Abstract
Nontypeable Haemophilus influenzae (NTHi) is a common cause of respiratory infections in adults, who are frequently treated with fluoroquinolones. The aims of this study were to characterize the genotypes of fluoroquinolone-resistant NTHi isolates and their mechanisms of resistance. Among 7,267 H. influenzae isolates collected from adult patients from 2000 to 2013, 28 (0.39%) were ciprofloxacin resistant according to Clinical and Laboratory Standards Institute (CLSI) criteria. In addition, a nalidixic acid screening during 2010 to 2013 detected five (0.23%) isolates that were ciprofloxacin susceptible but nalidixic acid resistant. Sequencing of their quinolone resistance-determining regions and genotyping by pulse-field gel electrophoresis and multilocus sequence typing of the 25 ciprofloxacin-resistant isolates available and all 5 nalidixic acid-resistant isolates were performed. In the NTHi isolates studied, two mutations producing changes in two GyrA residues (Ser84, Asp88) and/or two ParC residues (Ser84, Glu88) were associated with increased fluoroquinolone MICs. Strains with one or two mutations (n = 15) had ciprofloxacin and levofloxacin MICs of 0.12 to 2 μg/ml, while those with three or more mutations (n = 15) had MICs of 4 to 16 μg/ml. Long persistence of fluoroquinolone-resistant strains was observed in three chronic obstructive pulmonary disease patients. High genetic diversity was observed among fluoroquinolone-resistant NTHi isolates. Although fluoroquinolones are commonly used to treat respiratory infections, the proportion of resistant NTHi isolates remains low. The nalidixic acid disk test is useful for detecting the first changes in GyrA or in GyrA plus ParC among fluoroquinolone-susceptible strains that are at a potential risk for the development of resistance under selective pressure by fluoroquinolone treatment.
INTRODUCTION
Haemophilus influenzae is a human-restricted pathogen that forms part of the normal nasopharyngeal microbiota. It is classified either as encapsulated or as nontypeable H. influenzae (NTHi), depending on the presence of a polysaccharide capsule. Before the introduction of the conjugate vaccine against H. influenzae type b (Hib), this serotype was the most common cause of meningoencephalitis in young children. Since the introduction of the Hib vaccine, strain replacement has been observed and NTHi has become the predominant species among both invasive and noninvasive diseases such as otitis media, sinusitis, conjunctivitis, chronic bronchitis, and pneumonia (1–4). Fluoroquinolones (FQs) are frequently used as antimicrobial therapy in respiratory tract infections in adults and have shown good activity against respiratory pathogens such as H. influenzae, Streptococcus pneumoniae, Pseudomonas aeruginosa, and Moraxella catarrhalis (5, 6). Since their first description in 1993, FQ-resistant H. influenzae isolates have been detected all over the world (7–16). Although resistance in this bacterial pathogen remains low (8, 17), treatment failure with ofloxacin or levofloxacin (LVX) has already been described (15, 18). FQ resistance in H. influenzae is due mainly to chromosomal point mutations in the quinolone resistance-determining regions (QRDRs) of the genes encoding DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) (19). As in other Gram-negative bacteria, DNA gyrase is a primary target and topoisomerase IV is a secondary target for FQs. Mutations in the H. influenzae QRDRs have been shown to occur in a stepwise manner: a first mutation in gyrA produces reduced susceptibility to quinolones, but MICs remain in the susceptible range according to currently established breakpoints (20, 21). Strains susceptible to ciprofloxacin (CIP) or LVX could harbor first alterations in the QRDRs (22). Corkill et al. found that nalidixic acid (NAL) was a good indicator of reduced CIP susceptibility (23), and it has been proposed as a useful indicator for testing of low- and high-level quinolone resistance (24). Double mutations in both FQ targets generate a resistant phenotype that is detectable by using the current Clinical and Laboratory Standards Institute (CLSI) and European Society of Clinical Microbiology and Infectious Diseases (EUCAST) breakpoint interpretations (20, 21).
The aims of this study were (i) to analyze the genotypes of FQ-resistant NTHi clinical isolates collected in our hospital over a 14-year period (2000 to 2013), (ii) to detect CIP-susceptible isolates with NAL resistance that could harbor a first mutation in their QRDRs, and (iii) to characterize the mechanisms of resistance to FQs in clinical isolates.
MATERIALS AND METHODS
Hospital settings and bacterial strains.
This laboratory-based study was carried out at the Hospital Universitari de Bellvitge (HUB), a tertiary-care center for adult patients located in Barcelona, Spain. CIP susceptibility data on all of the H. influenzae isolates collected from patients who attended the HUB during a 14-year period (2000 to 2013) were recorded as part of the normal laboratory routine activity. These data were analyzed to determine the proportion of CIP-resistant isolates over the period studied. CIP susceptibility was determined by disk diffusion on Haemophilus test medium (HTM) plates and was interpreted by following CLSI criteria (20, 25). CIP-resistant isolates were stored at −80°C. Isolates were grown on chocolate agar plates and incubated at 37°C with 5% CO2. Informed consent was not required, as this process was part of the normal microbiological routine; patient confidentiality was protected in all cases. To detect isolates harboring first mutations in the QRDRs, isolates collected from 2010 to 2013 that showed susceptibility to CIP and had an inhibitory zone diameter of 21 to 28 mm were screened with NAL disks (30 μg). NAL screening was done by disk diffusion on HTM plates and interpreted according to EUCAST criteria, considering any isolate with an inhibitory zone diameter of <23 mm resistant, since CLSI has not defined breakpoints for NAL (21).
Identification.
H. influenzae was identified by conventional methodology (26). Isolate identification was confirmed by mass spectrometry with a matrix-assisted laser desorption ionization–time of flight Biotyper, version 3.0 (Bruker). Differentiation between H. influenzae and H. haemolyticus was performed by amplification of the fucK, iga, and lgtC genes (27); an isolate was considered H. influenzae when positive amplification of the genes tested was detected.
Serotyping, susceptibility testing, and sequencing of QRDRs.
Capsular serotypes were determined by PCR with the primers and under the conditions previously described (28). MICs were determined by the microdilution method with HTM and commercial panels (STRHAE2; Sensititre, West Sussex, England) and interpreted by following CLSI guidelines (20, 25). H. influenzae ATCC 49247 was used as a susceptible control strain. β-Lactamase activity was screened for by the chromogenic cephalosporin method (nitrocefin disks; BD, Madrid, Spain). QRDRs were amplified with specific oligonucleotide pairs as described previously (19, 29). PCR fragments that included gyrA nucleotides 137 to 546, parC nucleotides 129 to 547, gyrB nucleotides 1094 to 1539, and parE nucleotides 1002 to 1473 were purified with a GeneJET PCR purification kit (Thermo Scientific) and sequenced on both strands with the same oligonucleotides used for the PCRs and an Applied Biosystems 3730 XL DNA analyzer.
Molecular typing.
For molecular typing, genomic DNA was digested with SmaI and the fragments were separated by pulsed-field gel electrophoresis (PFGE) as reported previously (30). PFGE band patterns were analyzed with the Fingerprinting II Software 3.0 (Bio-Rad). The similarity of the PFGE banding patterns was estimated with the Dice coefficient. Isolates with ≥80% relatedness were considered highly genetically related. For multilocus sequence typing (MLST), DNA sequencing of internal fragments of seven housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA) was performed as previously described (31). Allele numbers and sequence types (STs) were assigned by using the H. influenzae MLST website (http://haemophilus.mlst.net). The STs were analyzed with e-BURST v3 in order to define the clonal relationship between the isolates.
RESULTS
Patients and CIP-resistant NTHi isolates.
Twenty-eight (0.39%) CIP-nonsusceptible isolates (CLSI criteria) were detected among 7,267 H. influenzae isolates collected in our Laboratory from 2000 to 2013. The proportion of CIP resistance over the period studied was low and remained stable over time at 0.58% from 2000 to 2004, 0.26% from 2005 to 2009, and 0.36% from 2010 to 2013.
Unfortunately, only 25 out of 28 isolates were available for molecular analysis. All of these 25 were NTHi isolates collected from 19 patients (16 [84%] males) with different episodes of respiratory disease. The mean age of these patients was 72.7 (range, 52 to 88) years. The main underlying diseases were chronic obstructive pulmonary disease (COPD) (n = 10, 52.7%) and bronchiectasis (n = 2, 10.5%), whereas no underlying disease was reported for 7 patients (36.8%).
NAL disk screening of isolates with CIP susceptibility.
Among 2,201 isolates collected from 2010 to 2013, 7 (0.32%) had a CIP inhibitory zone dimater of 21 to 28 mm. Five of these were resistant to NAL (inhibitory zone diameter of <23 mm). All of them were NTHi isolates from sputum samples of five patients (three females and two males) with a mean age of 69.4 years suffering from lung cancer, ischemic heart disease, bronchial asthma, COPD, and bronchiectasis.
Genotyping.
Thirty NTHi isolates (25 CIP resistant and 5 CIP susceptible but NAL resistant) were grouped into 15 different PFGE patterns (Table 1). Nine genotypes were unique, and the remaining 21 isolates were grouped into six small clusters. MLST results showed 16 different STs, 8 previously described in the MLST database and 8 described as new. Eleven STs were unique, and six clusters were detected (Table 1). The clusters were grouped as follows: cluster 1, isolates 16.1, 18.1, and 24.1, PFGE pattern B and ST 1281; cluster 2, isolates 3.1 and 7.1, PFGE pattern D and ST New4; cluster 3, isolates 15.1 and 23.1, PFGE pattern F and STs 159 and 485, a double-locus variant; cluster 4, isolates 20.1, 20.2, and 22.1, PFGE pattern A and ST New3; cluster 5, isolates 2.1 to 2.5 and 11.1, PFGE pattern C and ST 519; cluster 6, isolates 4.1, 5.1, 10.1, 10.2, and 14.1, PFGE pattern E and ST New5. No relationship between the patients in each cluster could be demonstrated.
TABLE 1.
Characteristics of 24 patients and molecular characterization of NTHi clinical isolatesa
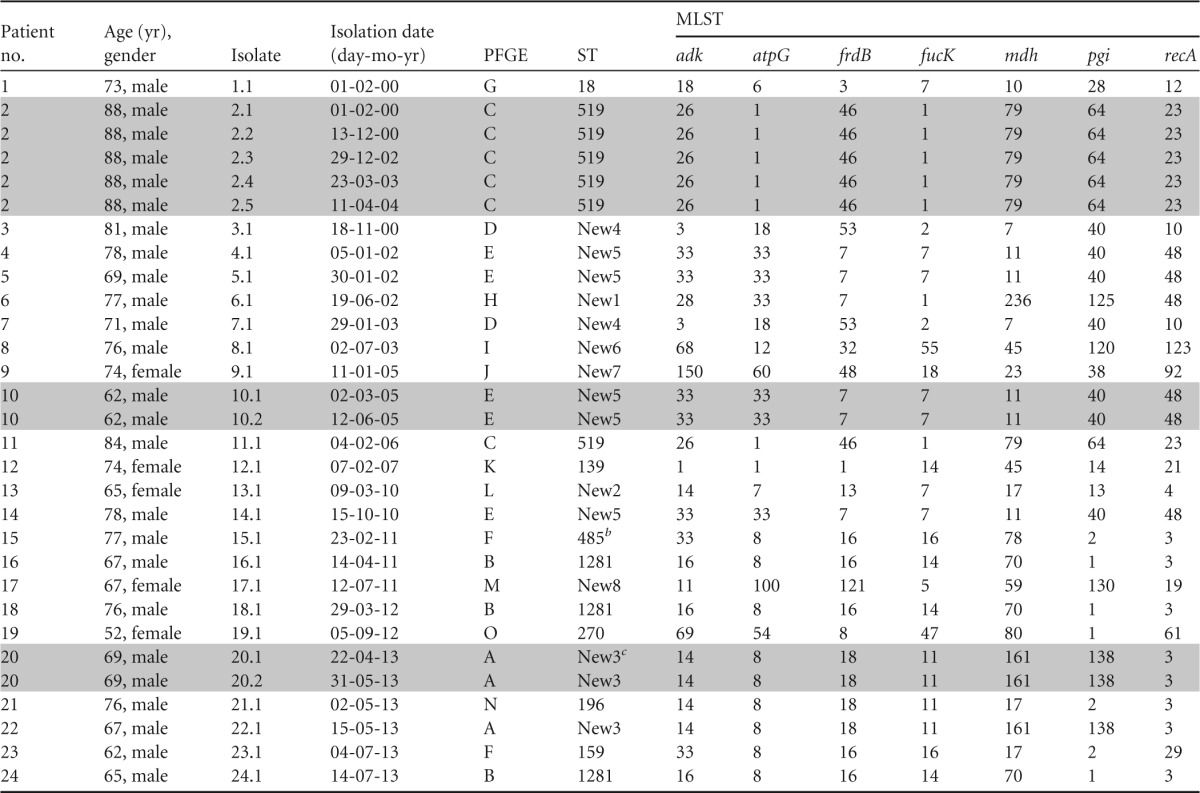
Gray-shaded lines indicate patients with persistent isolates.
eBURST analysis showed that ST 485 is a double-locus variant of ST 159.
ST New3 is a double-locus variant of ST 196 (eBURST analysis).
Three COPD patients were persistently infected with a particular NTHi strain (ST 519, New 3, or New 5) with a median persistence time of 17.6 (range, 1.33 to 48) months (Table 1).
Antimicrobial susceptibility.
NTHi isolates resistant to CIP (n = 25) were also resistant to other antimicrobial agents. The highest proportion of resistance was to cotrimoxazole (56%), followed by resistance to azithromycin (20%). Regarding β-lactams, 4% were ampicillin resistant because of β-lactamase production and 4% were β-lactamase-negative ampicillin-resistant (BLNAR) isolates with intermediate susceptibility to ampicillin and cefuroxime. All isolates were fully susceptible to cefotaxime, imipenem, tetracycline, and chloramphenicol. As for FQs, 22 isolates had CIP resistance (MIC, >1 μg/ml). The remaining three isolates had a CIP MIC of 1 μg/ml, which is considered susceptible according to the CLSI breakpoints, although all of them had a CIP disk diffusion inhibitory zone diameter inside the resistance range and had mutations in their QRDRs. Fifteen (60%) isolates were resistant to LVX (MIC, >2 μg/ml).
Seven CIP-resistant isolates with a CIP MIC of 2 μg/ml were considered low-level CIP resistant whereas 15 isolates were defined as high-level CIP resistant (MICs, 8 to 16 μg/ml). Although low-level CIP-resistant isolates were susceptible to LVX according to CLSI criteria, these isolates had gyrA and parC mutations that would favor the development of high-level resistance. All of the high-level CIP-resistant isolates showed cross-resistance to LVX (Table 2).
TABLE 2.
Mutations in the QRDR of and CIP and LVX MICs for 30 NTHi clinical isolates resistant to NAL
| Isolate(s)a | Disk diffusion inhibitory zone diam (mm) |
MIC (μg/ml) |
Mutation(s) in QRDRb |
||||
|---|---|---|---|---|---|---|---|
| NAL | CIP | CIP | LVX | GyrA | ParC | ParE | |
| 13.1 | 12 | 28 | 0.12 | 0.12 | Ser84Leu | — | — |
| 17.1 | 10 | 26 | 0.5 | 0.5 | Ser84Leu | — | — |
| 15.1, 23.1 | 6 | 24 | 0.5 | 0.5 | Ser84Leu | Ser84Ile | — |
| 1.1 | 9 | 20 | 0.5 | 0.5 | Ser84Tyr | Glu88Lys | — |
| 21.1 | 6 | 22 | 1 | 0.5 | Ser84Leu | Ser84Ile | — |
| 6.1 | 10 | 20 | 1 | 0.5 | Ser84Leu | Ser84Ile | — |
| 3.1 | 6 | 20 | 1 | 1 | Ser84Leu | Glu88Lys | — |
| 12.1 | 6 | 20 | 2 | 1 | Ser84Leu | Glu88Lys | — |
| 5.1 | 10 | 16 | 2 | 1 | Asp88Tyr | Glu88Lys | — |
| 2.1–2.5 | 6 | 20 | 2 | 1 | Ser84Tyr, Asp88Tyr | Asp83Gly, Ser84Ala | — |
| 11.1 | 6 | 15 | 8 | 4 | Ser84Tyr, Asp88Tyr | Ser84Asn | — |
| 7.1 | 6 | 6 | 8 | 4 | Ser84Leu, Asp88Tyr | Glu88Lys | — |
| 10.1, 10.2, 14.1 | 6 | 6 | 16 | 16 | Ser84Leu, Asp88Tyr | Glu88Lys | — |
| 8.1 | 6 | 6 | 16 | 16 | Ser84Leu, Asp88Asn | Gly82Asp | — |
| 19.1 | 6 | 6 | 16 | 16 | Ser84Leu, Asp88Asn | Ser84Arg | — |
| 16.1 | 6 | 6 | 16 | 8 | Ser84Tyr, Asp88Gly | Ser84Ile | Asp420Asn |
| 18.1 | 6 | 6 | 16 | 16 | Ser84Tyr, Asp88Gly | Ser84Ile | Asp420Asn |
| 24.1 | 6 | 6 | 16 | 16 | Ser84Leu, Asp88Gly | Ser84Ile | Asp420Asn |
| 20.1, 20.2, 22.1 | 6 | 6 | 16 | 16 | Ser84Leu, Asp88Asn | Ser84Ile | Asp420Asn |
| 4.1 | 6 | 6 | 16 | 16 | Ser84Leu, Asp88Tyr | Gly82Cys, Glu88Lys | — |
| 9.1 | 6 | 6 | 16 | 16 | Ser84Tyr, Asp88Tyr | Ser84Ile, Glu88Lys | — |
Isolates are numbered accord to their origins of isolation as defined in Table 1.
Changes at positions classically involved in resistance are shown in bold. Additional amino acid changes, found also in susceptible isolates, were ParC S133A (six isolates), ParC N138S (eight isolates), ParE R368H and S458L (one isolate), and GyrB A400V (three isolates). No amino acid changes have been detected in GyrB. —, no changes.
Of the five isolates susceptible to CIP but resistant to NAL, four were resistant to cotrimoxazole and one was considered intermediately resistant to ampicillin.
FQ resistance and amino acid substitutions in the QRDRs.
Determination of susceptibility to FQ antimicrobials (by microdilution) and characterization (by DNA sequencing) of the QRDRs of gyrA, gyrB, parC, and parE were performed. All of the isolates studied had nonsynonymous polymorphisms leading to amino acid substitutions in their QRDRs (Table 2), including the isolates resistant to CIP (n = 25) and those that were NAL resistant but CIP susceptible (n = 5). Two isolates presenting the single GyrA Ser84Leu change with CIP and LVX MICs of 0.12 to 0.5 μg/ml were considered susceptible. Thirteen isolates had two changes in equivalent positions of GyrA (Ser84 or Asp88) and ParC (Ser84 or Glu88) and had CIP MICs of 0.5 to 2 μg/ml and LVX MICs of 0.5 to 1 μg/ml. We also assessed five isolates from the same patient (isolates 2.1 to 2.5) with identical QRDR mutations which, despite having two GyrA (Ser84Tyr, Asp88Tyr) and two ParC (Asp83Gly, Ser84Ala) changes, had a CIP MIC of 2 μg/ml. As far as we know, the ParC Asp83Gly and Ser84Ala changes have not been implicated in resistance in H. influenzae or other bacteria and are probably polymorphisms. The mutations in GyrA (Ser84Tyr and Asp88Tyr) may have been the sole causes of the CIP and LVX MICs of the five isolates. Finally, 15 isolates harbored three (n = 13) or four (n = 2) amino acid substitutions and were fully resistant to CIP (Table 2). Four isolates with high-level resistance showed the Asp420Asn change in ParE. However, the involvement of this change in FQ resistance has not been demonstrated by genetic transformation.
DISCUSSION
Since its first description in 1993, FQ-resistant H. influenzae has been isolated mainly from elderly patients with chronic lung diseases who received frequent antimicrobial treatments, including quinolones (7, 15, 16, 32). Resistance to FQs in H. influenzae remains very low worldwide. A global study (SENTRY) performed by American and European institutions found that 0.15% of H. influenzae isolates were resistant to FQs (8). In Hong Kong, Japan, and South Korea, H. influenzae-resistant isolates were first described in 2009, when Hirakata et al. reported 0.1% CIP resistance in Japan (33, 34); by 2014, the level had increased to 1.3% (29). A recent surveillance study published in Taiwan showed a major increase in LVX resistance from 2% in 2004 to 24.3% in 2010 (35). In the United States in 2006, the percentage of FQ resistance was 0.1% (36), similar to the percentage found in Spain in 2011 (0.2%) (17). In our study, we found a low percentage of FQ-resistant H. influenzae isolates (0.39%), in accordance with the data published in other parts of the world but higher than previous reports in Spain (17). However, the proportion of H. influenzae isolates resistant to CIP may depend on the criteria used (CLSI or EUCAST) because of the difference in the breakpoints. In the disk diffusion method, the current EUCAST breakpoint for CIP susceptibility is ≥26 mm, whereas the CLSI breakpoint is ≥21 mm. This difference is also observed when using susceptibility microdilution breakpoints (CIP MICs of ≤1 μg/ml for CLSI and ≤0.5 μg/ml for EUCAST (20, 21). The use of current CLSI criteria underrecognizes a proportion of the low-level CIP-resistant strains with first-step mutations in gyrA and parC (22, 37, 38). In the present study, we identified five isolates with CIP MICs of 0.12 to 0.5 μg/ml and three strains with a MIC of 1 μg/ml that had at least one amino acid alteration in a position of the FQ targets involved in resistance. According to the current EUCAST resistance breakpoints for disk diffusion, 28 of the 30 isolates studied could be considered resistant. In order to detect nonsusceptible CIP strains with first-step mutations, the EUCAST guidelines have proposed screening with NAL as an indicator of resistance (22–24). In our study, five (0.23%) isolates collected from 2010 to 2013 with CIP inhibitory zone diameters of 21 to 28 mm were resistant to NAL and had changes in GyrA and/or ParC. Other authors have recommended general screening with NAL in order to identify these strains and to avoid therapeutic failures (22–24). Although FQs present good activity against H. influenzae, their use in respiratory tract infections merits special attention. To the best of our knowledge, two reports of FQ treatment failure have been published to date (15, 18). In 1999, Vila et al. reported a case of FQ resistance after treatment with ofloxacin in a patient with recurrent respiratory infections (15), and in 2003, Bastida et al. reported a case of LVX treatment failure in a patient with community-acquired pneumonia who had previously been treated with LVX and moxifloxacin (18).
Some specific mutations involved in FQ resistance in H. influenzae were originally described in 1996 (19). Subsequent studies have confirmed the mutations described by Georgiou et al. and described new mutations involved in resistance (15, 16, 18, 19, 22, 24, 29, 33, 37, 39). FQ resistance is acquired gradually with increasing numbers of mutations. Strains harboring one or two mutations in gyrA and parC have low-level resistance to FQs, while those with three or more mutations in gyrA, parC, and parE show high-level resistance (19, 40). In our study, strains with a single change in GyrA or one change in GyrA plus one in ParC had CIP MICs of 0.12 to 2 μg/ml, while those with three or four mutations (in GyrA, ParC, and ParE) had higher MICs (8 to 16 μg/ml). In the present study, the most common changes in GyrA were Ser84 to Leu or Tyr and Asp88 to Tyr, Asn, or Gly, which have been reported to contribute to resistance in H. influenzae (15, 16, 18, 19, 22, 29, 33, 37, 39). In ParC, the most common changes were Ser84Ile and Glu88Lys, which have been widely described in the literature (16, 18, 19, 22, 29, 33, 39), and Ser84Arg, a change also reported by other authors as an alteration involved in resistance (15, 16, 22, 24, 29, 33, 39). In addition, our study presents two strains harboring two new previously unidentified ParC changes, Ser84Asn and Ser84Ala. The strain carrying Ser84Asn had two additional changes in GyrA and had a CIP MIC of 8 μg/ml, suggesting its involvement in FQ resistance. In contrast, the strain carrying the ParC Ser84Ala change had a CIP MIC of 2 μg/ml, suggesting that this change would not be involved in resistance. Besides these changes at residues 84 and 88 of ParC, two strains had previously described changes in Gly82 (to Asp or Cys) (16, 22, 29, 39) and an Asp83Gly change that was not linked to any increase in the CIP MIC. This change (Asp83Gly) was already described by Pérez-Vázquez et al., but its involvement in quinolone resistance has not yet been established (22). Only one change was detected in ParE, Asp420Asn, which has been previously reported (11, 29, 33), although its role in resistance has not been proved by genetic transformation.
In spite of a clonal spread previously described in long-term care facilities (11, 12, 41), our study reports a high genetic diversity among FQ-resistant NTHi isolates. Although we found six small clusters, the majority of the strains were isolated from patients who had no relationship to each other. In a recent publication from Taiwan, regional clonal emergence was found in different areas of the country (35). None of the STs published in Taiwan were found in our study, suggesting that the evolution of FQ-resistant strains is regional (35).
A relevant finding in our study was the persistence of genotypically identical quinolone-resistant isolates in COPD patients. During the period studied, these three patients had more than one isolate that had the same PFGE pattern and ST. It is well known that COPD patients have several impairments in innate lung defenses, facilitating microorganism persistence (42). Groeneveld et al. found COPD patients persistently infected with the same H. influenzae strain for up to 23 months, and their antibiotic treatment was not effective in eradicating the strains (43). In addition, Sethi et al. reported that a quarter of the acute exacerbations of COPD were caused by a persistent strain when bacterial pathogens were present in sputum (44). Long persistence of NTHi was also described in patients with cystic fibrosis (32).
In conclusion, although FQs are commonly used to treat respiratory infections, the proportion of FQ-resistant NTHi isolates during the period studied remained low. Long persistence of FQ-resistant isolates was identified in three COPD patients. The NAL test is recommended to detect FQ-susceptible strains with first mutations in the QRDRs that may acquire full resistance under selective pressure with FQ therapy and cause treatment failure.
ACKNOWLEDGMENTS
This work was supported by grants from the Fondo de Investigaciones Sanitarias de la Seguridad Social (PI 0901904) and the Plan Nacional de I+D+I of Ministerio de Ciencia e Innovación (BIO2011-25343) and by CIBER de Enfermedades Respiratorias, CIBERES; (CB06/06/0037), run by the Instituto de Salud Carlos III, Madrid, Spain. C.P. was supported by FPU grant AP2010-3202 (Formación de Profesorado Universitario, Ministerio de Educación, Spain). S.M. was supported by Sara Borrell Postdoctoral contract CD10/00298 from the Instituto de Salud Carlos III, Madrid, Spain.
We have no conflict of interest to declare.
REFERENCES
- 1.Agrawal A, Murphy TF. 2011. Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J Clin Microbiol 49:3728–3732. doi: 10.1128/JCM.05476-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eldika N, Sethi S. 2006. Role of nontypeable Haemophilus influenzae in exacerbations and progression of chronic obstructive pulmonary disease. Curr Opin Pulm Med 12:118–124. doi: 10.1097/01.mcp.0000208451.50231.8f. [DOI] [PubMed] [Google Scholar]
- 3.Erwin AL, Smith AL. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol 15:355–362. doi: 10.1016/j.tim.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J 28:43–48. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- 5.Brueggemann AB, Kugler KC, Doern GV. 1997. In vitro activity of BAY 12-8039, a novel 8-methoxyquinolone, compared to activities of six fluoroquinolones against Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Antimicrob Agents Chemother 41:1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lode H, Allewelt M. 2002. Role of newer fluoroquinolones in lower respiratory tract infections. J Antimicrob. Chemother 49:709–712. doi: 10.1093/jac/dkf024. [DOI] [PubMed] [Google Scholar]
- 7.Barriere SL, Hindler JA. 1993. Ciprofloxacin-resistant Haemophilus influenzae infection in a patient with chronic lung disease. Ann Pharmacother 27:309–310. [DOI] [PubMed] [Google Scholar]
- 8.Biedenbach DJ, Jones RN. 2003. Five-year analysis of Haemophilus influenzae isolates with reduced susceptibility to fluoroquinolones: prevalence results from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 46:55–61. doi: 10.1016/S0732-8893(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 9.Bootsma HJ, Troelstra A, van Veen-Rutgers A, Mooi FR, de Neeling AJ, Overbeek BP. 1997. Isolation and characterization of a ciprofloxacin-resistant isolate of Haemophilus influenzae from The Netherlands. J Antimicrob. Chemother 39:292–293. doi: 10.1093/jac/39.2.292. [DOI] [PubMed] [Google Scholar]
- 10.Elliott E, Oosthuizen D, Johnson MM, Piddock LJ. 2003. Fluoroquinolone resistance in Haemophilus influenzae. J Antimicrob Chemother 52:734–735. doi: 10.1093/jac/dkg420. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Mariano N, Rahal JJ, Urban CM, Drlica K. 2004. Quinolone-resistant Haemophilus influenzae in a long-term-care facility: nucleotide sequence characterization of alterations in the genes encoding DNA gyrase and DNA topoisomerase IV. Antimicrob. Agents Chemother 48:3570–3572. doi: 10.1128/AAC.48.9.3570-3572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazir J, Urban C, Mariano N, Burns J, Tommasulo B, Rosenberg C, Segal-Maurer S, Rahal JJ. 2004. Quinolone-resistant Haemophilus influenzae in a long-term care facility: clinical and molecular epidemiology. Clin Infect Dis 38:1564–1569. doi: 10.1086/420820. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Vázquez M, Roman F, Garcia-Cobos S, Campos J. 2007. Fluoroquinolone resistance in Haemophilus influenzae is associated with hypermutability. Antimicrob Agents Chemother 51:1566–1569. doi: 10.1128/AAC.01437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Martinez JM, Lopez L, Garcia I, Pascual A. 2006. Characterization of a clinical isolate of Haemophilus influenzae with a high level of fluoroquinolone resistance. J Antimicrob. Chemother 57:577–578. doi: 10.1093/jac/dki488. [DOI] [PubMed] [Google Scholar]
- 15.Vila J, Ruiz J, Sanchez F, Navarro F, Mirelis B, de Anta MT, Prats G. 1999. Increase in quinolone resistance in a Haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob Agents Chemother 43:161–162. doi: 10.1093/jac/43.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokota S, Ohkoshi Y, Sato K, Fujii N. 2008. Emergence of fluoroquinolone-resistant Haemophilus influenzae strains among elderly patients but not among children. J Clin Microbiol 46:361–365. doi: 10.1128/JCM.01561-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Trallero E, Martin-Herrero JE, Mazon A, Garcia-Delafuente C, Robles P, Iriarte V, Dal-Re R, Garcia-de-Lomas J. 2010. Antimicrobial resistance among respiratory pathogens in Spain: latest data and changes over 11 years (1996-1997 to 2006-2007). Antimicrob Agents Chemother 54:2953–2959. doi: 10.1128/AAC.01548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastida T, Pérez-Vázquez M, Campos J, Cortes-Lletget MC, Roman F, Tubau F, de la Campa AG, Alonso-Tarres C. 2003. Levofloxacin treatment failure in Haemophilus influenzae pneumonia. Emerg Infect Dis 9:1475–1478. doi: 10.3201/eid0911.030176. [DOI] [PubMed] [Google Scholar]
- 19.Georgiou M, Muñoz R, Roman F, Canton R, Gomez-Lus R, Campos J, de la Campa AG. 1996. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of GyrA and ParC. Antimicrob Agents Chemother 40:1741–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. CLSI M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.EUCAST. 2014. Clinical breakpoints. European Society of Clinical Microbiology and Infectious Diseases (EUCAST) Basel, Switzerland: http://www.eucast.org/clinical_breakpoints. [Google Scholar]
- 22.Pérez-Vázquez M, Roman F, Aracil B, Canton R, Campos J. 2004. Laboratory detection of Haemophilus influenzae with decreased susceptibility to nalidixic acid, ciprofloxacin, levofloxacin, and moxifloxacin due to GyrA and ParC mutations. J Clin Microbiol 42:1185–1191. doi: 10.1128/JCM.42.3.1185-1191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corkill JE, Percival A, McDonald P, Bamber AI. 1994. Detection of quinolone resistance in Haemophilus spp. J Antimicrob Chemother 34:841–844. doi: 10.1093/jac/34.5.841. [DOI] [PubMed] [Google Scholar]
- 24.Brenwald NP, Andrews JM, Jevons G, Wise R. 2003. Detection of ciprofloxacin resistance in Haemophilus influenzae using nalidixic acid and BSAC methodology. J Antimicrob. Chemother 51:1311–1312. doi: 10.1093/jac/dkg200. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard CLSI M7-A9, 8th. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (ed). 2003. Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington DC. [Google Scholar]
- 27.Binks MJ, Temple B, Kirkham LA, Wiertsema SP, Dunne EM, Richmond PC, Marsh RL, Leach AJ, Smith-Vaughan HC. 2012. Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS One 7:e34083. doi: 10.1371/journal.pone.0034083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. 1994. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol 32:2382–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoji H, Shirakura T, Fukuchi K, Takuma T, Hanaki H, Tanaka K, Niki Y. 2014. A molecular analysis of quinolone-resistant Haemophilus influenzae: validation of the mutations in quinolone resistance-determining regions. J Infect Chemother 20:250–255. doi: 10.1016/j.jiac.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Puig C, Calatayud L, Marti S, Tubau F, Garcia-Vidal C, Carratala J, Linares J, Ardanuy C. 2013. Molecular epidemiology of nontypeable Haemophilus influenzae causing community-acquired pneumonia in adults. PLoS One 8:e82515. doi: 10.1371/journal.pone.0082515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campos J, Roman F, Georgiou M, Garcia C, Gomez-Lus R, Canton R, Escobar H, Baquero F. 1996. Long-term persistence of ciprofloxacin-resistant Haemophilus influenzae in patients with cystic fibrosis. J Infect Dis 174:1345–1347. doi: 10.1093/infdis/174.6.1345. [DOI] [PubMed] [Google Scholar]
- 33.Hirakata Y, Ohmori K, Mikuriya M, Saika T, Matsuzaki K, Hasegawa M, Hatta M, Yamamoto N, Kunishima H, Yano H, Kitagawa M, Arai K, Kawakami K, Kobayashi I, Jones RN, Kohno S, Yamaguchi K, Kaku M. 2009. Antimicrobial activities of piperacillin-tazobactam against Haemophilus influenzae isolates, including beta-lactamase-negative ampicillin-resistant and beta-lactamase-positive amoxicillin-clavulanate-resistant isolates, and mutations in their quinolone resistance-determining regions. Antimicrob Agents Chemother 53:4225–4230. doi: 10.1128/AAC.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue M, Lee NY, Hong SW, Lee K, Felmingham D. 2004. PROTEKT 1999-2000: a multicentre study of the antibiotic susceptibility of respiratory tract pathogens in Hong Kong, Japan and South Korea. Int J Antimicrob Agents 23:44–51. doi: 10.1016/j.ijantimicag.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Kuo SC, Chen PC, Shiau YR, Wang HY, Lai JF, Huang W, Lauderdale TL. 2014. Levofloxacin-resistant Haemophilus influenzae, Taiwan, 2004–2010. Emerg Infect Dis 20:1386–1390. doi: 10.3201/eid2008.140341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Critchley IA, Brown SD, Traczewski MM, Tillotson GS, Janjic N. 2007. National and regional assessment of antimicrobial resistance among community-acquired respiratory tract pathogens identified in a 2005-2006 U.S. Faropenem surveillance study. Antimicrob Agents Chemother 51:4382–4389. doi: 10.1128/AAC.00971-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho PL, Chow KH, Mak GC, Tsang KW, Lau YL, Ho AY, Lai EL, Chiu SS. 2004. Decreased levofloxacin susceptibility in Haemophilus influenzae in children, Hong Kong. Emerg Infect Dis 10:1960–1962. doi: 10.3201/eid1011.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim IS, Lee NY, Kim S, Ki CS, Kim SH. 2011. Reduced levofloxacin susceptibility in clinical respiratory isolates of Haemophilus influenzae is not yet associated with mutations in the DNA gyrase and topoisomerase II genes in Korea. Yonsei Med J 52:188–191. doi: 10.3349/ymj.2011.52.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura S, Yanagihara K, Morinaga Y, Izumikawa K, Seki M, Kakeya H, Yamamoto Y, Kamihira S, Kohno S. 2009. Melting curve analysis for rapid detection of topoisomerase gene mutations in Haemophilus influenzae. J Clin Microbiol 47:781–784. doi: 10.1128/JCM.01645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Mariano N, Rahal JJ, Urban CM, Drlica K. 2004. Quinolone-resistant Haemophilus influenzae: determination of mutant selection window for ciprofloxacin, garenoxacin, levofloxacin, and moxifloxacin. Antimicrob Agents Chemother 48:4460–4462. doi: 10.1128/AAC.48.11.4460-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CM, Lauderdale TL, Lee HC, Lee NY, Wu CJ, Chen PL, Lee CC, Chen PC, Ko WC. 2010. Colonisation of fluoroquinolone-resistant Haemophilus influenzae among nursing home residents in southern Taiwan. J Hosp Infect 75:304–308. doi: 10.1016/j.jhin.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 42.Sethi S, Murphy TF. 2008. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med 359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 43.Groeneveld K, van Eijk ALPP, Visschers G, Jansen HM, Zanen HC. 1990. Endogenous and exogenous reinfections by Haemophilus influenzae in patients with chronic obstructive pulmonary disease: the effect of antibiotic treatment on persistence. J Infect Dis 161:512–517. doi: 10.1093/infdis/161.3.512. [DOI] [PubMed] [Google Scholar]
- 44.Sethi S, Evans N, Grant BJ, Murphy TF. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 347:465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]


