:695–702. doi: 10.1111/jam.12099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.