Abstract
The Belgian National Reference Centre for Salmonella received 16,544 human isolates of Salmonella enterica between January 2009 and December 2013. Although 377 different serotypes were identified, the landscape is dominated by S. enterica serovars Typhimurium (55%) and Enteritidis (19%) in a ratio which is inverse to European Union averages. With outbreaks of Salmonella serotypes Ohio, Stanley, and Paratyphi B variant Java as prime examples, 20 serotypes displayed significant fluctuations in this 5-year period. Typhoid strains account for 1.2% of Belgian salmonellosis cases. Large-scale antibiotic susceptibility analyses (n = 4,561; panel of 12 antibiotics) showed declining resistance levels in S. Enteritis and Typhimurium isolates for 8 and 3 tested agents, respectively. Despite low overall resistance to ciprofloxacin (4.4%) and cefotaxime (1.6%), we identified clonal lineages of Salmonella serotypes Kentucky and Infantis displaying rising resistance against these clinically important drugs. Quinolone resistance is mainly mediated by serotype-specific mutations in GyrA residues Ser83 and Asp87 (92.2% not wild type), while an additional ParC_Ser80Ile mutation leads to ciprofloxacin resistance in 95.5% S. Kentucky isolates, which exceeds European averages. Plasmid-mediated quinolone resistance (PMQR) alleles qnrA1 (n = 1), qnrS (n = 9), qnrD1 (n = 4), and qnrB (n = 4) were found in only 3.0% of 533 isolates resistant to nalidixic acid. In cefotaxime-resistant isolates, we identified a broad range of Ambler class A and C β-lactamase genes (e.g., blaSHV-12, blaTEM-52, blaCTX-M-14, and blaCTX-M-15) commonly associated with members of the family Enterobacteriaceae. In conclusion, resistance to fluoroquinolones and cefotaxime remains rare in human S. enterica, but clonal resistant serotypes arise, and continued (inter)national surveillance is mandatory to understand the origin and routes of dissemination thereof.
INTRODUCTION
Although there has been a steady decrease in the number of cases of human salmonellosis in the European Union, it remains the second most frequently reported zoonosis (20.4 confirmed cases per population of 100,000) and the most frequent cause of foodborne outbreaks (1). Nontyphoid salmonellosis is usually self-limiting and normally resolves without the need of antimicrobial agents, but life-threatening invasive infections may occur in vulnerable patients (2). The causative agent, Salmonella enterica subsp. enterica, is subdivided into 1,531 serovars based on antigenic differences in the lipopolysaccharide O antigen and two flagellin structures (3). These serotypes differ greatly in their natural reservoirs, their ability to provoke infections, and their resistance to antimicrobials (4, 5). The severity of inflicted disease also differs substantially among serotypes, with case fatality rates ranging over 100-fold and proportions of hospitalizations varying between 14 and 67% for nontyphoidal strains (6). Typhoidal Salmonella enterica serotypes Typhi and Paratyphi A and B are notorious pathogens, restricted to humans and causing 13.5 million annual episodes of typhoid fever, especially in low- and middle-income countries (7). In developed countries, outbreaks of typhoid fever account for <1% of all infections and are mainly associated with travel (8).
Invasive Salmonella infections mandate the need for chemotherapy. The increasing rates of resistance against traditional agents (aminopenicillins, trimethoprim-sulfamethoxazole, and chloramphenicol) caused a shift to fluoroquinolones (FQ) and third-generation cephalosporins (CSP) in empirical treatment (9, 10). In Europe, average resistance rates against ciprofloxacin (FQ) and cefotaxime (CSP) remain relatively low for S. enterica at <6% and <2%, respectively (1). However, a plethora of studies report high-level FQ and CSP resistance emerging in different parts of the world (11–15), and global travel and food trade increase the likelihood of acquiring infection from nondomestic sources. As such, it is crucial to closely monitor trends in resistance and the prevalence of mobile and nonmobile genetic determinants underlying resistance to these first-line drugs.
Genetic determinants of resistance against FQ and CSP are well documented. FQ resistance (FQR) is primarily associated with accumulation of chromosomal mutations in the targeted bacterial enzymes, i.e., DNA gyrases A and B (GyrA and GyrB) and DNA topoisomerase IV (ParC). Additional low-level resistance can be acquired by plasmid-encoded proteins (plasmid-mediated quinolone resistance [PMQR]) including Qnr-type proteins which protect the DNA gyrases from quinolones, the Aac(6′)-Ib-cr acetyltransferase, and OqxAB and QepA efflux pumps that extrude quinolones (16, 17). Resistance against CSP (CSPR) is predominantly associated with plasmid-borne β-lactamases, the most prevalent belonging to the Ambler class A extended-spectrum β-lactamases (ESBLs) of the CTX-M, TEM, and SHV families and the acquired class C cephalosporinases (AmpCs) (18). These ESBLs are capable of hydrolyzing penicillins, cephalosporins (except cephamycins), and monobactams and are inhibited by clavulanic acid or cloxacillin (19). Although resistance to carbapenems, often considered the antibiotic of last resort (20), is extremely rare in S. enterica, isolates expressing acquired carbapenemases have been reported (21–23).
Here, we describe the serotype distribution and antimicrobial resistance patterns of all human Salmonella isolates sent to the Belgian Reference Centre for Salmonella (NRCS) between 2009 and 2013. By unraveling resistance determinants in isolates displaying reduced susceptibility to FQ and CSP, we compiled a blueprint of the current resistance landscape in Belgian human Salmonella isolates, which at the moment remains only marginally influenced by plasmid-encoded mechanisms.
MATERIALS AND METHODS
Bacterial strains, serotyping, and antimicrobial susceptibility testing.
In Belgium, peripheral clinical laboratories collect Salmonella isolates from human patients and send them voluntarily to the Belgian Reference Centre for Salmonella (NRCS) for serotyping. The full collection of 16,544 S. enterica strains which were received between January 2009 and December 2013 was subjected to serotyping by slide agglutination with commercial antisera, according to the Kauffmann-White scheme (24). A subset of 4,561 S. enterica strains (for selection criteria, see Table S1 in the supplemental material) was subjected to antimicrobial resistance typing using the disk diffusion (Kirby-Bauer) method and Mueller-Hinton agar as culture medium. They represent the most prevalent serological forms and the Salmonella serotypes typically associated with invasive infections: Typhimurium (n = 2,434), Enteritidis (n = 838), Kentucky (n = 206), Infantis (n = 193), Derby (n = 164), Paratyphi B dT+ (n = 116), Typhi (n = 112), Newport (n = 105), Virchow (n = 79), Brandenburg (n = 76), Dublin (n = 62), Hadar (n = 44), Paratyphi A (n = 32), and Paratyphi B (n = 13). Susceptibilities to nalidixic acid (Nal), ciprofloxacin (Cip), cefotaxime (Ctx), ampicillin (Amp), amoxicillin (Amx), chloramphenicol (Chl), tetracycline (Tet), gentamicin (Gen), streptomycin (Str), trimethoprim (Tmp), sulfonamides (Sul), and trimethoprim-sulfamethoxazole (Stx) were determined. Strains that were suspected to have extended-spectrum cephalosporin-hydrolyzing β-lactamases were subjected to three additional β-lactams (cefepime ([Cep], Ctx, and ceftazidime [Caz] in the presence or absence of clavulanic acid) and meropenem (Mem). All antibiotic disks were purchased from Bio-Rad (Nazareth, Belgium). MICs for Nal and Cip were determined using the Etest macromethod (bioMérieux). Inhibition zones were interpreted according to the EUCAST guidelines (document version 4.0, 2014) (25) when available, or CLSI guidelines M100-S24 (26) in the absence of EUCAST breakpoints (see Table S2 in the supplemental material). Multidrug resistance was defined as acquired nonsusceptibility to at least one agent in three or more classes of antimicrobials (27).
Identification of antibiotic resistance (ABR) determinants.
Total DNA was extracted from strains displaying resistance to nalidixic acid and/or cefotaxime, indicated by inhibition zones of <13 and <17 mm, respectively (see Table S2 in the supplemental material). To investigate the molecular determinants for FQ resistance, relevant regions in chromosomal gyrA, gyrB, and parC genes were amplified by PCR and sequenced using previously optimized primers and conditions (28, 29). Using the same DNA preparations, a screen for plasmid-mediated FQR genes (qnrA to qnrD and qnrS) was carried out by PCR amplification (30, 31). Likewise, the amplification of OXA, TEM, CTX-M, and SHV alleles was performed in CSP-resistant isolates using previously described primers and protocols (32, 33). The exact sequences of all resulting PCR products were determined by direct Sanger sequencing.
Determination of genetic relationships.
For a subset of S. Typhimurium isolates, multiple-locus variable-number tandem-repeat analysis (MLVA) was performed essentially as described elsewhere (34). Briefly, a DNA lysate was prepared by heating a single colony in 300 μl Milli-Q (mQ) water at 100°C for 10 min. Using the supernatant as the template, five loci were amplified by multiplex PCR, and the resulting products were subjected to capillary electrophoresis on an ABI 3130xl genetic analyzer (Life Technologies). The size of the PCR products was determined with GeneMapper software v.1.0, using GeneScan 600 LIZ (Life Technologies) as the standard. The calibration strains were chosen from a previous study (35). MLVA profiles were reported as a string of five numbers (STTR5-STTR5-STTR6-STTR10-STTR3) representing the number of repeats at the corresponding locus following the nomenclature proposed by Larsson and colleagues (36). A dendrogram was generated using the categorical coefficient and unweighted-pair group method with arithmetic means (UPGMA). The minimum spanning tree was generated using the categorical coefficient and no priority rules for the algorithm with BioNumerics 6.5 (Applied Maths).
For less common serotypes, clonal relatedness was determined by XbaI macrorestriction followed by pulsed-field gel electrophoresis (PFGE). The resulting fragments were interpreted according to the PulseNet protocol (37) using the BioNumerics software package. The dendrogram was constructed based on Dice coefficient of similarity and UPGMA algorithm with 1.5% band position tolerance.
RESULTS AND DISCUSSION
Serotype distribution.
In the 5-year period spanning this study, the Belgian Reference Centre for Salmonella (NRCS) received 16,544 human S. enterica strains mainly isolated from feces (94.4%), blood (2.4%), and urine (1.5%) samples. The number of annually submitted strains remained fairly constant, varying between 3,182 and 3,668 isolates (Fig. 1A). In this vast collection, we identified 377 different serotypes (see Table S3 in the supplemental material). The landscape is clearly dominated by Salmonella serotypes Typhimurium and Enteritidis which were retrieved in approximately 54.2% and 19.2% of all samples, respectively (Fig. 1A). S. Typhimurium has a well-characterized ability to infect various species (38) and can survive for a long time in the environment (39), enhancing its ability to be one of the most common causes of salmonellosis. The observed ratio for Salmonella serotypes Typhimurium and Enteritidis (∼3/1) contrasts to the ratio in other European Union (EU)/European Economic Area (EEA) countries where Salmonella Typhimurium and Enteritidis account for 25% and 44% of all reported serotypes, respectively (1). An explanation of this discrepancy can be found in the national vaccination program in layer flocks at the beginning of the millennium, which caused a drastic reduction in S. Enteritidis (40).
FIG 1.
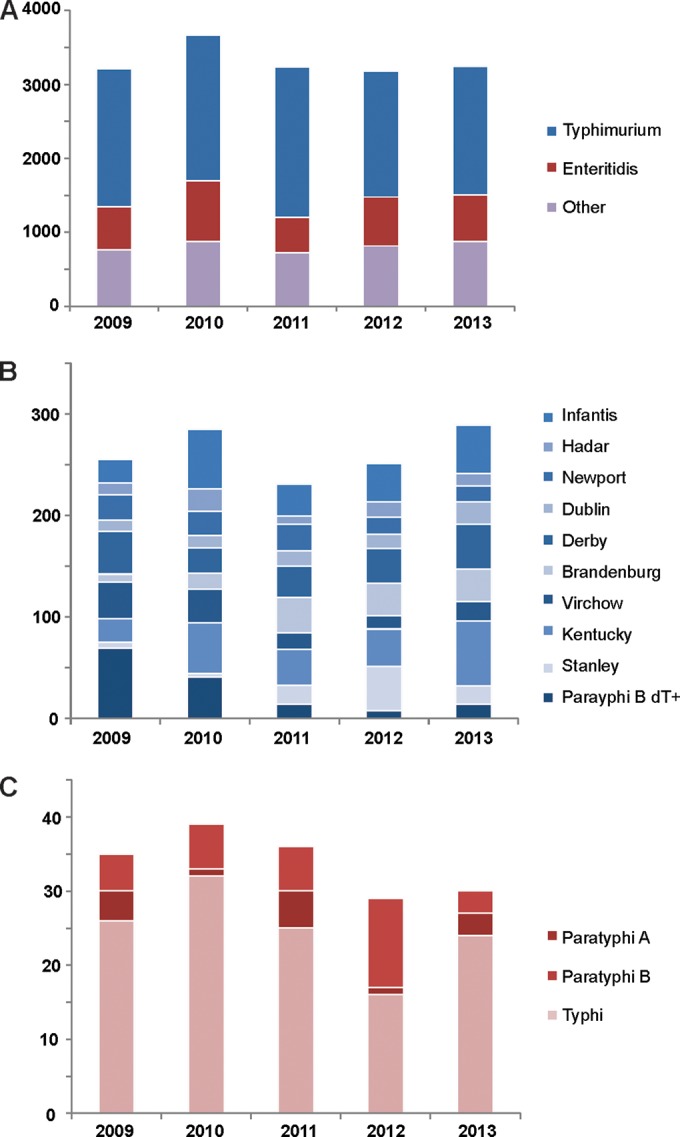
Epidemiology of human salmonellosis in Belgium, 2009 to 2013. Serotype distribution of S. enterica in the 5-year period under study. Results are shown grouped by the total number of isolates (with the contribution of the two major Salmonella serotypes indicated) (A), prevalence of 10 important nontyphoid Salmonella serotypes (B), and three typhoid Salmonella serotypes (C).
Other nontyphoid Salmonella strains are far less commonly encountered and account for a maximum of 2.1% isolates/year (Fig. 1B; see Table S3 in the supplemental material). Chi-square trend analysis (including Bonferroni's correction) showed significant fluctuations in 20 serovars, with outbreaks of Salmonella serovars Ohio (2009), Paratyphi B dT+ (2009 to 2010), and Manhattan (2012) being the most significant (Table S3). The outbreak of Salmonella serotype Stanley in 2012 was traced back to Hungary by joint European Centre for Disease Prevention and Control (ECDC) efforts (41). Also notable is the stable, low prevalence of Salmonella serotype Newport (annually between 0.49 and 0.80%), which is comparable to the European average (1) but 10-fold lower than the global average (42, 43).
In the past 5 years, on average 1.23% of all Belgian salmonellosis cases involved typhoid infections (Fig. 1C). The large majority of cases involved Salmonella serotype Typhi. In 2013, only 0.09% of human Belgian Salmonella isolates were positively identified as Salmonella serotypes Paratyphi A and B.
Serotype-dependent patterns of antimicrobial resistance.
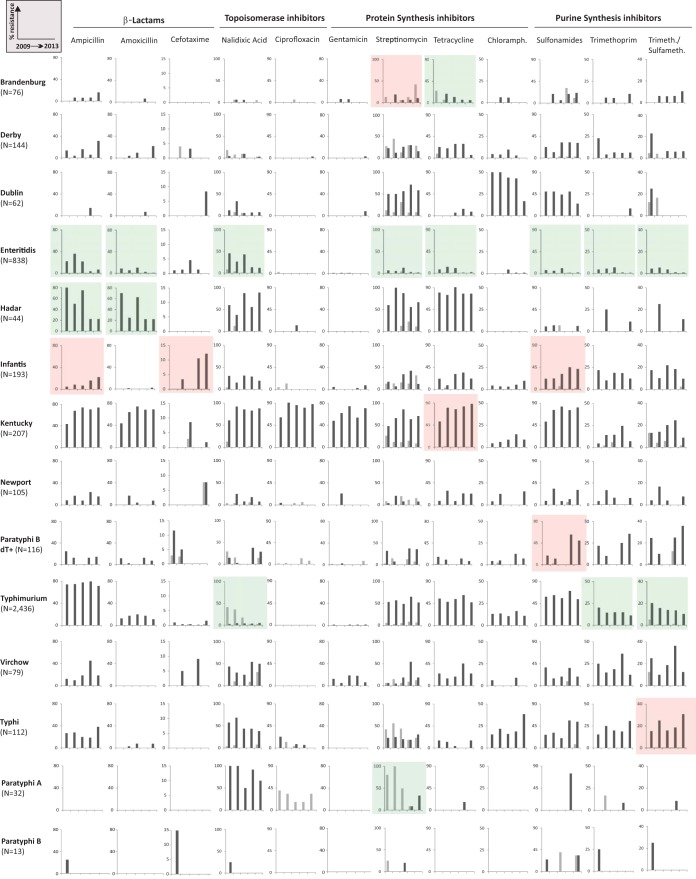
Based on selection criteria formulated in Table S1 in the supplemental material, the susceptibility of 4,561 Salmonella isolates against a panel of 12 antibiotics was determined (Fig. 2). We found that 67.0% of all isolates were resistant against at least one antibiotic, while 32.4% were multidrug resistant (MDR). In total, 4.4% and 1.6% of all Salmonella strains are resistant to ciprofloxacin and oxyimino-CSPs cefotaxime and ceftazidime, respectively (Tables S4 and S5), which is comparable to recently published European data (i.e., 5.1 and 1.1%) (1). One must take into account that this might be an underestimation of the actual degree of FQR, as it now established that 5 μg ciprofloxacin will not reliably detect low-level resistance in Salmonella spp., justifying the switch to pefloxacin disks in routine screens from 2014 onwards (25).
FIG 2.
Antimicrobial resistance in human salmonellosis in Belgium, 2009 to 2013. Antibiotics are grouped according to class. The graphs show percentage of intermediate (gray) and full (black) resistance against the antibiotic in question per serotype (n) and per year. Linear trend analyses were performed using the STATCALC X2 module with the extended Mantel-Haenszel method (EpiInfo; CDC, Atlanta, GA). Significantly (P < 0.01) increasing and decreasing susceptibilities are indicated by red and green shading, respectively. Chloramph., chloramphenicol; Trimeth./Sulfameth., trimethoprim-sulfamethoxazole.
Of the 2,435 S. Typhimurium strains tested, the majority displayed resistance against ampicillin (75.6%), streptomycin (55.2%), tetracycline (53.2%), and sulfonamides (60.1%), which is linked to the presence of Salmonella genomic island 1 (44). Although this particular phenotype remained stable during the last 5 years, resistance to nalidixic acid (NalR), trimethoprim-sulfamethoxazole, and sulfonamides declined significantly in this period (P < 0.01; Fig. 2). Clonal relatedness of 75 random isolates was investigated using 5-loci MLVA. We detected 44 different profiles clustering in 10 subgroups, with conservation of resistance profiles within each group although some exceptions are noticed (see Fig. S1A in the supplemental material). S. Enteritidis isolates are generally more susceptible to antibiotics (4) and show declining levels of resistance to 8/12 tested agents (Fig. 2). S. Dublin, one of the most invasive serotypes, generally displays resistance against only streptomycin and chloramphenicol. However, in 2013, a MDR isolate (S13BD03130) displaying resistance to cefotaxime was identified.
More disturbing trends appear in three specific serotypes. First, S. Hadar isolates are highly resistant to tetracycline, streptomycin, aminopenicillins, and nalidixic acid but remain sensitive to fluoroquinolones (FQ) and cephalosporins (CSP) (Fig. 2). Second, S. Infantis displays an unusually high incidence in clinical resistance against third-generation CSP, increasing from 0% in 2009 to 12.2% in 2013. Fifteen isolates revealed nine XbaI PFGE profiles (85.5% minimal similarity), with clear clustering of the CSP-resistant isolates in three specific profiles (see Fig. S1B in the supplemental material). Third, FQR in S. Kentucky isolates reaches 95.5% (Fig. 2), which is higher than the European average of 73.4% (45). Moreover, a clear trend toward complete resistance to aminopenicillins, tetracycline, aminoglycosides, and co-trimoxazole is observed in the 2009-2013 period, leaving third-generation CSP as the drug of choice (Fig. 2). Notably, one S. Kentucky isolate (11-4366) was pan-resistant against the entire panel of tested antibiotics.
All Belgian typhoid Salmonella isolates remain susceptible to CSP. S. Typhi isolates display above average resistance to purine synthesis inhibitors, while NalR is encountered in the majority of both S. Typhi (58/112) and Paratyphi A (27/32) isolates. Moreover, 25% of the S. Paratyphi A strains show intermediate ciprofloxacin resistance, which might complicate treatment outcome (8).
Molecular mechanisms of FQ resistance.
Since it requires less chromosomal substitutions in the quinolone resistance-determining regions (QRDR) of gyrA, gyrB, and parC to establish NalR (46), this phenotype is widely used as indicator for upcoming FQR. Therefore, we screened NalR isolates in our collection (representing 16.4% of the 4,561 strains) for mutations in the QRDR of GyrA, GyrB, and ParC.
First, we observed that not a single Salmonella NalR strain has a mutated GyrB protein, but a minority (8.1%) have wild-type GyrA (Fig. 3). Contrasting Asian studies reporting a plethora of mutations in both GyrA and ParC (47), the two main mutational GyrA hot spots in Belgian isolates are clearly Ser83 and Asp87, each displaying serotype-specific patterns. For example, the Asp87Asn mutation dominates in S. serotype Hadar, while Ser83Phe is typical for S. serotype Paratyphi A. Notably, 50% of the S. Typhi GyrA proteins carry the Glu133Gly mutation not found in any other serovar (Fig. 3), shedding further doubt on which amino acid confines the wild-type GyrA_133 residue in S. Typhi (48). This mutation is often found in combination with a mutated Ser83 residue, but additional mutations in ParC are necessary to confine full FQ resistance, exemplified by S. Typhi isolate 09-02031 (see Table S4 in the supplemental material).
FIG 3.
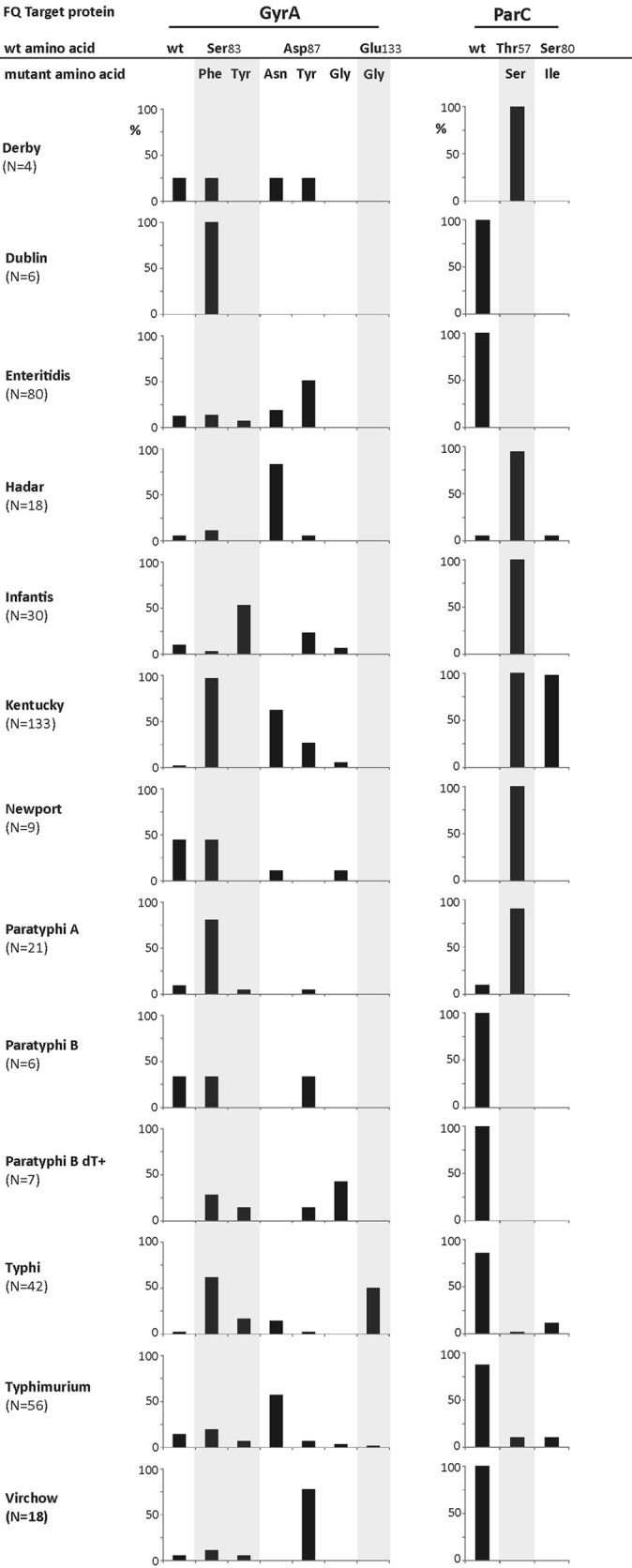
Chromosomal mutations conferring fluoroquinolone (FQ) resistance. Sequencing results of the quinolone resistance- determining regions (QRDR) of 460 nalidixic acid-resistant (NalR) S. enterica isolates. For each serotype (number sequenced [n]), the percentage of isolates displaying a QRDR-related mutation in their gyrA or parC genes is shown. wt, wild-type.
Many Salmonella serovars (including Newport, Kentucky, Infantis, and Paratyphi A) carry the Thr57Ser mutation in ParC, but this mutation has been shown not to be implicated in FQ resistance (49). Apart from this mutation, only 3.0% of the non-Kentucky strains carry a mutated ParC_Ser80 residue, which was solely found in S. Hadar (5.5%), Typhi (11.9%), and Typhimurium (10.7%) strains (Fig. 3). The FQR phenotype of the Kentucky isolates can be explained by a triple QRDR mutation (GyrA_Ser83/Asp87 and ParC_Ser80) found in no less than 97.4% of the NalR isolates. The recently reported dissemination of this serotype in the European continent (1) has clearly reached Belgium, with poultry as the most likely vehicle for infection (22). Its clonal spread is reflected in the XbaI PFGE profiles of 7 randomly chosen isolates, which are >92% identical (see Fig. S1C in the supplemental material).
Next, we also screened 533 NalR isolates for the presence of PMQR alleles, finding positive hits in only 16 (3.0%) strains, which is comparable to other recent surveys (47, 49). As such, NalR might not effectively detect plasmid-mediated mechanisms. Dominant alleles were qnrS1 (n = 8), qnrD1 (n = 4), and qnrB (n = 4) (variations of qnrB), while qnrS2 and qnrA1 were identified once (see Table S4 in the supplemental material). Notably, statistical analysis showed that within our data set, qnr alleles are more prevalent among strain carrying wild-type QRDR (χ2 = 33.5; df = 1; P < 0.05). Prime examples are strains 12-2428 and 12-2447, which are the sole S. Kentucky isolates with wild-type QRDR, yet carry either one (QnrD1) or two (QnrB1 and QnrD1) PMQR alleles. In this particular case, the additional presence of QnrB1 led to a 4-fold increase in MIC.
Resistance against β-lactam antibiotics.
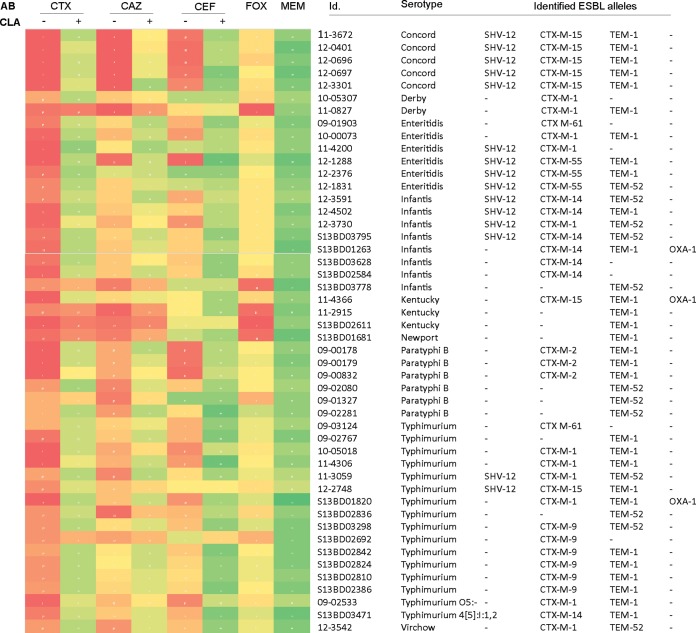
For 48 strains which fell below the respective EUCAST thresholds for CSP resistance, we set up a confirmatory ESBL screen using a combination disk diffusion test using 10 μg clavulanic acid (CLA). In all but six cases, addition of the β-lactamase inhibitor restored the susceptibility to the tested CSP (Fig. 4), phenotypically confirming the presence of Ambler class A ESBL genes (50). In contrast to all other tested strains, the CLA-insensitive strains displayed resistance against the 7-α-methoxygroup-containing cefoxitin (Fig. 4) which indicates the presence of Ambler class C-type cephalosporinases (51). In none of the Salmonella isolates in our study, resistance against the indicator carbapenem meropenem was detected.
FIG 4.
Resistance of S. enterica against β-lactam antibiotics. Heat map of the resistance patterns of Salmonella isolates resistant against third-generation cephalosporins and indicator carbapenems, based on (double) disk diffusion tests and according to EUCAST guidelines. Red and orange indicate full and intermediate resistance, respectively. For each isolate, the sample identifier (Id.), Salmonella serotype, and identified ESBL alleles are shown. AB, antibiotic; CLA, clavulanic acid (10 μg); Fox, cefoxitin, MEM, meropenem.
In each cefotaxime-resistant isolate, we found at least one β-lactamase. The SHV-12 (n = 12) enzymes, always found in combination with at least one other ESBL (Fig. 4), have recently been reported in human S. enterica isolates in the United States, India, and United Kingdom (52–55). The most common alleles were blaTEM and blaCTX-M which were identified in 41 and 39 isolates, respectively. The relatively innocuous penicillinase blaTEM-1 (n = 29) was the most prevalent gene. Twelve strains carried the ESBL variant blaTEM-52, specified by the E104K/G238S mutations and widely found in European isolates (56). Sequencing of the blaCTX-M alleles revealed the presence of the most globally widespread ESBLs, CTX-M-14 (n = 7) and CTX-M-15 (n = 7). The most prevalent allele, blaCTX-M-1 (n = 11), is the most common food animal-associated CTX-M enzyme in Europe and is circulating on IncN plasmids through Escherichia coli and Salmonella spp. from human, animal, and environmental sources (53, 57). In contrast, blaCTX-M-55 has mainly been reported from Asian enterobacterial isolates (58). Other blaCTX-M genes identified in this study were confined to a specific serotype, like blaCTX-M-2 to S. Paratyphi B (15) and blaCTX-M-9 to S. Typhimurium (Fig. 4). Two isolates, S. Enteritidis 09-1903 and S. Typhimurium 09-3124 contained the blaCTX-M-61 gene. This is, to our knowledge, the first description of this gene in S. enterica. Notable, five of the seven ESBL genes found in human Belgian Salmonella isolates (i.e., CTX-M-1, -2, -9, TEM-52, and SHV-12) were recently retrieved from fecal Salmonella isolates from healthy Belgian pigs and broiler chickens (58).
Last, although the cooccurrence of PMQR genes and CSP resistance has been reported (59), it remains a very rare phenomenon in human salmonellosis. In this study, only a single isolate of S. Infantis (S13BD03795) carried a plasmid containing both qnrB, shv-12, blaCTX-M-14, and blaTEM-52 alleles (see Tables S4 and S5 in the supplemental material).
Conclusions.
In this work, we provide a blueprint of current serotype prevalence and antibiotic resistance among Belgian S. enterica strains isolated from humans with salmonellosis. While the serotype landscape has remained largely stable for the past 5 years apart from specific outbreaks, serotype-dependent trends of antibiotic resistance are emerging. A particular threat for public health are circulating clonal lineages of CSPR and FQR S. Infantis and Kentucky strains, respectively, and intermediate FQR S. Paratyphi A isolates. While the influence of PMQR on FQR seems marginal in S. enterica, plasmid-encoded β-lactamases which are commonly identified among members of the family Enterobacteriaceae are found in all tested serotypes. This confirms previous observations that the epidemiology of resistance may involve spread of multidrug resistance plasmids between enterobacterial strains and species (53, 57). It is mandatory that these genetic determinants are monitored closely in the coming years.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gabriela Serrano Becerra and Frédéric Fux for excellent technical assistance. Capillary electrophoresis for MLVA was performed at the Platform Biotechnology and Molecular Biology at the Scientific Institute of Public Health (WIV-ISP).
The national reference center is partially supported by the Belgian Ministry of Social Affairs through a fund within the health insurance system.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04203-14.
REFERENCES
- 1.European Food Safety Authority, European Centre for Disease Prevention and Control. 2013. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J 11:3129. doi: 10.2903/j.efsa.2013.3129. [DOI] [Google Scholar]
- 2.Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, Shallow S, Smith K, Angulo FJ, Emerging Infections Program FoodNet Working Group . 2004. Invasive Salmonella infections in the United States, FoodNet, 1996–1999: incidence, serotype distribution, and outcome. Clin Infect Dis 38(Suppl 3):S149–S156. doi: 10.1086/381581. [DOI] [PubMed] [Google Scholar]
- 3.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PA, Weill FX. 2010. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Lauderdale TL, Aarestrup FM, Chen PC, Lai JF, Wang HY, Shiau YR, Huang IW, Hung CL, TSAR hospitals . 2006. Multidrug resistance among different serotypes of clinical Salmonella isolates in Taiwan. Diagn Microbiol Infect Dis 55:149–155. doi: 10.1016/j.diagmicrobio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Parry CM, Threlfall EJ. 2008. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr Opin Infect Dis 21:531–538. doi: 10.1097/QCO.0b013e32830f453a. [DOI] [PubMed] [Google Scholar]
- 6.Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D'Angelo M, Hurd S, Medus C, Cronquist A, Angulo FJ. 2008. Salmonellosis outcomes differ substantially by serotype. J Infect Dis 198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- 7.Buckle GC, Walker CLF, Black RE. 2012. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health 2:010401. doi: 10.7189/jogh.02.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassing RJ, Menezes GA, van Pelt W, Petit PL, van Genderen PJ, Goessens WH. 2011. Analysis of mechanisms involved in reduced susceptibility to ciprofloxacin in Salmonella enterica serotypes Typhi and Paratyphi A isolates from travellers to Southeast Asia. Int J Antimicrob Agents 37:240–243. doi: 10.1016/j.ijantimicag.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Chen HM, Wang Y, Su LH, Chiu CH. 2013. Nontyphoid salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr Neonatol 54:147–152. doi: 10.1016/j.pedneo.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Kim DM, Neupane GP, Jang SJ, Kim SH, Lee BK. 2010. In vitro efficacy of the combination of ciprofloxacin and cefotaxime against nalidixic acid resistant Salmonella enterica serotype Typhi. Int J Antimicrob Agents 36:155–158. doi: 10.1016/j.ijantimicag.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Olsen SJ, DeBess EE, McGivern TE, Marano N, Eby T, Mauvais S, Balan VK, Zirnstein G, Cieslak PR, Angulo FJ. 2001. A nosocomial outbreak of fluoroquinolone-resistant salmonella infection. N Engl J Med 344:1572–1579. doi: 10.1056/NEJM200105243442102. [DOI] [PubMed] [Google Scholar]
- 12.Hsueh PR, Teng LJ, Tseng SP, Chang CF, Wan JH, Yan JJ, Lee CM, Chuang YC, Huang WK, Yang D, Shyr JM, Yu KW, Wang LS, Lu JJ, Ko WC, Wu JJ, Chang FY, Yang YC, Lau YJ, Liu YC, Liu CY, Ho SW, Luh KT. 2004. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg Infect Dis 10:60–68. doi: 10.3201/eid1001.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassing RJ, Goessens WH, van Pelt W, Mevius DJ, Stricker BH, Molhoek N, Verbon A, van Genderen PJ. 2014. Salmonella subtypes with increased MICs for azithromycin in travelers returned to the Netherlands. Emerg Infect Dis 20:705–708. doi: 10.3201/eid2004.131536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abgottspon H, Zurfluh K, Nüesch-Inderbinen M, Hächler H, Stephan R. 2014. Quinolone resistance mechanisms in Salmonella enterica serovars Hadar, Kentucky, Virchow, Schwarzengrund, and 4,5,12:i:−, isolated from humans in Switzerland, and identification of a novel qnrD variant, qnrD2, in S. Hadar. Antimicrob Agents Chemother 58:3560–3563. doi: 10.1128/AAC.02404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doublet B, Praud K, Nguyen-Ho-Bao T, Argudín MA, Bertrand S, Butaye P, Cloeckaert A. 2014. Extended-spectrum β-lactamase- and AmpC β-lactamase-producing D-tartrate-positive Salmonella enterica serovar Paratyphi B from broilers and human patients in Belgium, 2008–10. J Antimicrob Chemother 69:1257–1264. doi: 10.1093/jac/dkt504. [DOI] [PubMed] [Google Scholar]
- 16.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 18.Gniadkowski M. 2008. Evolution of extended-spectrum beta-lactamases by mutation. Clin Microbiol Infect 14(Suppl 1):11–32. doi: 10.1111/j.1469-0691.2007.01854.x. [DOI] [PubMed] [Google Scholar]
- 19.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livermore DM. 2012. Fourteen years in resistance. Int J Antimicrob Agents 39:283–294. doi: 10.1016/j.ijantimicag.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Savard P, Gopinath R, Zhu W, Kitchel B, Rasheed JK, Tekle T, Roberts A, Ross T, Razeq J, Landrum BM, Wilson LE, Limbago B, Perl TM, Carroll KC. 2011. First NDM-positive Salmonella sp. strain identified in the United States. Antimicrob Agents Chemother 55:5957–5958. doi: 10.1128/AAC.05719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, Guibert V, Zerouali K, Weill FX. 2013. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infect Dis 13:672–679. doi: 10.1016/S1473-3099(13)70124-5. [DOI] [PubMed] [Google Scholar]
- 23.Seiffert SN, Perreten V, Johannes S, Droz S, Bodmer T, Endimiani A. 2014. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob Agents Chemother 58:2446–2449. doi: 10.1128/AAC.02417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimond PAD, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella, World Health Organization, Institut Pasteur, Paris, France. [Google Scholar]
- 25.European Committee on Antimicrobial Susceptibility Testing. 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0, 2014. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf.
- 26.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 28.Chiu CH, Wu TL, Su LH, Chu C, Chia JH, Kuo AJ, Chien MS, Lin TY. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype choleraesuis. N Engl J Med 346:413–419. doi: 10.1056/NEJMoa012261. [DOI] [PubMed] [Google Scholar]
- 29.Yue L, Jiang HX, Liao XP, Liu JH, Li SJ, Chen XY, Chen CX, Lü DH, Liu YH. 2008. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet Microbiol 132:414–420. doi: 10.1016/j.vetmic.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Cavaco LM, Frimodt-Møller N, Hasman H, Guardabassi L, Nielsen L, Aarestrup FM. 2008. Prevalence of quinolone resistance mechanisms and associations to minimum inhibitory concentrations in quinolone-resistant E. coli isolated from humans and swine in Denmark. Microb Drug Resist 14:163–169. doi: 10.1089/mdr.2008.0821. [DOI] [PubMed] [Google Scholar]
- 31.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 32.Olesen I, Hasman H, Aarestrup FM. 2004. Prevalence of beta-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb Drug Resist 10:334–340. doi: 10.1089/mdr.2004.10.334. [DOI] [PubMed] [Google Scholar]
- 33.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. 2005. Beta-lactamases among extended spectrum beta-lactamase resistant (ESBL) Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother 56:115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 34.Wuyts V, Mattheus W, De Laminne de Bex G, Wildemauwe C, Roosens NH, Marchal K, De Keersmaecker SC, Bertrand S. 2013. MLVA as a tool for public health surveillance of human Salmonella Typhimurium: prospective study in Belgium and evaluation of MLVA loci stability. PLoS One 8:e84055. doi: 10.1371/journal.pone.0084055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindstedt BA, Vardund T, Aas L, Kapperud G. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J Microbiol Methods 59:163–172. doi: 10.1016/j.mimet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Larsson JT, Torpdahl M, Petersen RF, Sorensen G, Lindstedt BA, Nielsen EM. 2009. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill 14(15):pii=19174 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19174. [PubMed] [Google Scholar]
- 37.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol 43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschäpe H, Adams LG, Bäumler AJ. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun 70:2249–2255. doi: 10.1128/IAI.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baudart J, Lemarchand K, Brisabois A, Lebaron P. 2000. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl Environ Microbiol 66:1544–1552. doi: 10.1128/AEM.66.4.1544-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collard JM, Bertrand S, Dierick K, Godard C, Wildemauwe C, Vermeersch K, Duculot J, Van Immerseel F, Pasmans F, Imberechts H, Quinet C. 2008. Drastic decrease of Salmonella Enteritidis isolated from humans in Belgium in 2005, shift in phage types and influence on foodborne outbreaks. Epidemiol Infect 136:771–781. doi: 10.1017/S095026880700920X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinross P, van Alphen L, Martinez Urtaza J, Struelens M, Takkinen J, Coulombier D, Mäkelä P, Bertrand S, Mattheus W, Schmid D, Kanitz E, Rücker V, Krisztalovics K, Pászti J, Szögyényi Z, Lancz Z, Rabsch W, Pfefferkorn B, Hiller P, Mooijman K, Gossner C. 2014. Multidisciplinary investigation of a multicountry outbreak of Salmonella Stanley infections associated with turkey meat in the European Union, August 2011 to January 2013. Euro Surveill 19(19):pii=20801 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20801. [DOI] [PubMed] [Google Scholar]
- 42.Johnson LR, Gould LH, Dunn JR, Berkelman R, Mahon BE, FoodNet Travel Working Group . 2011. Salmonella infections associated with international travel: a Foodborne Diseases Active Surveillance Network (FoodNet) study. Foodborne Pathog Dis 8:1031–1037. doi: 10.1089/fpd.2011.0854. [DOI] [PubMed] [Google Scholar]
- 43.Centers for Disease Control and Prevention. 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 1996–2010. MMWR Morb Mortal Wkly Rep 60:749–755. [PubMed] [Google Scholar]
- 44.Boyd D, Cloeckaert A, Chaslus-Dancla E, Mulvey MR. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob Agents Chemother 46:1714–1722. doi: 10.1128/AAC.46.6.1714-1722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westrell T, Monnet DL, Gossner C, Heuer O, Takkinen J. 2014. Drug-resistant Salmonella enterica serotype Kentucky in Europe. Lancet Infect Dis 14:270–271. doi: 10.1016/S1473-3099(14)70703-0. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins KL, Davies RH, Threlfall EJ. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents 25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Jeong HS, Kim JA, Shin JH, Chang CL, Jeong J, Cho JH, Kim MN, Kim S, Kim YR, Lee CH, Lee K, Lee MA, Lee WG, Shin JH, Lee JN. 2011. Prevalence of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes in Salmonella isolated from 12 tertiary-care hospitals in Korea. Microb Drug Resist 17:551–557. doi: 10.1089/mdr.2011.0095. [DOI] [PubMed] [Google Scholar]
- 48.Walther-Rasmussen J, Høiby N. 2011. Salmonella enterica serovar Typhi and S. Paratyphi A: need to expand the QRDR region? Epidemiol Infect 139:1281–1283. doi: 10.1017/S0950268810002487. [DOI] [PubMed] [Google Scholar]
- 49.Wasyl D, Hoszowski A, Zając M. 2014. Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Vet Microbiol 171:307–314. doi: 10.1016/j.vetmic.2014.01.040. [DOI] [PubMed] [Google Scholar]
- 50.Towne TG, Lewis JS Jr, Herrera M, Wickes B, Jorgensen JH. 2010. Detection of SHV-type extended-spectrum beta-lactamase in Enterobacter isolates. J Clin Microbiol 48:298–299. doi: 10.1128/JCM.01875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whichard JM, Gay K, Stevenson JE, Joyce KJ, Cooper KL, Omondi M, Medalla F, Jacoby GA, Barrett TJ. 2007. Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg Infect Dis 13:1681–1688. doi: 10.3201/eid1311.061438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burke L, Hopkins KL, Meunier D, de Pinna E, Fitzgerald-Hughes D, Humphreys H, Woodford N. 2014. Resistance to third-generation cephalosporins in human non-typhoidal Salmonella enterica isolates from England and Wales, 2010-12. J Antimicrob Chemother 69:977–981. doi: 10.1093/jac/dkt469. [DOI] [PubMed] [Google Scholar]
- 54.Menezes GA, Khan MA, Harish BN, Parija SC, Goessens W, Vidyalakshmi K, Baliga S, Hays JP. 2010. Molecular characterization of antimicrobial resistance in non-typhoidal salmonellae associated with systemic manifestations from India. J Med Microbiol 59:1477–1478. doi: 10.1099/jmm.0.022319-0. [DOI] [PubMed] [Google Scholar]
- 55.Boyle F, Healy G, Hale J, Kariuki S, Cormican M, Morris D. 2011. Characterization of a novel extended-spectrum β-lactamase phenotype from OXA-1 expression in Salmonella Typhimurium strains from Africa and Ireland. Diagn Microbiol Infect Dis 70:549–553. doi: 10.1016/j.diagmicrobio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Cloeckaert A, Praud K, Doublet B, Bertini A, Carattoli A, Butaye P, Imberechts H, Bertrand S, Collard JM, Arlet G, Weill FX. 2007. Dissemination of an extended-spectrum-beta-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob Agents Chemother 51:1872–1875. doi: 10.1128/AAC.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D, Peixe L, Poirel L, Schuepbach-Regula G, Torneke K, Torren-Edo J, Torres C, Threlfall J. 2013. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis 56:1030–1037. doi: 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 58.de Jong A, Smet A, Ludwig C, Stephan B, De Graef E, Vanrobaeys M, Haesebrouck F. 2014. Antimicrobial susceptibility of Salmonella isolates from healthy pigs and chickens (2008–2011). Vet Microbiol 171:298–306. doi: 10.1016/j.vetmic.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 59.Herrera-León S, González-Sanz R, Herrera-León L, Echeita MA. 2011. Characterization of multidrug-resistant Enterobacteriaceae carrying plasmid-mediated quinolone resistance mechanisms in Spain. J Antimicrob Chemother 66:287–290. doi: 10.1093/jac/dkq423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.