Abstract
Therapeutic nonequivalence of generic antibiotics may lead to treatment failure and enrichment of resistance. However, there has been no demonstration that an equivalent generic displays the same resistance selection profile as the innovator drug. We aimed to test this hypothesis with five generic versions of ciprofloxacin by assessing their pharmaceutical equivalence with microbiological assays and their efficacy against Pseudomonas aeruginosa PAO1 in the neutropenic murine thigh infection model. One equivalent generic was selected for analysis by high-pressure liquid chromatography–tandem mass spectrometry (LC-MS/MS), to confirm chemical identity, and resistance selection experiments in a hollow-fiber (HF) system simulating two clinical dosing regimens. Total and resistant populations were measured, and the MICs of the resistant cells with and without an efflux pump inhibitor were determined. LC-MS/MS found no differences between products, and the innovator and the generic selected resistance with the same magnitude and mechanism after 7 days of treatment in the HF system, supporting the fact that a generic with demonstrated equivalence in vivo is also equivalent regarding resistance selection.
INTRODUCTION
The interchangeability of innovator drugs and equivalent generic drugs has been promoted by the World Health Organization (WHO) and drug regulatory agencies on all continents. Equivalence implies the demonstration of good manufacturing practices, chemical identity, and bioequivalence. However, the latter requirement is waived for intravenously administered drugs, and no additional in vivo or clinical tests are mandatory (1).
For anti-infective agents, concerns have arisen after the demonstration of therapeutic failures of pharmaceutically equivalent products in a validated animal model of infection, especially with complex, difficult-to-purify antibiotics such as vancomycin (2) and gentamicin (3). Generic versions of drugs without manufacturing problems, such as metronidazole (4) and fluconazole (5), are more likely to be therapeutically equivalent under current regulations.
A consequence of the clinical use of nonequivalent products is treatment failure (6); even more worrisome, however, is the potential selection of resistance. For instance, we demonstrated that therapeutically nonequivalent (but bioequivalent) generic versions of vancomycin significantly enriched resistant subpopulations of Staphylococcus aureus after 12 days of exposure in the mouse thigh infection model; bacteria exposed to these generics grew at 1, 2, and 3 mg/liter vancomycin, while those exposed to the innovator product were eliminated (7). Although this evidence shows that nonequivalent generics of vancomycin select for resistant microorganisms in vivo, there has been no experimental demonstration that a therapeutically equivalent generic of any antimicrobial is as efficient as the innovator in preventing (or promoting) resistance. The importance of this demonstration is that such a product would not require further resistance testing, and its interchangeability would be ensured. Thus, we aimed to determine whether a pharmaceutically and therapeutically equivalent generic product of ciprofloxacin displays the same magnitude and mechanism of resistance as the innovator with Pseudomonas aeruginosa.
(Preliminary results of this work were presented at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy, 2012 [8].)
MATERIALS AND METHODS
Antimicrobials.
The study included the innovator ciprofloxacin (Cipro, lots BXC5SW1, BXFAHK1, and BXFNAV1, manufactured by Bayer) and five generics of ciprofloxacin lactate for intravenous use, marketed in Colombia by Blaskov Laboratories (Cifloblas, lot B1909, manufactured by Arbofarma), Chalver (Quinopron, lot L1508), Corpaul (lot 5CI090917-1), Ryan (lot CPF-150CH), and Vitalis (lot V110996, manufactured by Vitrofarma). All products were purchased in authorized drug stores and used before the expiration date.
Strains.
Pseudomonas aeruginosa PAO1 was used for susceptibility studies, in vivo studies, and hollow-fiber (HF) experiments. The reference strain for susceptibility testing was P. aeruginosa ATCC 27853. We employed Mueller-Hinton broth (MHB) and Mueller-Hinton agar (MHA) (Difco Laboratories) as culture media in all experiments.
In vitro activity.
The MIC and minimal bactericidal concentration (MBC) of each product were determined in duplicate by broth microdilution according to CLSI protocol M100-S23. The differences between products were assessed with the Kruskal-Wallis (KW) test (GraphPad Prism 6.05).
Pharmaceutical equivalence.
The concentration and potency of the products were determined with a validated microbiological assay (9), using Klebsiella pneumoniae ATCC 10031 as the seeding strain in antibiotic medium 3 (Difco). The polynomial equation describing the assay error of this method, calculated with Pmetrics, was standard deviation (SD) (in mg/liter) = −0.18 + 0.84x + 0.02x2 (10). To test equivalence, the inhibition zones (y axis, in mm) induced by at least five concentrations per product (x axis, in log10 mg/liter), ranging from 0.25 to 16 mg/liter, were fitted to a straight-line equation by the least-squares method, weighting the data by the inverse of the variance (1/SD2). As we described elsewhere (9), with this method the slope of the regression tests the potency of the drug and the intercept measures its concentration. Then, slopes and intercepts for innovator and generic drugs were compared by curve-fitting analysis (CFA) (GraphPad Prism 6.05). The same microbiological assay was used to quantify serum concentrations in mice after a single dose of ciprofloxacin.
Pharmacokinetics.
Twelve neutropenic and infected mice, weighing 25 g on average, received a single 16-mg/kg dose of innovator ciprofloxacin, administered subcutaneously. The animals were divided into 3 groups of 4 mice each. The first group was sampled by retro-orbital puncture at 5, 60, and 180 min, the second at 15, 90, and 240 min, and the third at 30, 120, and 300 min postdose. The samples were centrifuged to separate serum and frozen at −70°C. Ciprofloxacin concentrations were determined with the microbiological assay described above. The parametric population software SADAPT-TRAN (11) was used to fit the data to different models (1 or 2 compartments, linear, or Michaelis-Menten elimination). Selection of the best-performing model was based on the objective function (−2 log likelihood), the observed versus predicted plots, and the residual analysis. Between-subject variability was expressed as an apparent coefficient of variation, and the residual error included both additive (SDintercept) and proportional (SDslope) terms. The simulation module of the ADAPT 5 program (12) was then used to calculate the area under the concentration-time curve (AUC) of the different doses used in vivo.
Therapeutic equivalence in the neutropenic murine thigh infection model.
The University of Antioquia Animal Experimentation Ethics Committee reviewed and approved the experimental protocol. Female, murine-pathogen-free, Udea:ICR(CD-2) mice, weighing 23 to 27 g, were rendered neutropenic with two intraperitoneal doses of cyclophosphamide (150 and 100 mg/kg, injected 4 days and 1 day before infection, respectively) (13). A log-phase culture containing ∼7.0 log10 CFU/ml of P. aeruginosa PAO1 was inoculated in both thighs. Treatment began 2 h after infection, with a range of subcutaneous doses of the innovator and the Corpaul, Vitalis, Ryan, and Blaskov generics from minimal to maximal efficacy (3 to 128 mg/kg per day, divided into injections every 3 h), and lasted for 24 h. At the end of the treatment period, the mice were euthanized, their thighs were aseptically removed and homogenized, and the cells were diluted, plated on MHA, and incubated at 37°C for 18 h. Antimicrobial effects were calculated by subtracting the CFU/g in infected thighs from the values for untreated controls. Dose-effect data were analyzed by nonlinear regression fitting Hill's sigmoidal model to estimate the maximum effect (Emax), 50% effective dose (ED50), and slope (N) for each ciprofloxacin product (SigmaPlot 12.3), according to the equation effect = (−Emax × doseN)/(ED50N + doseN), and data were compared by CFA (GraphPad Prism 6.05). Nonlinear regressions were assessed by the adjusted coefficient of determination (adjusted R2), the standard error of estimate (Sy|x), and the fulfillment of normality and homoscedasticity (constant variance). We checked for the absence of multicolinearity by measuring the variance inflation factor (VIF). To test the impact of a smaller inoculum, an additional experiment was performed by infecting mice with ∼6 log10 CFU/ml of P. aeruginosa PAO1 and comparing the innovator and the Chalver generic. For pharmacokinetic (PK)/pharmacodynamic (PD) analysis, the exposures (in terms of AUC for the free fraction [fAUC]/MIC) required for stasis (bacteriostatic dose [BD]) and 3.0-log10 killing (3-log killing dose [3LKD]) were estimated using modified Hill's equations (3). In order to assess the reliability of the model, the repeatability of the results for the innovator was determined in two independent experiments performed on different days. In the event that all generics were pharmaceutically and therapeutically equivalent, one product was to be selected randomly (the first in alphabetical order) for chemical analysis and resistance selection experiments. This was eventually the case, and the Blaskov product was chosen for further testing.
Chemical analysis.
Six standard concentrations of the innovator, the internal standard (levofloxacin), and the selected equivalent generic were dissolved in sterile deionized water and analyzed by high-pressure liquid chromatography–tandem mass spectrometry (LC-MS/MS). The LC-MS/MS system consisted of an Agilent 1100 liquid chromatograph coupled to a mass spectrometer-electrospray ionization VL system. A Thermo Scientific Hypersil Gold analytical column (150 mm by 4.6 mm; particle diameter, 5 μm) was used as the stationary phase, with each ciprofloxacin product having its own column. The mobile phase consisted of 90:10 water-acetonitrile, at a flow rate of 0.5 ml/min. Each sample was run for 10 min. Data analysis was performed using the manufacturer's software. The scan mode was used to gather the mass spectra from m/z 150 to m/z 1,000, and the abundance of each mass was compared between the innovator and the generic, to assess chemical identity.
Resistance enrichment.
A hollow-fiber system was used to simulate the human 24-h fAUC attained with two clinical doses, i.e., 200 mg and 400 mg administered intravenously twice a day (24-h fAUC values of ∼11 and ∼23 mg · h/liter, respectively) for 7 days, against P. aeruginosa PAO1 (MIC = 0.125 mg/liter), comparing the innovator and the selected generic. The expected fAUC/MIC values were 88 and 184, respectively. The central compartment was filled with 238 ml of MHB, and then ∼8.0 log10 CFU/ml of P. aeruginosa PAO1 was inoculated into 10-ml cellulosic cartridges (FiberCell Systems). The flow was adjusted to 0.5 ml/min, for a half-life of 5.5 h. Ciprofloxacin concentrations in the central compartment were measured by LC-MS/MS, and the actual fAUC values were estimated with SADAPT-TRAN by integration (11). Total and resistant subpopulations were determined at daily intervals by plating samples from the cartridges on MHA without ciprofloxacin and with 2.5× MIC (0.3125 mg/liter). Resistance frequencies were calculated by dividing the resistant subpopulation by the total population, and the significance of the difference was assessed with the Mann-Whitney (MW) test (GraphPad Prism 6.05). Additionally, the ciprofloxacin MICs for 3 colonies randomly picked from the resistant population plates were measured at 24 and 72 h by agar dilution, in the absence or presence of the Mex-Opr inhibitor phenylalanine–arginine–β-naphthylamide (PAβN) (20 mg/liter; Sigma-Aldrich). A 4-fold reduction in the MIC value with the inhibitor was considered indicative of active efflux. The difference between the two products was assessed with the MW test.
RESULTS
In vitro activity.
There were no differences in the MIC or MBC values of the ciprofloxacin products against P. aeruginosa PAO1. For all products, the MIC duplicates were 0.125 mg/liter and the MBC ranged from 0.125 to 0.50 mg/liter (P = 0.36, KW test).
Pharmaceutical equivalence.
The straight-line regressions from microbiological assays overlapped, indicating that all data could be described by a single curve, with indistinguishable slopes (potency) and intercepts (concentration) for the innovator and generics (P > 0.15, CFA). The goodness of fit to the model for the global curve was indicated by an adjusted R2 of >0.9653 and a Sy|x of <3.79 mm.
Pharmacokinetics.
The one-compartment model with linear elimination and first-order absorption exhibited the best performance. The parameters clearance (CL), volume of distribution (V), and absorption rate constant (Ka) are presented in Table 1 with their respective between-subject variability and standard errors. Because all generics were pharmaceutically equivalent, the pharmacokinetics were determined only with the innovator, and its parameters were used for the PK/PD analysis of all products. The raw data, modeled concentrations, individual and population predicted versus observed plots, and comparison of 1-compartment and 2-compartment models are presented in Fig. S1, S2, and S3 and Table S1 in the supplemental material.
TABLE 1.
Population pharmacokinetics of ciprofloxacin in mice
| Parameter | Mean (CV [%]; SE [%])a |
|---|---|
| CL (liter/h) | 0.11 (10.2; 3.31) |
| V (liters) | 0.06 (19.6; 9.83) |
| Ka (h−1) | 1.88 (20.0; 9.81) |
| SDintercept | 0.003 |
| SDslope | 0.059 |
For the raw data, model fit, and diagnostic data, see the supplemental material. CV, coefficient of variation; SE, standard error.
Therapeutic equivalence.
The mean growth of P. aeruginosa PAO1 in untreated controls was 4.37 log10 CFU/g. Innovator ciprofloxacin (Bayer) exhibited overlapping dose-response curves in two independent experiments (P = 0.57, CFA), confirming the model's repeatability. All nonlinear regressions passed the normality and homoscedasticity tests, and the highest VIF for any parameter was 5.14, indicating negligible multicolinearity. Figure 1 displays the exposure-response curves for the innovator and the four generics tested against an inoculum of 7 log10 CFU/ml, and Table 2 displays their primary and secondary pharmacodynamic parameters. CFA showed that all data belonged to the same population and were better described by a single curve, i.e., the generics were therapeutically equivalent to the innovator (P = 0.54, CFA). In the experiment with the smaller inoculum, bacterial growth in controls was higher (4.9 log10 CFU/g) and the maximal efficacy was reduced (Emax values of 7.27 ± 0.34 and 7.18 ± 2.45 log10 CFU/g for the innovator and the generic, respectively); however, the two curves overlapped (P = 0.93, CFA), indicating that the Chalver product was therapeutically equivalent.
FIG 1.
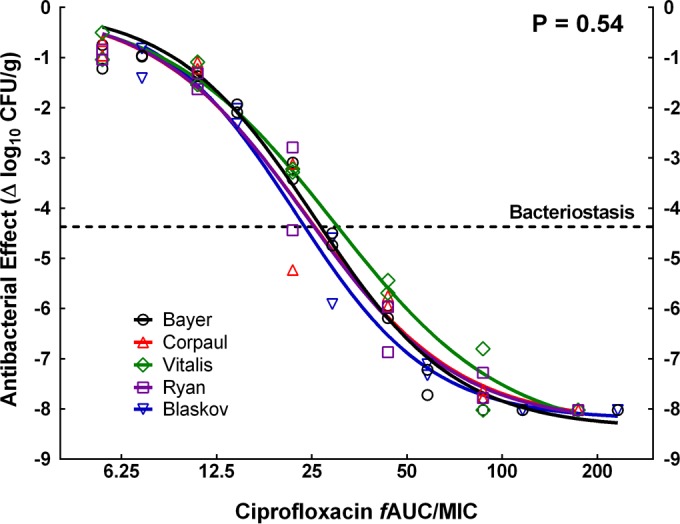
In vivo exposure-response relationship of ciprofloxacin against P. aeruginosa PAO1, comparing the innovator and four generic products. Global CFA indicated that all data belonged to the same population and could be described by a single curve, confirming the therapeutic equivalence of the generics. Stasis was achieved with a fAUC/MIC value of ∼27 and 99.9% kill with a fAUC/MIC value of ∼75.
TABLE 2.
Pharmacodynamic parameters from the neutropenic murine thigh infection model for innovator and generic ciprofloxacin against P. aeruginosa PAO1a
| Product | Emax (log10 CFU/g) | ED50 (mg/kg/day) | N | Adjusted R2 | Sy|x (log10) |
fAUC/MIC |
|
|---|---|---|---|---|---|---|---|
| BD | 3LKD | ||||||
| Bayer | 8.39 ± 0.17 | 14.1 ± 0.59 | 1.95 ± 0.13 | 0.988 | 0.33 | 26.7 ± 0.82 | 70.5 ± 4.20 |
| Corpaul | 8.28 ± 0.51 | 13.3 ± 1.69 | 1.80 ± 0.33 | 0.966 | 0.62 | 28.4 ± 1.19 | 76.1 ± 5.10 |
| Vitalis | 8.56 ± 0.39 | 16.2 ± 1.53 | 1.62 ± 0.18 | 0.987 | 0.37 | 29.5 ± 1.60 | 86.9 ± 8.16 |
| Ryan | 8.29 ± 0.43 | 13.2 ± 1.41 | 1.82 ± 0.28 | 0.974 | 0.53 | 24.9 ± 1.83 | 71.2 ± 10.2 |
| Blaskov | 8.21 ± 0.25 | 12.3 ± 0.83 | 2.03 ± 0.24 | 0.980 | 0.41 | 24.1 ± 1.30 | 68.0 ± 8.24 |
The P value from global curve-fitting analysis was 0.5351. Emax, maximum effect; ED50, 50% effective dose; N, Hill's slope; adjusted R2, adjusted coefficient of determination; Sy|x, standard error of estimate; BD, bacteriostatic dose; 3LKD, 3-log killing dose.
Chemical analysis.
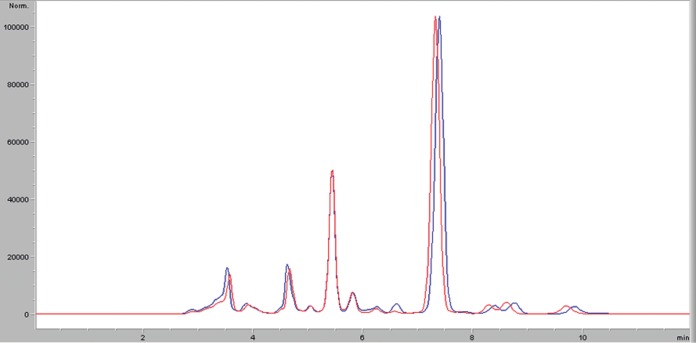
The innovator and the Blaskov generic had the same signal for the analyte in LC-MS/MS. In the mass spectral analysis in scan mode, the two products displayed the same major peaks (Fig. 2), confirming their chemical equivalence.
FIG 2.
LC-MS/MS analysis. The innovator (blue spectrum) and the generic (red spectrum) displayed the same major peaks at almost identical times. The peak corresponding to ciprofloxacin appeared between 7 and 8 min, with equal abundances for the two products.
Ciprofloxacin selection of resistant mutants in the HF system.
Measured ciprofloxacin concentrations in the central compartment were very close to predicted values. The actual fAUC/MIC values for the innovator were 90.8 and 181 for the low-dose and high-dose groups, respectively; in the case of the generic, the fAUC/MIC values were 90.4 and 182, respectively. The basal frequency of resistance to 2.5 × MIC of ciprofloxacin for P. aeruginosa PAO1 was 10−7.1, and the value reached 10−7.5 after 7 days in the untreated arm. In the 800-mg/day group, the initial decrease in the total bacterial population was 3 log10 CFU/ml, and the population was completely composed of resistant bacteria after 24 h of exposure (∼6.0 log10 CFU/ml) (Fig. 3A). The medians of the resistance frequency from day 1 to day 7 were 100.0 and 10−0.03 for the Bayer and Blaskov generics, respectively (exact P = 0.85). In the 400-mg/day group, there was an initial decrease in the total population of approximately 2 log10 CFU/ml with a concomitant increase in resistant cells, which had overtaken the whole population by day 1 of treatment (∼8.0 log10 CFU/ml) (Fig. 3B). The medians of the resistance frequency from day 1 to day 7 were 10−0.04 and 10−0.025 for the innovator and the generic, respectively (exact P = 1.0).
FIG 3.
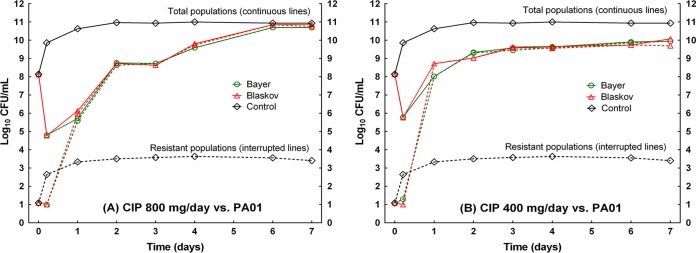
Resistance selection in the hollow-fiber system during 7 days of exposure to innovator and generic ciprofloxacin. Solid lines, total populations; dashed lines, resistant subpopulations; green lines, group exposed to the innovator; red lines, group exposed to the generic; black lines, untreated controls. (A) 800-mg/day group. (B) 400-mg/day group. In both cases, the resistant cells took over the population after day 1, with overlapping lines until day 7.
Regarding MIC changes, only low-level resistance was observed in the 400-mg/day group (MICs of 2 mg/liter after both 24 and 96 h of exposure) and it was reversed completely by PAβN (to 0.25 mg/liter, 1 dilution above the baseline MIC), indicating active efflux as the only responsible mechanism. In the 800-mg/day group, low-level resistance from active efflux was seen on day 1 (MIC of 2 mg/liter, reduced to 0.25 mg/liter with PAβN), but high-level resistance appeared on day 4 (MIC of 8 to 32 mg/liter) and was only partially reversible with PAβN (to 2 to 4 mg/liter), suggesting selection of additional mechanisms, such as gyrAB or parCE mutations (Table 3).
TABLE 3.
MICs after 24 and 96 h of exposure in the hollow-fiber system, equivalent to human dosing regimens of 400 and 800 mg/day, with or without an efflux pump inhibitor
| Treatment group and time | MIC (mg/liter)a |
|||
|---|---|---|---|---|
| Exposure equivalent to 400 mg/day |
Exposure equivalent to 800 mg/day |
|||
| CIP | CIP + PAβN | CIP | CIP + PAβN | |
| 24 h | ||||
| Bayer | 2/2/2 | 0.25/0.25/0.25 | 2/2/2 | 0.25/0.25/0.25 |
| Blaskov | 2/2/2b | 0.25/0.25/0.25b | 2/2/2b | 0.25/0.25/0.25b |
| 96 h | ||||
| Bayer | 2/2/2 | 0.25/0.25/0.25 | 32/32/32 | 4/4/4 |
| Blaskov | 2/2/2b | 0.25/0.25/0.25b | 8/8/32c | 2/2/4c |
MICs are duplicate geometric means from assays performed three times. CIP, ciprofloxacin only; CIP + PAβN, ciprofloxacin plus phenylalanine–arginine–β-naphthylamide (20 mg/liter).
P = 1.00, MW test.
P = 0.188, MW test.
DISCUSSION
There has been an intense debate worldwide, involving the scientific community, drug regulatory agencies, and the public, about the therapeutic equivalence of generic antibiotics, especially vancomycin (14, 15). A source of the debate is data published by our group in the past 10 years, demonstrating that pharmaceutical equivalence is a necessary but not sufficient condition to ensure therapeutic equivalence. We have also presented evidence that the use of nonequivalent generic antibiotics may lead to clinical failure in debilitated patients (6) and, more concerning, to the selection of resistance (7), a problem with serious public health, environmental, and economic consequences.
Here we evaluated five generic products of ciprofloxacin with the standard tests for pharmaceutical equivalence and in vitro activity, in which they performed in the same way as the innovator. Based on our previous results with other antibiotics, indicating that these tests did not predict in vivo equivalence, we tested the products against P. aeruginosa in the extensively validated mouse thigh infection model (16) and found that their pharmacodynamic profiles were also undistinguishable from that of the innovator. Our hypothesis was that generic products with demonstrated (not assumed) equivalence should select for resistance with the same frequency and mechanism as the innovator. For this purpose, we compared resistance outcomes in the hollow-fiber system, an in vitro dynamic model that allows longer treatment courses and the simulation of human dosing regimens, with an excellent correlation with in vivo data (17, 18).
A previous study demonstrated that exposure to therapeutically nonequivalent generics of vancomycin causes enrichment of resistant subpopulations of S. aureus in vivo (7). This suggests the obvious, i.e., the use of agents that kill smaller numbers of microorganisms, allowing survival of more microorganisms after drug exposure, creates a favorable environment for the selection of resistant mutants (“dead bugs do not mutate”) (19). In contrast, a therapeutically equivalent generic product that displays the same pharmacodynamic profile as the innovator should not differ in terms of the frequency and mechanisms of resistance.
This is the case for ciprofloxacin, a synthetic antibiotic developed by Bayer in the 1980s. It is a simple (chemical formula, C17H18FN3O3) and stable small molecule, with a mass of 331.4, that is available for oral and parenteral use. Its U.S. patent expired in 2003, and many generics are marketed worldwide. Ciprofloxacin has no solubility issues, is easily synthetized from 2,4-dichloro-5-fluorobenzoyl chloride in a six-step procedure that is now available in the public domain, and does not require complex purification processes (as do other antibiotics, such as vancomycin), features that may facilitate the manufacturing of identical copies of the drug.
In conclusion, we have demonstrated the pharmaceutical and therapeutic equivalence, in terms of in vitro activity and in vivo pharmacodynamics, of five ciprofloxacin generics. Moreover, we have shown that a pharmaceutically and therapeutically equivalent product has no differences regarding the magnitude and mechanisms of resistance selection at clinically relevant drug exposures. These results support our hypothesis that generics with demonstrated equivalent efficacy in a validated animal model of infection display the same resistance profile as the innovator. We are now seeking to expand the therapeutic nonequivalence-resistance hypothesis for generic antibiotics, which was initially shown with vancomycin and Staphylococcus aureus, to other antimicrobial agents and microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank George Drusano for kindly allowing us to perform the hollow-fiber experiments at the Ordway Research Institute (Albany, NY), with the valuable support of Brian VanScoy and Robert Kulawy. We also express our gratitude to Antonio Oliver for the donation of the PAO1 strain.
Funding for this project came from the University of Antioquia (CODI and Estrategia de Sostenibilidad 2013-2014) and the Rodrigo Vesga-Meneses Scientific Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03633-14.
REFERENCES
- 1.Gauzit R, Lakdhari M. 2012. Generic antibiotic drugs: is effectiveness guaranteed? Med Mal Infect 42:141–148. doi: 10.1016/j.medmal.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Vesga O, Agudelo M, Salazar BE, Rodriguez CA, Zuluaga AF. 2010. Generic vancomycin products fail in vivo despite being pharmaceutical equivalents of the innovator. Antimicrob Agents Chemother 54:3271–3279. doi: 10.1128/AAC.01044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuluaga AF, Agudelo M, Cardeno JJ, Rodriguez CA, Vesga O. 2010. Determination of therapeutic equivalence of generic products of gentamicin in the neutropenic mouse thigh infection model. PLoS One 5:e10744. doi: 10.1371/journal.pone.0010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agudelo M, Vesga O. 2012. Therapeutic equivalence requires pharmaceutical, pharmacokinetic, and pharmacodynamic identities: true bioequivalence of a generic product of intravenous metronidazole. Antimicrob Agents Chemother 56:2659–2665. doi: 10.1128/AAC.06012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez JM, Agudelo M, Leiva LM, Rodriguez CA, Vesga O. 2012. Standardization of a murine model of Candida albicans infection to study therapeutic equivalence (TE) of fluconazole (FCZ) generics, abstr A-1945. Abstr 52nd Intersci Conf Antimicrob Agents Chemother, San Francisco, CA. [Google Scholar]
- 6.Rodriguez CA, Agudelo M, Catano JC, Zuluaga AF, Vesga O. 2009. Potential therapeutic failure of generic vancomycin in a liver transplant patient with MRSA peritonitis and bacteremia. J Infect 59:277–280. doi: 10.1016/j.jinf.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez CA, Agudelo M, Zuluaga AF, Vesga O. 2012. Generic vancomycin enriches resistant subpopulations of Staphylococcus aureus after exposure in a neutropenic mouse thigh infection model. Antimicrob Agents Chemother 56:243–247. doi: 10.1128/AAC.05129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez CA, Agudelo M, Perez JA, Gonzalez J, Vesga O. 2012. Pharmaceutical equivalence (PE), therapeutic equivalence (TE) and resistance (R) selection of 5 generic products of ciprofloxacin (CIP) compared with the innovator against Pseudomonas aeruginosa, abstr A-1968. Abstr 52nd Intersci Conf Antimicrob Agents Chemother, San Francisco, CA. [Google Scholar]
- 9.Zuluaga AF, Agudelo M, Rodriguez CA, Vesga O. 2009. Application of microbiological assay to determine pharmaceutical equivalence of generic intravenous antibiotics. BMC Clin Pharmacol 9:1. doi: 10.1186/1472-6904-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jelliffe RW, Tahani B. 1993. Pharmacoinformatics: equations for serum drug assay error patterns; implications for therapeutic drug monitoring and dosage. Proc Annu Symp Comput Appl Med Care 1993:517–521. [PMC free article] [PubMed] [Google Scholar]
- 11.Bulitta JB, Bingolbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 13:201–211. doi: 10.1208/s12248-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]
- 13.Zuluaga AF, Salazar BE, Rodriguez CA, Zapata AX, Agudelo M, Vesga O. 2006. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect Dis 6:55. doi: 10.1186/1471-2334-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadwiger ME, Sommers CD, Mans DJ, Patel V, Boyne MT Jr. 2012. Quality assessment of U.S. marketplace vancomycin for injection products using high-resolution liquid chromatography-mass spectrometry and potency assays. Antimicrob Agents Chemother 56:2824–2830. doi: 10.1128/AAC.00164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jong GW. 2012. In vivo inferiority of generic product compared with branded vancomycin: a paradigm shift. Ther Drug Monit 34:2–3. doi: 10.1097/FTD.0b013e318243e739. [DOI] [PubMed] [Google Scholar]
- 16.Zuluaga AF, Rodriguez CA, Agudelo M, Vesga O. 2014. About the validation of animal models to study the pharmacodynamics of generic antimicrobials. Clin Infect Dis 59:459–461. doi: 10.1093/cid/ciu306. [DOI] [PubMed] [Google Scholar]
- 17.Louie A, VanScoy BD, Brown DL, Kulawy RW, Heine HS, Drusano GL. 2012. Impact of spores on the comparative efficacies of five antibiotics for treatment of Bacillus anthracis in an in vitro hollow fiber pharmacodynamic model. Antimicrob Agents Chemother 56:1229–1239. doi: 10.1128/AAC.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumbe N, Louie A, Leary R, Liu W, Deziel MR, Tam VH, Bachhawat R, Freeman C, Kahn JB, Bush K, Dudley MN, Miller MH, Drusano GL. 2003. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J Clin Invest 112:275–285. doi: 10.1172/JCI200316814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratton CW. 2003. Dead bugs don't mutate: susceptibility issues in the emergence of bacterial resistance. Emerg Infect Dis 9:10–16. doi: 10.3201/eid0901.020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.