Abstract
In Candida albicans, the ERG11 gene encodes lanosterol demethylase, the target of the azole antifungals. Mutations in ERG11 that result in an amino acid substitution alter the abilities of the azoles to bind to and inhibit Erg11, resulting in resistance. Although ERG11 mutations have been observed in clinical isolates, the specific contributions of individual ERG11 mutations to azole resistance in C. albicans have not been widely explored. We sequenced ERG11 in 63 fluconazole (FLC)-resistant clinical isolates. Fifty-five isolates carried at least one mutation in ERG11, and we observed 26 distinct positions in which amino acid substitutions occurred. We mapped the 26 distinct variant positions in these alleles to four regions in the predicted structure for Erg11, including its predicted catalytic site, extended fungus-specific external loop, proximal surface, and proximal surface-to-heme region. In total, 31 distinct ERG11 alleles were recovered, with 10 ERG11 alleles containing a single amino acid substitution. We then characterized 19 distinct ERG11 alleles by introducing them into the wild-type azole-susceptible C. albicans SC5314 strain and testing them for susceptibilities to FLC, itraconazole (ITC), and voriconazole (VRC). The strains that were homozygous for the single amino acid substitutions Y132F, K143R, F145L, S405F, D446E, G448E, F449V, G450E, and G464S had a ≥4-fold increase in FLC MIC. The strains that were homozygous for several double amino acid substitutions had decreased azole susceptibilities beyond those conferred by any single amino acid substitution. These findings indicate that mutations in ERG11 are prevalent among azole-resistant clinical isolates and that most mutations result in appreciable changes in FLC and VRC susceptibilities.
INTRODUCTION
Candida albicans is the most prevalent cause of fungal disease (1). Clinical manifestations of infections with Candida species can range from superficial mucosal infections to deep organ involvement usually resulting from hematogenous spread of infection. Despite the significant progress that has been made in the management of patients with fungal infections, the emergence of antifungal-resistant isolates creates a significant problem with regard to antifungal prophylaxis and empirical treatment strategies (2, 3). The azole antifungal class has been the so-called “work horse” of antifungal pharmacotherapy for the past 30 years, defined by its efficacy against Candida species and paucity of side effects. As the only oral option available for systemic antifungal treatment, the azoles are the most suitable option for the long treatment periods sometimes required for antifungal prophylaxis and therapy. As the azoles are fungistatic against Candida species, lengthy and repeated treatment courses have resulted in azole-resistant clinical isolates, resulting in treatment failure.
Sterols are essential components that function to maintain fluidity in eukaryotic membranes. The azole class of antifungals inhibits ergosterol biosynthesis and allows for the accumulation of toxic methylated sterol precursors (4). The primary sterol in the fungal cell membrane is ergosterol, and Cyp51 in C. albicans is a critical part of this biosynthetic process. Cyp51 in C. albicans (CaCyp51) catalyzes a three-step reaction that ultimately results in the demethylation of lanosterol. Each step requires one molecule of oxygen and NADPH. The azoles inhibit lanosterol demethylase by binding the nucleophilic N-4 atom of the azole ring to the heme iron at its sixth coordinate position (5). The normal substrate for this enzyme is lanosterol, and azole derivatives sit in the same binding pocket. Biochemical analysis shows that all azoles bind selectively to CaCyp51; however, the Kd (dissociation constant) values show a 2-fold to 4-fold lower affinity to fluconazole than that to itraconazole or voriconazole (5).
In C. albicans, the modulation of the ERG11 gene in the ergosterol biosynthetic pathway and the alteration of the Erg11 protein targeted by azole antifungals have been shown to contribute to azole resistance. The overexpression of ERG11 transcripts, either by gain-of-function mutations (GOF) in the transcriptional regulator, Upc2, or increased chromosome 5 copy number (on which ERG11 resides), result in reduced azole susceptibility (6–8). Mutations in the Erg11 protein mediating lanosterol demethylation have been shown to alter the ability of azole antifungals to bind to and inhibit its activity and to result in enhanced resistance to this class of antifungal agents (9–11). Previous reports of mutations in ERG11 have defined three hot spot regions corresponding to amino acids 105 to 165, 266 to 287, and 405 to 488, which are particularly permissive to amino acid substitutions (12). In order to show that ERG11 mutations can contribute to azole resistance, investigators have used several approaches, including heterologous expression of mutant ERG11 alleles in other microbial species (including Saccharomyces cerevisiae and Pichia pastoris), enzyme inhibition with fluconazole (FLC) in cell extracts, and biochemical analysis (10, 11, 13, 14). While a number of different amino acid substitutions have been associated with azole resistance (13), the majority of the previously analyzed mutations have not been studied in C. albicans.
Advances in the sequencing of the C. albicans genome and transformation of this pathogen have now allowed the study of ERG11 variations in the pathogen itself. For this, we examined the prevalence and variance of ERG11 mutations in a group of 63 characterized clinical C. albicans isolates with reduced FLC susceptibilities. We then expressed a select group of these ERG11 mutant alleles in an azole-susceptible C. albicans strain to determine the relative contributions of the individual ERG11 mutations to azole susceptibility, and we mapped the critical positions of variation on the predicted structure of CaErg11 (Cyp51).
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains were cultured on YPD (1% yeast extract, 2% peptone, and 2% dextrose) agar plates at 30°C and stored as frozen stock in 40% glycerol at −80°C (Table 1). YPD liquid medium was used for the routine growth of the strains. Nourseothricin (200 μg/ml or 25 μg/ml) was added to YPD agar plates to select strains containing the SAT1 flipper cassette (15). For plasmid construction and propagation, One Shot Escherichia coli TOP10 chemically competent cells (Invitrogen, Carlsbad, CA) were used and grown in Luria-Bertani (LB) broth or on LB agar plates supplemented with 50 μg/ml kanamycin (Fisher BioReagents, Fair Lawn, NJ) or 100 μg/ml ampicillin (Sigma).
TABLE 1.
C. albicans strains carrying mutant ERG11 alleles used in this study
| C. albicans strain | Relevant characteristic(s) or genotype | Source or reference |
|---|---|---|
| SC5314 | ERG11-1/ERG11-2 | ATCC |
| Clinical isolate | ||
| 10-72 | Azole resistant | University of Iowa |
| Constructed laboratory strainsa | ||
| ERG11WT A1A50A | ERG11WT::FRT/ERG11WT::FRT | This study |
| ERG11WT B37A30A | ERG11WT::FRT/ERG11WT::FRT | This study |
| 5A2A43A | ERG11Q21L::FRT/ERG11Q21L::FRT | This study |
| 5B6A19A | ERG11Q21L::FRT/ERG11Q21L::FRT | This study |
| 20E1II1G1 | ERG11Y132F::FRT/ERG11Y132F::FRT | This study |
| 20B5A14A | ERG11Y132F::FRT/ERG11Y132F::FRT | This study |
| 10C1B1M1 | ERG11K143R::FRT/ERG11K143R::FRT | This study |
| 10B1A32A | ERG11K143R::FRT/ERG11K143R::FRT | This study |
| 2A1A18A | ERG11F145L::FRT/ERG11F145L::FRT | This study |
| 2B1A51A | ERG11F145L::FRT/ERG11F145L::FRT | This study |
| 21C1M1A1 | ERG11S405F::FRT/ERG11S405F::FRT | This study |
| 21B12A61B | ERG11S405F::FRT/ERG11S405F::FRT | This study |
| 22AABA56A | ERG11D446E::FRT/ERG11D446E::FRT | This study |
| 22B12A58A | ERG11D446E::FRT/ERG11D446E::FRT | This study |
| 20NA11A57A | ERG11G448E::FRT/ERG11G448E::FRT | This study |
| 20NB16A10A | ERG11G448E::FRT/ERG11G448E::FRT | This study |
| 7A5A5A | ERG11F499V::FRT/ERG11F499V::FRT | This study |
| 7B4A29A | ERG11F449V::FRT/ERG11F449V::FRT | This study |
| 15A3A108A | ERG11G450E::FRT/ERG11G450E::FRT | This study |
| 16A14A47A | ERG11G450E::FRT/ERG11G450E::FRT | This study |
| 19A1A1C1 | ERG11G464S::FRT/ERG11G464S::FRT | This study |
| 19B1A71A | ERG11G464S::FRT/ERG11G464S::FRT | This study |
| 6A1A47A | ERG11K143R,E266D::FRT/ERG11K143R,E266D::FRT | This study |
| 6B18A101A | ERG11K143R,E266D::FRT/ERG11K143R,E266D::FRT | This study |
| 27A5A33A | ERG11Y132F,F145L::FRT/ERG11Y132F,F145L::FRT | This study |
| 27B7A63A | ERG11Y132F,F145L::FRT/ERG11Y132F,F145L::FRT | This study |
| 8A4A1A | ERG11G450E,I483V::FRT/ERG11G450E,I483V::FRT | This study |
| 8B4A47A | ERG11G450E,I483V::FRT/ERG11G450E,I483V::FRT | This study |
| 9A14A21 | ERG11Y132F,K143R::FRT/ERG11Y132F,K143R::FRT | This study |
| 9B4B34A | ERG11Y132F,K143R::FRT/ERG11Y132F,K143R::FRT | This study |
| 7NA35A40A | ERG11F145L,E266D::FRT/ERG11F145L,E266D::FRT | This study |
| 7NB44A56A | ERG11F145L,E266D::FRT/ERG11F145L,E266D::FRT | This study |
| 13A1A57A | ERG11D278N,G464S::FRT/ERG11D278N,G464S::FRT | This study |
| 13B6A16A | ERG11D278N,G464S::FRT/ERG11D278N,G464S::FRT | This study |
| 29NA24A23A | ERG11E266D,G464S::FRT/ERG11E266D,G464S::FRT | This study |
| 29NB30A22A | ERG11E266D,G464S::FRT/ERG11E266D,G464S::FRT | This study |
| 30A5A53A | ERG11M258L,G464S::FRT/ERG11M258L,G464S::FRT | This study |
| 30B5A57A | ERG11M258L,G464S::FRT/ERG11M258L,G464S::FRT | This study |
| 8OA31A17A | ERG11G307S,G450E::FRT/ERG11G307S,G450E::FRT | This study |
| 8OB37A4A | ERG11G307S,G450E::FRT/ERG11G307S,G450E::FRT | This study |
All laboratory strains have SC5314 as background.
Plasmid construction for allele sequencing.
The CaERG11 coding sequences were amplified by PCR (Pfu DNA polymerase; Stratagene) from C. albicans genomic DNA using the primers ERG11-A and ERG11-E (Table 2). The products were cloned into pCR-BluntII-TOPO using a Zero Blunt TOPO PCR cloning kit (Invitrogen) and transferred into E. coli TOP10 cells, with selection on LB agar plates containing 50 μg/ml kanamycin. Plasmid DNA was purified (QIAprep; Qiagen, Germantown, MD) and sequenced on an ABI 3130XL genetic analyzer using the ERG11 sequencing primers (Table 2), resulting in full-length sequence from both strands of the CaERG11 gene. The sequencing was performed using six sets of clones derived from three independent PCRs for each strain/isolate sequenced.
TABLE 2.
ERG11 primers used in this study
| Primer name by purpose | Sequencea |
|---|---|
| ERG11 mutant construction | |
| ERG11-A | 5′-GGGCCCGGGTTATTTGAGAACAGCC-3′ |
| ERG11-B | 5′-ATCCGTTCTCGAGCACTAAGGGACAA-3′ |
| ERG11-C | 5′-GTAATCAATTGAGCTCTTTTAACTTT-3′ |
| ERG11-D | 5′-GATTATAGTTCCGCGGTGGTTTTACC-3′ |
| ERG11-E | 5′-TGATGGTTTTTGTCCACTGGCTCGAG-3′ |
| ERG11 sequencing | |
| T7 | 5′-TAATACGACTCACTATAGGG-3′ |
| ERG11seqB | 5′-TATTTTCACTGCTTCAAGATCT-3′ |
| ERG11seqC | 5′-CCAAAAGGTCATTATGTTTTAG-3′ |
| M13R | 5′-CAGGAAACAGCTATGACC-3′ |
| ERG11seqE | 5′-CATTTAGGTGAAAAACCTCATT-3′ |
| ERG11seqF | 5′-TACTCCAGTTTTCGGTAAAGGG-3′ |
Underlined sequences reflect the introduction of a restriction site sequence.
Sequenced plasmids of the ERG11 open reading frames (ORFs) whose predicted translations indicated an amino acid substitution were digested with the restriction enzymes ApaI and XhoI, which excised the full-length ORF from the plasmid, and these ERG11 coding sequences were cloned upstream of the SAT1 flipper cassette into the ApaI and XhoI sites of plasmid pSFS2 (15). The ERG11 downstream segments were amplified with Ex Taq (TaKaRa) using primers ERG11-C and ERG11-D and cloned downstream of the SAT1 flipper cassette in pSFS2 using the NotI and SacII sites. This process generated plasmids (and their substitutions) pERG11-2 (F145L), pERG11-5 (Q21L), pERG11-6 (K143R+E266D), pERG11-7 (F449V), pERG11-7N (F145L+E266D), pERG11-8 (G450E+I483V), pERG11-8O (G307S+G450E), pERG11-9 (Y132F+K143R), pERG11-10 (K143R), pERG11-13 (D278N+G464S), pERG11-15/16 (G450E), pERG11-19 (G464S), pERG11-20 (Y132F), pERG11-20N (G448E), pERG11-21 (S405F), pERG11-22 (D446E), pERG11-27 (Y132F+F145L), pERG11-29N (E266D+G464S), and pERG11-30 (M258L+G464S).
Construction of strains carrying specific ERG11 alleles.
C. albicans strain SC5314 was transformed by electroporation with gel-purified inserts from each pSF2-derived plasmid that contained the SAT1 flipper disruption cassette developed by Reuss et al. (15). In these, the SAT1 selectable marker that confers resistance to nourseothricin and the FLP flipper recombinase gene are both flanked by flipper recombinase target (FRT) sites, allowing for the direct selection of nourseothricin-resistant transformants carrying only the ERG11 allele with a downstream FRT left in the ERG11 locus, as previously described (15). The integration of the constructs was confirmed by Southern hybridization.
Azole susceptibility testing.
MICs were obtained by using a modified CLSI protocol outlined in document M27-A3 (16), using RPMI medium. Overnight cultures grown at 30°C were streaked onto Sabouraud's agar and then grown for 24 h at 30°C. Individual colonies were suspended in sterile water until an optical density at 600 nm of 0.1 was reached. The working colony concentration was made by making a 1:50 dilution and a 1:20 dilution sequentially in medium. One hundred microliters from the working stock was used to inoculate a series of azole/RPMI medium dilutions, with the highest being 64 μg/ml for fluconazole. Similar procedures were used for the voriconazole and itraconazole dilutions; however, the highest concentration used for these agents was 8 μg/ml. The cultures were incubated at 35°C for 48 h, and the MICs were recorded.
Homology modeling.
The construction of the Cyp51 homology model was done according to the single-template approach outlined by Baudry, Rupasinghe, and Schuler (17) and Rupasinghe and Schuler (18) using functions within MOE (version 2011; Chemical Computing Group, Inc., Montreal, Canada). Based on the sequence identities returned by the BLOSUM62 scoring matrix (19) within the ALIGN function in MOE for the wild-type C. albicans sequence (GenBank accession no. XM_711668), S. cerevisiae Cyp51 (Genpept accession no. 4LXJ_A), and other structurally defined Cyp51 proteins, the lanosterol-bound S. cerevisiae Cyp51 (PDB 4LKJ [20]), sharing 66% amino acid identity, was chosen as the template, with no replacements in the variable regions. A second C. albicans allele (GenBank accession no. XM_711729) characterized in this study as wild type contains two variant positions (D116E and K128T) that do not affect azole resistance levels.
The predicted structures for the wild-type C. albicans Cyp51 were constructed with this single substrate-bound template using the homology function in MOE to generate 10 coarsely energy-minimized models. The model generated with the best packing score was energy minimized using the CHARMM22 force field (21) in MOE. The predicted structures were then inspected to ensure that all major P450 structural motifs (FG loop, I-helix, and the substrate access channel) were intact. Ramachandran plots were used to evaluate any torsional outliers in the final energy-minimized model, and the models generated with torsional outliers within a SAG1-related sequence (SRS) site or near the active site cavity were scrapped.
The DOCK function in MOE was used to predict the binding mode of the inhibitor FLC in the Cyp51 catalytic site identified using the Site Finder function in MOE. Simulations were run using the MMFF94x force field to optimize the binding configuration by scoring spatial contacts and electrostatic interactions (22, 23). With 100 configurations set as an upper limit for the docking, the populated list of configurations was searched for docking modes that placed these inhibitors within close proximity to the heme-bound oxygen in orientations consistent with azole inhibition. Promising configurations were then taken through another round of energy minimization under the MMFF94x force field, with a flexible protein backbone and ligand, rigid heme, and a target energy gradient of 0.1 kcal/mol · Å.
The interaction energy between each potential inhibitor and Cyp51 was calculated as the difference between the total potential energy of the minimized complex and the sum of the individual protein and ligand components of the minimized complex. The potential energy function contains the sum of the ligand/protein internal energy, van der Waals, and electrostatic energy terms. The conformer with the lowest calculated interaction energy was selected as the most probable binding interaction.
Nucleotide sequence accession numbers.
The coding sequences of the ERG11 alleles described in this study have been deposited in GenBank under the accession numbers KM875712, KM875713, KM875714, KM875715, KM875716, KM875717, KM875718, KM875719, KM875720, KM875721, KM875722, KM875723, KM875724, KM875725, KM875726, KM875727, KM875728, KM875729, and KM881482.
RESULTS
Many clinical isolates with reduced fluconazole susceptibilities carry mutations in ERG11.
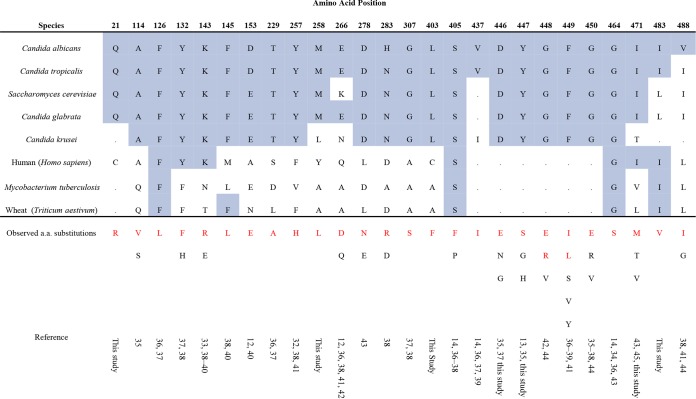
Of the 63 isolates that were determined to be resistant to fluconazole (i.e., MIC, ≥8 μg/ml), 55 carried a mutation in ERG11 that led to at least one amino acid substitution. Although silent mutations were observed in the ERG11 alleles tested (data not shown), we recovered 26 distinct positions in which mutations occurred that resulted in an amino acid substitution either alone or in combination with other mutations (Table 3 and Fig. 1). With the exception of isolate 16, which harbored a Q21R substitution in only one ERG11 allele, other isolates were homozygous for mutations in the ERG11 allele. Nine of these substitutions, due to either their position (Q21R, M258L, L403F, and I483V) or the specific amino acid substitution (A114V, D446E, Y447S, F449I, and I471M), have not been described in previous reports. Among the isolates that carry amino acid substitutions in the Erg11 protein, the number of substitutions varied between the isolates and ranged between 1 (n = 28) and 4 (n = 3), with most isolates in this collection carrying a single amino acid substitution. In total, 31 unique ERG11 alleles were recovered from sequence analysis (Table 3), with the most common polymorphisms detected at positions E266 (n = 10), Y132 (n = 9), G464 (n = 8), and K143 (n = 7).
TABLE 3.
Occurrence of Erg11 amino acid (aa) substitutions in the predicted translated sequence in fluconazole-resistant clinical ERG11-overexpressing isolates
| Erg11 aa substitution(s) (ERG11 mutation) | IDa | Zygosity | FLC MIC (μg/ml) | Upc2 aa substitutionb | Other resistance mechanism(s)b |
|---|---|---|---|---|---|
| None | 36 | 64 | ↑ERG11, CDR1, CDR2 | ||
| 37 | 64 | ↑ERG11, CDR1, CDR2 | |||
| 53 | >256 | ↑ERG11 | |||
| 55 | >256 | G648S | ↑ERG11 | ||
| 56 | >256 | G648S | ↑ERG11, CDR1, CDR2 | ||
| 57 | >256 | G648S | ↑ERG11, CDR1, CDR2 | ||
| 58 | >256 | G648S | ↑ERG11, CDR1, CDR2 | ||
| 60 | >256 | ↑ERG11 | |||
| Q21R (A62G) | 16 | Heterozygous | 16 | ↑MDR1 | |
| Y132F (A395T) | 48 | Homozygous | 256 | ↑ERG11, CDR1, CDR2 | |
| K143R (A428G) | 13 | Homozygous | 16 | A643T | ↑ERG11, CDR1, CDR2 |
| 12 | Homozygous | 16 | ↑ERG11, CDR1, CDR2 | ||
| 14 | Homozygous | 16 | ↑ERG11, CDR1, CDR2 | ||
| F145L (T435G) | 22 | Homozygous | 32 | ↑ERG11 | |
| S405F (C1214T) | 18 | Homozygous | 32 | ↑ERG11, CDR1, CDR2 | |
| 19 | Homozygous | 32 | ↑ERG11, CDR1, CDR2 | ||
| 20 | Homozygous | 32 | ↑ERG11, CDR1, CDR2 | ||
| 41 | Homozygous | 128 | ↑ERG11, MDR1 | ||
| D446E (T1338A) | 25 | Homozygous | 32 | G648Sc | ↑ERG11, MDR1 |
| 35 | Homozygous | 64 | G648Sc | ↑ERG11 | |
| 44 | Homozygous | 128 | ↑ERG11, CDR1, CDR2, MDR1 | ||
| G448E (G1341A) | 31 | Homozygous | 64 | ↑ERG11, CDR1, CDR2 | |
| 65 | Homozygous | >256 | G648Sc | ↑ERG11, CDR1, CDR2 | |
| 66 | Homozygous | >256 | ↑CDR1, CDR2 | ||
| 67 | Homozygous | >256 | G648Sc | ↑ERG11, CDR1, CDR2 | |
| 69 | Homozygous | >256 | G648Sc | ↑ERG11, CDR1, CDR2 | |
| F449V (T1345G) | 40 | Homozygous | 128 | ↑ERG11, CDR1, CDR2 | |
| G450E (G1349A) | 24 | Homozygous | 32 | ↑CDR1, CDR2, MDR1 | |
| 34 | Homozygous | 64 | Y642F | ↑CDR1, CDR2, MDR1 | |
| 63 | Homozygous | >256 | ↑ERG11, CDR1, CDR2, MDR1 | ||
| 64 | Homozygous | >256 | ↑ERG11, CDR1, CDR2, MDR1 | ||
| G464S (G1390A) | 32 | Homozygous | 64 | ↑CDR1, CDR2, MDR1 | |
| 38 | Homozygous | 64 | G648Dc | ↑ERG11, CDR1, CDR2 | |
| 39 | Homozygous | 64 | ↑ERG11, CDR1, CDR2 | ||
| 42 | Homozygous | 128 | ↑ERG11, CDR1, CDR2 | ||
| 52 | Homozygous | 256 | G648Dc | ↑ERG11 | |
| A114S (G340T), Y257H (T769C) | 17 | Homozygous | 32 | ↑CDR1 | |
| F126L (C378A), Y132F (A395T) | 30 | Homozygous | 64 | A643Vc | ↑ERG11 |
| Y132F (A395T), K143R (A428G) | 50 | Homozygous | 256 | ↑CDR1, CDR2 | |
| 61 | Homozygous | >256 | ↑CDR1, CDR2 | ||
| Y132F (A395T), F145L (T435G) | 29 | Homozygous | 64 | ↑CDR1, CDR2 | |
| K143R (A428G), E266D (A798C) | 10 | Homozygous | 16 | ↑ERG11, CDR1 | |
| F145L (T435G), E266D (A798C) | 33 | Homozygous | 64 | Y642F | ↑CDR1, CDR2 |
| M258L (A772T), G464S (G1390A) | 70 | Homozygous | >256 | A646Vc | ↑ERG11 |
| E266D (A798C), G464S (G1390A) | 45 | Homozygous | 128 | A646Vc | ↑ERG11, CDR1, CDR2 |
| 15 | Homozygous | 16 | W478Cc | ↑ERG11, CDR1, CDR2 | |
| 27 | Homozygous | 64 | W478Cc | ↑ERG11, CDR1, CDR2 | |
| E266D (A798C), V488I (G1312A) | 11 | Homozygous | 16 | ↑ERG11, CDR1, CDR2 | |
| 28 | Homozygous | 64 | W478Cc | ↑ERG11, CDR1, CDR2 | |
| D278N (G832A), G464S (G1390A) | 43 | Homozygous | 128 | ↑CDR1 | |
| 51 | Homozygous | 256 | ↑ERG11, CDR1, CDR2 | ||
| 62 | Homozygous | >256 | ↑CDR1, CDR2 | ||
| G307S (G919A), G450E (G1349A) | 26 | Homozygous | 32 | ||
| 54 | Homozygous | >256 | ↑ERG11, MDR1 | ||
| G450E (G1349A), I483V (A1309G) | 23 | Homozygous | 32 | ↑CDR1, CDR2, MDR1 | |
| A114V (C341T), E226D (A798C), H283R (A848G) | 21 | Homozygous | 32 | G648Dc | ↑ERG11, CDR1, CDR2 |
| Y132F (A395T), T229A (A685G), F449L (T1345A) | 59 | Homozygous | >256 | G648Dc | ↑ERG11, CDR1, CDR2 |
| Y132F (A395T), V437I (G1309A), F449L (T1345C) | 71 | Homozygous | >256 | G648Dc | ↑ERG11, CDR1, CDR2 |
| G307S (G919A), L403F (A1209T), G448R (G1342C) | 46 | Homozygous | 128 | ||
| G307S (G919A), V437I (G1309A), Y447S (A1340C) | 49 | Homozygous | 256 | ↑ERG11, CDR1, CDR2 | |
| A114V (C341T), D153E (T459G), E266D (A798C), G450E (G1349A) | 72 | Homozygous | >256 | ↑CDR1, CDR2 | |
| A114V (C341T), Y132F (A395T), E266D (A798C), V437I (G1309A) | 47 | Homozygous | 128 | G648Dc | ↑ERG11, CDR1, CDR2 |
| Y132F (A395T), E266D (A798C), I471M (T1413G), I483V (A1309G) | 68 | Homozygous | >256 | T273A, A643V | ↑ERG11, CDR1, CDR2, MDR1 |
ID, identification.
Data were previously published by Flowers et al (6).
Amino acid substitution recovered in one of two UPC2 alleles.
FIG 1.
Observed amino acid substitutions in Erg11 in C. albicans compared to those in other medically important Candida species and organisms in other kingdoms. The alignment was generated by using the UniProt alignment function (www.uniprot.org). The amino acid numbering is based on the C. albicans sequence. The conserved amino acids are highlighted in blue. The amino acid substitutions in red type are those observed in this study. The substitutions in black type have been noted in the literature (13). The following GenBank accession numbers were used: C. albicans, P1613; Candida tropicalis, P14263; S. cerevisiae, P1614; Candida glabrata, P50859; C. krusei, Q02315; human (Homo sapiens), Q16850; Mycobacterium tuberculosis, P0A512; Triticum aestivum, P93596. a.a., amino acid.
Single mutations in ERG11 contribute to azole resistance in C. albicans.
In order to assess the contribution of each individual mutant ERG11 allele to azole antifungal resistance, we expressed each ERG11 allele homozygously in each constructed strain. Each strain was constructed in duplicate, and its susceptibilities against fluconazole, itraconazole, and voriconazole were tested and compared to those of the wild-type susceptible parent SC5314 strain at 48 h (Fig. 2). Two independent strains were constructed to carry two ERG11 wild-type alleles, which did not show a change in MIC compared to that of the parent strain for any of the azoles tested. We initially examined the effects of 10 mutant ERG11 alleles containing a single amino acid substitution in their predicted protein sequence, including Q21L, Y132F, K134R, F145L, S405F, D446E, G448E, F449V, G450E, and G464S. With the exception of the amino acid substitution Q21L, most ERG11 mutations resulted in decreased susceptibility to fluconazole compared to that of SC5314 (Fig. 2). Strains homozygously expressing K143R resulted in the strongest decrease in fluconazole susceptibility. The strains that were homozygous for alleles containing single amino acid substitutions Y132F, F145L, S405F, D446E, D448E, F449V, G464S, or G450E had a ≥4-fold increase in their fluconazole MIC. No single amino acid substitution affected itraconazole or voriconazole MICs more than 2-fold. These data suggest that structural differences between the individual azoles affect their activities against specific mutant ERG11 alleles.
FIG 2.
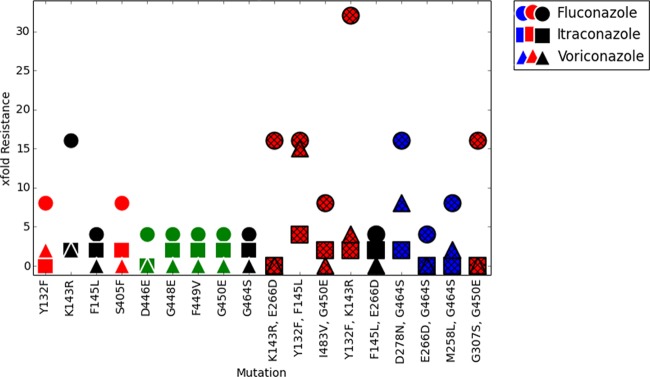
Azole resistance levels relative to amino acid variation(s) in predicted Cyp51 structure. The resistance levels of single and double mutants to FLC, ITC, and VRC are graphed, with red designating variations in or close to catalytic site residues, black designating variations in residues on the proximal side of the heme, green designating fungus-specific external loop residues, and blue designating surface residues. The strains that were constructed to carry two copies of the ERG11 wild-type allele showed no appreciable change in azole MICs over those of SC5314 (data not shown).
Multiple mutations in ERG11 can result in decreased azole susceptibility.
Previous observations showed that combinations of ERG11 mutations can lead to considerable increases in the MICs to fluconazole (14, 24). To examine the effects of multiple mutations on azole susceptibility, we selected a group of clinically occurring ERG11 alleles that carried two amino acid substitutions, with at least one substitution having been characterized alone (Fig. 2). Of those analyzed, the allele with the K143R and Y132F substitutions showed the strongest characterized combination effect, with 32-fold and 4-fold increased FLC and VRC MICs, respectively, over those observed for the azole-susceptible SC5314 strain. Interestingly, this combination did not affect ITC susceptibility. Other notable increases in the FLC MICs were detected for the Y132F+F145L and G307S+G450E combinations, with both increasing their FLC MICs by 16-fold over that observed for the SC5314 strain. Unlike all other ERG11 alleles we characterized, the Y132F+F145L combination also significantly increased ITC and VRC MICs by 4-fold and 16-fold, respectively.
Among the other combinations of polymorphisms present in our collection, the I483V+G450E combination increased the FLC MIC by 8-fold over that of the SC5314 strain and by 2-fold over that observed for the G450E allele. This combination did not significantly change either the ITC or VRC MIC. Similarly, combinations of M258L or D278N with G464S increased their FLC MICs by 2-fold and 4-fold, respectively, over that observed for the G464S allele.
Amino acid substitution E266D does not contribute to azole resistance.
The E266D substitution was the most prevalent polymorphism detected by sequence analysis, and it occurred only in combination with other amino acid substitutions. To investigate the contribution of E266D to azole resistance, we compared the susceptibilities of ERG11 alleles carrying one amino acid substitution alone to an allele containing an identical amino acid substitution combined with E266D. In our collection, the K143R, F145L, and G464S substitutions all occurred as single mutations and in combination with the E266D substitution. In all three combinations, the E266D substitution did not confer any additional effect on azole susceptibility beyond what was observed with the single amino acid substitution.
Molecular mapping of Cyp51 variant positions.
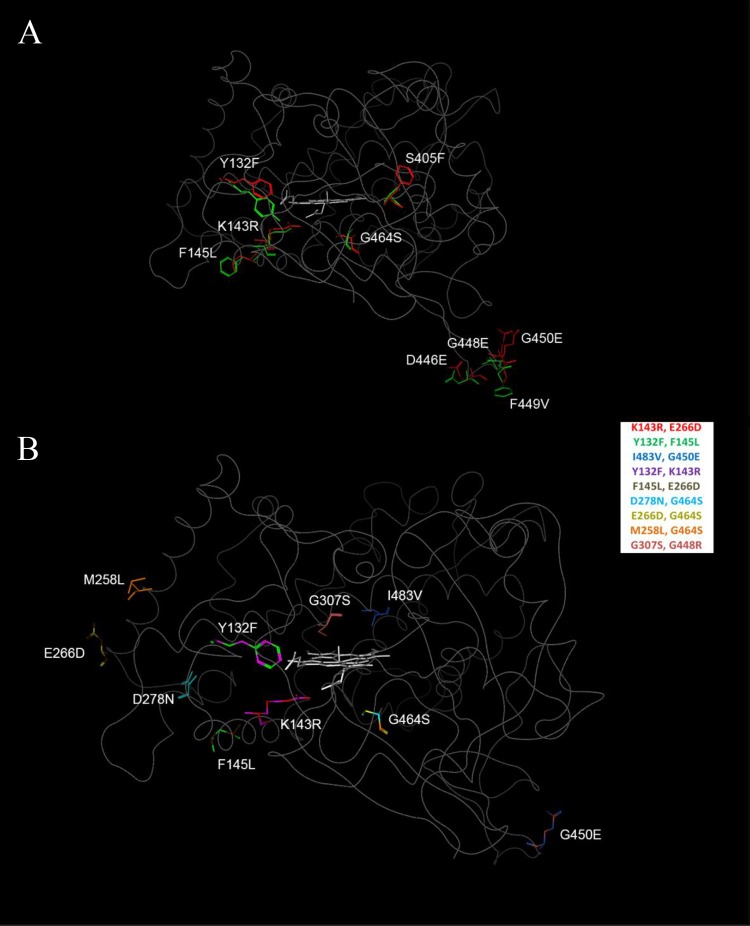
To better understand the positions of the variations enhancing FLC resistance, all were mapped on a predicted structure for ligand-free wild-type Cyp51 (GenBank accession no. XM_711668), derived using the modeling techniques described by Baudry, Rupasinghe, and Schuler (17) and Rupasinghe and Schuler (18), with lanosterol-bound S. cerevisiae Cyp51 (PDB 4LXJ [20]) as the backbone. As shown in Fig. 3A, single amino acid variations affecting resistance to one or more of these azole inhibitors occur in the predicted catalytic site (Y132F and S405F), the extended fungus-specific external loop (D446E, G448E, F449V, and G450E), on the proximal surface with potential loop interactions (F145L), and between the proximal surface and heme (K143R and G464S). As shown in Fig. 3B, double amino acid variations affecting resistance to one or more azole inhibitors exist, with additional changes occurring in the catalytic site (G307S and I483V) and on the surface (M258L, E266D, and D278N). A graph of the resistance levels in these mutants relative to the positions of these variations shown in Fig. 2 allows several conclusions to be made. First, the combination of Y132F+F145L variations mapping to the catalytic site and proximal surface maintain the resistance level for FLC as in the K143R single variant while moderately increasing the resistance level for ITC and dramatically increasing the resistance level for VRC. Second, the combination of Y132F+K143R variations mapping to the catalytic site and region below the heme dramatically increases the resistance level for FLC compared to that of the K143R single variant while moderately increasing the resistance level for VRC. Third, the combination of G307S+G450E mapping to the catalytic site and external loop dramatically increases the resistance level to FLC compared to that of the G450E single variant. To a lesser extent, the combination of I483V+G450E variations mapping to the catalytic site and external loop also significantly increases the resistance level for FLC compared to that of the G450E single variant while having no effect on the resistance levels of the other azoles. Fourth, the combination of D278N+G464S mapping to the surface and proximal regions significantly increases the resistance level for VRC and dramatically increases the resistance level for FLC compared to that with the G464S single variant. To a lesser extent, the combination of the M258L+G464S variations mapping to the surface and proximal region also increases the resistance level to FLC while having little effect on the resistance levels to the other azoles. Finally, the E266D variation occurring in a number of double variants and mapping to the surface has no effect on resistance levels.
FIG 3.
Mapping of mutant positions on the predicted structure for wild-type Cyp51. (A) The sequence variations in the single mutants characterized in this study are shown, with wild-type residues shown in green stick format, variant residues shown in red stick format, and heme shown in gray stick format. (B) The co-occurring sequence variations in the double mutants are shown with various colors, as identified in the legend box.
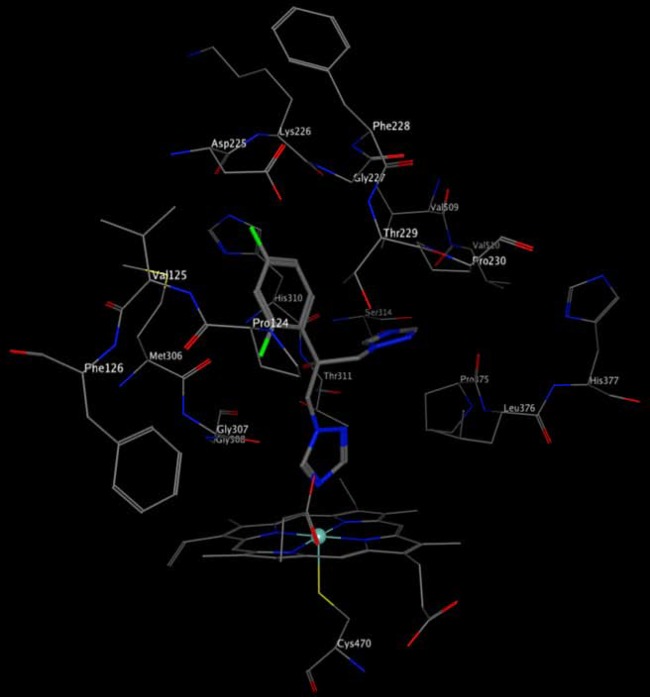
Low-throughput docking in the predicted wild-type Cyp51 catalytic site using the DOCK function in MOE with fluconazole (Fig. 4) suggests that it binds with one of its two azole rings coordinated with the heme. Based on this predicted binding mode, G307 and Y132 are within interaction distance of FLC, and their changes in the G307S and Y132F variants are predicted to interfere with FLC binding, consistent with their observed increases in FLC resistance. The other catalytic site residue whose variation significantly affects FLC resistance levels (S405) lies just beyond the set predicted to be within 4.5 Å of FLC. Additionally, the variations in the Y132F and S405F single mutants have the potential to alter hydrogen bonding and/or the dimensions of the catalytic site. The remaining catalytic site residue whose variation significantly affects FLC resistance levels (I483) occurs in another part of the catalytic site where the conservative nature of its variation to valine would not be expected to alter FLC binding. In contrast, K143, whose variation to arginine induces maximum resistance to FLC, lies in a region below the heme expected to participate in electron transfer from P450 reductase.
FIG 4.
Predicted docking modes of fluconazole in wild-type Cyp51. FLC docked in the Cyp51 site is shown in elemental stick format, with residues within 4.5 Å shown in elemental line format.
DISCUSSION
The cytochrome P450 (CYP) superfamily of enzymes contains >2,500 members that can be roughly placed into two groups stratified by their function (25). The members of the first group metabolize a wide variety of xenobiotics, while those in the second group generally participate in key biosynthetic processes, such as sterol biosynthesis. The substrate specificity for this second group of P450s is narrow. Sterol 14α-demethylase (Cyp51 or Erg11) is considered to be the most ancient of CYP family enzymes because it is the only P450 class that is found in different kingdoms, such as animals, plants, and bacteria (26, 27). The first virally encoded CYP450 gene identified in 2009 was shown to have low-level sequence identity with previously characterized CYP51 genes, but its function remains unknown (28). Despite the limited sequence identity (22 to 33%) shared among the Cyp51 proteins in different kingdoms (27), there are many structural similarities in the available Cyp51 crystal structures. The differences among these structures are most notable in C. albicans and other fungal Cyp51 proteins that contain an additional external loop between residues 428 and 459.
Recently, the Clinical Laboratory and Standards Institute (CLSI) redefined its standards for in vitro antifungal susceptibility testing. The current standards now consider species-specific clinical breakpoints (CBP) using established epidemiological cutoff values (ECV), defined as the upper cutoff value for wild-type MIC, pharmacokinetic (PK)-pharmacodynamic (PD) parameters, and the relationship between MIC and clinical outcome (29, 30). The current interpretation of in vitro susceptibility testing of C. albicans to fluconazole is susceptible with an MIC of ≤2 μg/ml, susceptible dose dependent with an MIC of 4 μg/ml, and resistant with an MIC of ≥8 μg/ml. The clinical breakpoints to voriconazole are defined as susceptible with an MIC of ≤0.12 μg/ml, susceptible dose dependent with an MIC of 0.25 to 0.5 μg/ml, and resistant with an MIC of ≥1 μg/ml.
In agreement with previous studies (13), the majority (87%) of our fluconazole-resistant clinical isolates carried point mutations in ERG11 that led to at least one amino acid substitution. The substitutions in our collection were recovered in 26 distinct positions, with 21 located in previously defined hot spot regions of CaERG11 mutations (Fig. 1) (12). Our homology model for the Erg11 protein demonstrates that these variations occur in the predicted catalytic site and extended fungus-specific external loop, as well as on the proximal surface and between the proximal surface and the heme.
In our constructed strains, most ERG11 alleles containing single-nucleotide changes resulted in meaningful changes in the fluconazole MIC but not in the itraconazole or voriconazole MIC (Fig. 2). Among the single amino substitutions, K143R, which demonstrated the strongest increase in fluconazole MIC in this study and is predicted to occur between the proximal surface and the heme, likely affects catalytic efficiency toward the lanosterol substrate of Erg11. Y132F, which demonstrated a strong increase in fluconazole MIC and is predicted to occur in the catalytic site, likely affects fluconazole binding. Interestingly, the presence of both in the Y132F+K143R double mutant yielded the strongest increase in fluconazole MIC of any allele tested. Notably, the K143 and Y123 positions are conserved among fungal species (Fig. 1).
Among the remaining single amino acid substitutions associated with fluconazole resistance, the S405F and G464S substitutions have been demonstrated to increase azole resistance when heterologously expressed in S. cerevisiae (14). In our collection, the S405F substitution, which was recovered in four distinct clinical isolates and as a single polymorphism only, is predicted to occur in the catalytic site and likely influences the binding of fluconazole. As documented by Kelly et al. (11) and as suggested by its position below the heme, the G464S substitution changes the heme environment and reduces its affinity for fluconazole without affecting the catalytic activity of the enzyme.
Several other single and double amino acid substitutions recovered in our collection occur in the fungus-specific external insertion loop. Sequence comparisons of this loop in different fungal species have indicated that the N-terminal portion of the loop is variable in sequence and length, while the C-terminal portion contains acidic residues followed by a more invariable portion with the motif DYG[FY]Gx[VI][ST]KG (31) corresponding to D446YGFGKVSKG455 in the CaCyp51 protein. All four amino acid substitutions recovered in this sequence, D446E, G448E, F449V, and G450E, occur within the invariable portion.
We also examined the collective effects of multiple mutations in ERG11 that were derived from clinical isolates. The K143R+Y132F combination resulted in the strongest increase in MIC by substantially increasing fluconazole and voriconazole MICs. This combination occurred independently in two isolates that were highly resistant to fluconazole (MIC, ≥256 μg/ml) and was accompanied by increased expression of ABC transporters CDR1 and CDR2 in both isolates. F145L and Y132F were the only amino acid substitutions that significantly affected MICs to all azoles. Previous modeling in Aspergillus fumigatus defined F145 as a position that interacts with posaconazole (32). Structurally, itraconazole is similar to posaconazole, and notably, this was the only combination of amino acid substitutions that significantly affected susceptibility to itraconazole. This combination of substitutions was also shown to catalytically impair Cyp51 when derived from a clinical C. albicans isolate. An investigation of Cyp51 function in A. fumigatus showed that positions G464S and G307 disturb the heme environment (32). This observation is in accordance with our prediction of azole binding, which indicates that G307 is located within interaction distance of FLC.
Other amino acid substitutions corresponding to I483V, M258L, and D278N occur only in combination with one of the previously mentioned single amino acid substitutions, and in all cases, they significantly increase fluconazole MICs. The I483V substitution that has not been described is predicted to be located in the catalytic site and, when combined with the G450E substitution, it increases the fluconazole MIC an additional 2-fold over that observed for the G450E substitution alone. Similar trends are observed for the M258L and D278N substitutions that in combination with the G464S substitution increase fluconazole MICs by 2-fold and 4-fold, respectively, over that observed for the G464S substitution alone. Notably, the D278E substitution previously shown to occur in azole-resistant isolates (33) is a more conserved substitution than the D278N substitution observed in our collection.
In this study, we sequenced only ERG11 alleles from isolates exhibiting decreased susceptibilities to fluconazole; it is likely that some mutations in ERG11 occurring in both azole-susceptible and -resistant isolates do not affect azole resistance. Another possibility is that mutations may arise in conjunction with mutations creating azole resistance that also affect fitness. Previous work has shown that specific ERG11 mutations affect the fitness and catalytic activity of the enzyme (5, 20, 24). Further investigation of how ERG11 mutations affect fitness is warranted.
Last, one heterozygous fluconazole-resistant isolate contained a Q21L amino acid substitution in one ERG11 mutant allele and a wild-type ERG11 allele. Because this substitution is located in the N-terminal transmembrane helix in membrane-bound lanosterol demethylases, anchoring this enzyme to the endoplasmic reticulum (34), its role in fluconazole resistance cannot be predicted from structural models. Our finding that the Q21L substitution has little effect on azole resistance levels when expressed in the azole-susceptible SC5314s strain indicates that mutations occurring in the transmembrane region of the Erg11 protein are unlikely to interfere with enzyme-substrate interactions.
Clearly, the susceptibility of clinical isolates is a product of the interplay of multiple mechanisms of resistance. Mutations in the ERG11 gene have been shown to be a significant and prevalent mechanism of resistance in C. albicans. Notably, in addition to carrying mutations in ERG11, 20 of the clinical isolates in this study also carried activating mutations in UPC2, which have been shown to have a combinatorial effect on azole susceptibility (24). Our data demonstrate that many ERG11 mutations result in fluconazole resistance, but most are not as significant when tested against voriconazole or itraconazole. Susceptibility to itraconazole in particular seems to be less affected by ERG11 mutations that produce significant resistance to fluconazole. Despite this general observation, we have identified a specific combination of amino acid substitutions that significantly reduces itraconazole and voriconazole susceptibilities.
ACKNOWLEDGMENTS
This research was supported by NIH NIAID grant R01AI058145 (to P.D.R.) and by the Society of Infectious Diseases Pharmacists/ASTELLAS Antifungal Research Award (to S.A.F.).
We thank Qing Zhang for her assistance in the laboratory. We also thank Daniel Diekema for providing the clinical isolates used in this study and Joachim Morschhäuser for pSFS2 and guidance.
REFERENCES
- 1.Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, Horn D. 2012. Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance) registry, 2004–2008. Diagn Microbiol Infect Dis 74:323–331. doi: 10.1016/j.diagmicrobio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Thompson GR III, Patel PK, Kirkpatrick WR, Westbrook SD, Berg D, Erlandsen J, Redding SW, Patterson TF. 2010. Oropharyngeal candidiasis in the era of antiretroviral therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 109:488–495. doi: 10.1016/j.tripleo.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel PK, Erlandsen JE, Kirkpatrick WR, Berg DK, Westbrook SD, Louden C, Cornell JE, Thompson GR, Vallor AC, Wickes BL, Wiederhold NP, Redding SW, Patterson TF. 2012. The changing epidemiology of oropharyngeal candidiasis in patients with HIV/AIDS in the era of antiretroviral therapy. AIDS Res Treat 2012:262471. doi: 10.1155/2012/262471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8,24(28)-dien-3 beta,6 alpha-diol. Biochem Biophys Res Commun 207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 5.Warrilow AG, Martel CM, Parker JE, Melo N, Lamb DC, Nes WD, Kelly DE, Kelly SL. 2010. Azole binding properties of Candida albicans sterol 14-alpha demethylase (CaCYP51). Antimicrob Agents Chemother 54:4235–4245. doi: 10.1128/AAC.00587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhauser J, Rogers PD. 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 11:1289–1299. doi: 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol 68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 9.Warrilow AG, Mullins JG, Hull CM, Parker JE, Lamb DC, Kelly DE, Kelly SL. 2012. S279 point mutations in Candida albicans sterol 14-alpha demethylase (CYP51) reduce in vitro inhibition by fluconazole. Antimicrob Agents Chemother 56:2099–2107. doi: 10.1128/AAC.05389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly SL, Lamb DC, Kelly DE. 1999. Y132H substitution in Candida albicans sterol 14alpha-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol Lett 180:171–175. doi: 10.1111/j.1574-6968.1999.tb08792.x. [DOI] [PubMed] [Google Scholar]
- 11.Kelly SL, Lamb DC, Loeffler J, Einsele H, Kelly DE. 1999. The G464S amino acid substitution in Candida albicans sterol 14alpha-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem Biophys Res Commun 262:174–179. doi: 10.1006/bbrc.1999.1136. [DOI] [PubMed] [Google Scholar]
- 12.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. 1999. Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701–2713. [DOI] [PubMed] [Google Scholar]
- 13.Morio F, Loge C, Besse B, Hennequin C, Le Pape P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis 66:373–384. doi: 10.1016/j.diagmicrobio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42:241–253. doi: 10.1093/jac/42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved method M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Baudry J, Rupasinghe S, Schuler MA. 2006. Class-dependent sequence alignment strategy improves the structural and functional modeling of P450s. Protein Eng Des Sel 19:345–353. doi: 10.1093/protein/gzl012. [DOI] [PubMed] [Google Scholar]
- 18.Rupashinghe SG, Schuler MA. 2006. Homology modeling of plant cytochrome P450s. Phytochem Rev 5:473–505. doi: 10.1007/s11101-006-9028-y. [DOI] [Google Scholar]
- 19.Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A 89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JD, JD O'Connell III, Cannon RD, McDonald JG, Rodriguez A, Finer-Moore JS, Stroud RM. 2014. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc Natl Acad Sci U S A 111:3865–3870. doi: 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKerell AD Jr, Bashford D, Bellot M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub K, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 22.Courcot B, Bridgeman AJ. 2011. Optimization of a molecular mechanics force field for type-II polyoxometalates focussing on electrostatic interactions: a case study. J Comput Chem 32:1703–1710. doi: 10.1002/jcc.21752. [DOI] [PubMed] [Google Scholar]
- 23.Halgren TA. 1996. Merck molecular force field. I. Basis, form, scope, parameterization and performance of MMFF94. J Comput Chem 17:490–519. [Google Scholar]
- 24.Sasse C, Dunkel N, Schäfer T, Schneider S, Dierolf F, Ohlsen K, Morschhäuser J. 2012. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol Microbiol 86:539–556. doi: 10.1111/j.1365-2958.2012.08210.x. [DOI] [PubMed] [Google Scholar]
- 25.Lepesheva GI, Waterman MR. 2004. CYP51: the omnipotent P450. Mol Cell Endocrinol 215:165–170. doi: 10.1016/j.mce.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. 2000. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem Biophys Res Commun 273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
- 27.Nelson DR. 1999. Cytochrome P450 and the individuality of species. Arch Biochem Biophys 369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 28.Lamb DC, Lei L, Warrilow AG, Lepesheva GI, Mullins JG, Waterman MR, Kelly SL. 2009. The first virally encoded cytochrome P450. J Virol 83:8266–8269. doi: 10.1128/JVI.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller MA, Andes D, Diekema DJ, Espinel-Ingroff A, Sheehan D, CLSI Subcommittee for Antifungal Susceptibility Testing . 2010. Wild-type MIC distributions, epidemiological cutoff values and species-specific clinical breakpoints for fluconazole and Candida: time for harmonization of CLSI and EUCAST broth microdilution methods. Drug Resist Updat 13:180–195. doi: 10.1016/j.drup.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Espinel-Ingroff A, Barchiesi F, Cuenca-Estrella M, Pfaller MA, Rinaldi M, Rodriguez-Tudela JL, Verweij PE. 2005. International and multicenter comparison of EUCAST and CLSI M27-A2 broth microdilution methods for testing susceptibilities of Candida spp. to fluconazole, itraconazole, posaconazole, and voriconazole. J Clin Microbiol 43:3884–3889. doi: 10.1128/JCM.43.8.3884-3889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez-Rueda N, Fleury A, Morio F, Pagniez F, Gastinel L, Le Pape P. 2011. Amino acid substitutions at the major insertion loop of Candida albicans sterol 14alpha-demethylase are involved in fluconazole resistance. PLoS One 6:e21239. doi: 10.1371/journal.pone.0021239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao L, Madison V, Chau AS, Loebenberg D, Palermo RE, McNicholas PM. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14alpha-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob Agents Chemother 48:568–574. doi: 10.1128/AAC.48.2.568-574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manavathu EK, Kallakuri S, Arganoza MT, Vázquez JA. 1999. Amino acid variations of cytochrome P-450 lanosterol 14 alpha-demethylase (CYP51A1) from fluconazole-resistant clinical isolates of Candida albicans. Rev Iberoam Micol 16:198–203. [PubMed] [Google Scholar]
- 34.Denisov IG, Shih AY, Sligar SG. 2012. Structural differences between soluble and membrane bound cytochrome P450s. J Inorg Biochem 108:150–158. doi: 10.1016/j.jinorgbio.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard D, Bille J. 2002. Action of and resistance to antifungal agents, p 370 In Calderone RA (ed), Candida and candidiasis. American Society for Microbiology, Washington, DC. [Google Scholar]
- 36.Favre B, Didmon M, Ryder NS. 1999. Multiple amino acid substitutions in lanosterol 14α-demethylase contribute to azole resistance in Candida albicans. Microbiology 145:2715–2725. [DOI] [PubMed] [Google Scholar]
- 37.Perea S, López-Ribot JL, Kirkpatrick WR, McAtee RK, Santillán RA, Martínez M, Calabrese D, Sanglard D, Patterson TF. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob Agents Chemother 45:2676–2684. doi: 10.1128/AAC.45.10.2676-2684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother 48:2124–2131. doi: 10.1128/AAC.48.6.2124-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MK, Williams LE, Warnock DW, Arthington-Skaggs BA. 2004. Drug resistance genes and trailing growth in Candida albicans isolates. J Antimicrob Chemother 53:217–224. doi: 10.1093/jac/dkh040. [DOI] [PubMed] [Google Scholar]
- 40.Manastir L, Ergon MC, Yücesoy M. 2011. Investigation of mutations in Erg11 gene of fluconazole resistant Candida albicans isolates from Turkish hospitals. Mycoses 54:99–104. doi: 10.1111/j.1439-0507.2009.01766.x. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Chen L, Li C. 2008. Susceptibility of clinical isolates of Candida species to fluconazole and detection of Candida albicans ERG11 mutations. J Antimicrob Chemother 61:798–804. doi: 10.1093/jac/dkn015. [DOI] [PubMed] [Google Scholar]
- 42.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother 46:1704–1713. doi: 10.1128/AAC.46.6.1704-1713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Brown N, Chau AS, López-Ribot JL, Ruesga MT, Quindos G, Mendrick CA, Hare RS, Loebenberg D, DiDomenico B, McNicholas PM. 2004. Changes in susceptibility to posaconazole in clinical isolates of Candida albicans. J Antimicrob Chemother 53:74–80. doi: 10.1093/jac/dkh027. [DOI] [PubMed] [Google Scholar]
- 44.Löffler J, Kelly SL, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett 151:263–268. doi: 10.1016/S0378-1097(97)00172-9. [DOI] [PubMed] [Google Scholar]
- 45.Kakeya H, Miyazaki Y, Miyazaki H, Nyswaner K, Grimberg B, Bennett JE. 2000. Genetic analysis of azole resistance in the Darlington strain of Candida albicans. Antimicrob Agents Chemother 44:2985–2990. doi: 10.1128/AAC.44.11.2985-2990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]