Abstract
The Acinetobacter baumannii clonal complex 113/79 (CC113/79) and class 2 integrons predominate in Latin America; a relationship between these characteristics was explored. The presence of integrases was determined in successive hospital Acinetobacter isolates (163 A. baumannii isolates and 72 Acinetobacter nosocomialis isolates). Most isolates had integrons, but class 1 and 2 integrons were present significantly more often in CC109/1 and CC113/79, respectively. The high prevalence of CC113/79 in Latin America may account for the predominance of class 2 integrons.
TEXT
The dissemination of multidrug-resistant (MDR) Acinetobacter baumannii international clones (IC) and Acinetobacter nosocomialis has challenged health care (1, 2). The MDR phenotype in Acinetobacter has been related to class 1 and 2 integrons (3, 4, 5); class 2 integrons predominate in A. baumannii from Latin America (5, 6). This finding is possibly related to local clonal groups (6), but a hypothesis has not yet been addressed. The class 2 integron structure seems less diverse than that of class 1 (7) and is usually embedded within transposon Tn7 (5, 8).
Four A. baumannii and one A. nosocomialis IC characterized by multilocus sequence typing (MLST) have been described and are spread in Rio de Janeiro, Brazil (9, 10). The purpose of the present study was to explore the association of class 2 integrons and gene cassettes with these clonal lineages. Between 2007 and 2008, 163 A. baumannii and 72 A. nosocomialis hospital isolates were investigated (10); 153 (94%) of the A. baumannii and 21 (29%) of the A. nosocomialis isolates were MDR. Among the A. nosocomialis isolates, antimicrobial resistance was highest for trimethoprim-sulfamethoxazole (66%), cefepime (23%), and ciprofloxacin (23%) (10).
The presence of class 1 and 2 integrons was screened in all isolates by multiplex PCR with intI1 and intI2 gene-specific primers (11). The intI amplicons of two A. baumannii isolates were sequenced as controls. The variable region for intI2 gene-positive isolates was characterized by amplification with the primers Hep74 and Hep51 (12) and by sequence analyses with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Isolates with no amplification of the variable region were further studied by PCR mapping with a Hep74 primer combined with the dfrA1, sat2, aadA1, and aadB reverse primers, as described previously (13). PCR was also performed for Tn7 transposition genes (tnsA, tnsB, tnsC, tnsD, and tnsE), where the 3′ conserved sequence region is located (7). One positive amplicon of each tns gene was sequenced as a control.
All isolates had been typed by randomly amplified polymorphic DNA (RAPD) PCR (10), and a few were typed by MLST (using University of Oxford [UO] and Institut Pasteur [IP] schemes) (9, 10). In the present study, MLST was performed for all A. baumannii isolates. The proportions were compared by the Fisher exact test or the chi-square test (see http://www.openepi.com); a P value of <0.05 was considered significant.
Integrase-encoding genes were found in 158 (67%) of the 235 isolates, with a slight predominance of intI1 in A. baumannii and intI2 in A. nosocomialis (Fig. 1). All A. baumannii isolates were clonal complex 113/79 (CC113/79) (according to UO/IP schemes), CC109/1, CC103/15, or CC110/25. A few A. baumannii isolates (5%) harbored both of the integrases, belonging to CC113/79 (n = 4) and CC110/25 (n = 1). The intI1 gene was predominant in CC109/1 (n = 30, 71%), and the intI2 gene was predominant in CC113/79 (n = 41, 41.7%) (P < 0.001 for CC109/1 versus CC113/79) (Fig. 2A). Altogether, CC109/1 accounted for 46% of class 1 integron-bearing isolates, and CC113/79 accounted for 89% of class 2 integron-bearing isolates. Among the 25 patients with intI2-positive CC113/79 isolates and the 11 patients with intI1-positive CC109/1 isolates, 7 (28%) and 6 (55%), respectively, were moved to the intensive care unit (ICU). In A. nosocomialis, integrase-encoding genes were found in 53 (73%) isolates and intI2 was found in 39 (54%) isolates (Fig. 1). In RAPD-type A (CC260/71), intI1 and intI2 genes were each present in ≥50% of the isolates (Fig. 2B).
FIG 1.
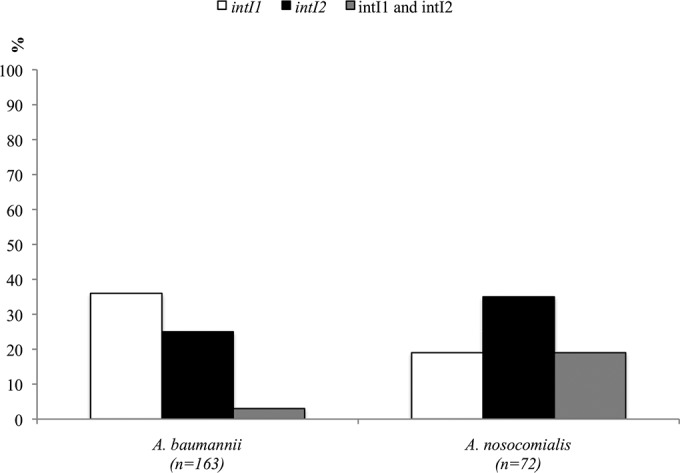
Distribution of intI genes among A. baumannii and A. nosocomialis isolates.
FIG 2.
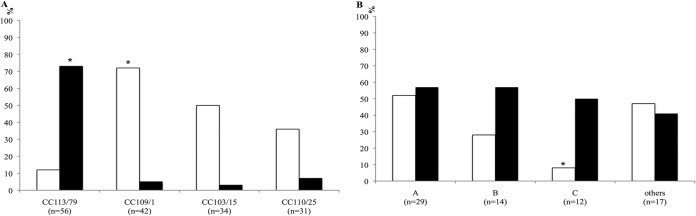
Frequency of intI genes in 163 Acinetobacter baumannii isolates according to MLST clonal complexes (CC) (A) and 72 Acinetobacter nosocomialis isolates according to RAPD types (B). The white and black columns are intI1- and intI2-positive isolates, respectively. The asterisk in panel A represents a P value of <0.001 (compared to A. baumannii isolates included in all other clonal types), and that in panel B represents a P value of 0.02 (for intI1 in A. nosocomialis type C compared to type A).
Five and three different cassette arrays were identified in class 2 integrons among A. baumannii and A. nosocomialis isolates, respectively (Table 1). Class 2 integrons In2-0, In2-1, In2-2, In2-4, and In2-6 were not particularly associated with any A. baumannii or A. nosocomialis clonal type. However, In2-4 was found in 20 (50%) of 41 CC113/79 isolates.
TABLE 1.
A. baumannii clonal complexes and A. nosocomialis RAPD genotypes, class 2 integrons, and the variable regions
| Species (no. of isolates) | Genotype (no. of isolates) | Class 2 integron variable region (no. of isolates) | Transposition gene(s) (no. of isolates) | Class 2 integron name |
|---|---|---|---|---|
| A. baumannii (46) | CC113/79 (41) | dfrA1-sat2-aadA1 (20) | tnsE, tnsD, tnsC, tnsB, tnsA (3) | Tn7::In2-4 |
| tnsE, tnsD, tnsB, tnsA (5) | ΔTn7::In2-4 | |||
| tnsE, tnsC, tnsB, tnsA (1) | ||||
| tnsE, tnsD, tnsA (2) | ||||
| tnsE, tnsC, tnsA (2) | ||||
| tnsE, tnsB, tnsA (2) | ||||
| tnsE, tnsC, tnsB (2) | ||||
| tnsE, tnsA (1) | ||||
| tnsE, tnsD (1) | ||||
| tnsE (1) | ||||
| sat2-aadA1 (11) | tnsE, tnsD, tnsC, tnsB, tnsA (3) | Tn7::In2-6 | ||
| tnsE, tnsC, tnsB, tnsA (1) | ΔTn7::In2-6 | |||
| tnsE, tnsD, tnsC, tnsA (1) | ||||
| tnsE, tnsC, tnsA (2) | ||||
| tnsE, tnsB, tnsA (1) | ||||
| tnsE, tnsC, tnsB (1) | ||||
| tnsE, tnsD, tnsB(1) | ||||
| tnsE, tnsB (1) | ||||
| sat2 (3) | tnsE, tnsD, tnsC, tnsA (1) | ΔTn7::In2-1 | ||
| tnsE, tnsD, tnsB, tnsA (1) | ||||
| tnsE, tnsD, tnsA (1) | ||||
| dfrA1 (4) | tnsE, tnsD, tnsC, tnsB, tnsA (1) | Tn7::In2-2 | ||
| tnsE, tnsD, tnsB, tnsA (1) | ΔTn7:In2-2 | |||
| tnsE, tnsD, tnsB (1) | ||||
| —a (1) | ||||
| — (2) | tnsE, tnsC, tnsB, tnsA (1) | ΔTn7::In2-0 | ||
| — (1) | ||||
| CC103/15 (1) | dfrA1 (1) | tnsE, tnsB (1) | ΔTn7::In2-2 | |
| CC109/1 (2) | dfrA1-sat2-aadA1 (1) | tnsE (1) | ΔTn7::In2-4 | |
| — | — (1) | In2-0 | ||
| CC110/25 (2) | dfrA1-sat2-aadA1 (1) | tnsE (1) | ΔTn7::In2-4 | |
| dfrA1 (1) | tnsE (1) | ΔTn7::In2-2 | ||
| A. nosocomialis (39) | A (17) | dfrA1 (5) | tnsB (1) | ΔTn7::In2-2 |
| — (4) | ||||
| — (12) | tnsB, tnsE (1) | ΔTn7::In2-0 | ||
| tnsB (1) | ||||
| — (10) | ||||
| B (8) | dfrA1-sat2-aadA1 (1) | — (1) | ΔTn7::In2-4 | |
| dfrA1 (3) | tnsD, tnsE (1) | ΔTn7::In2-2 | ||
| tnsB, tnsE (1) | In2-0 | |||
| — (1) | ||||
| Negative (4) | — (4) | In2-0 | ||
| C (7) | dfrA1-sat2-aadA1 (2) | tnsD (2) | ΔTn7::In2-4 | |
| dfrA1 (1) | — (1) | ΔTn7::In2-2 | ||
| — (4) | — (4) | In2-0 | ||
| D (2) | dfrA1 (1) | — (1) | In2-2 | |
| — (1) | — (1) | In2-0 | ||
| E (2) | — (2) | — (2) | In2-0 | |
| G (1) | — (1) | — (1) | In2-0 | |
| H (1) | — (1) | — (1) | In2-0 | |
| L (1) | — (1) | — (1) | In2-0 |
—, no gene found.
The dfrA1, sat2, and aadA1 genes were identified in 61%, 76%, and 73%, respectively, of A. baumannii isolates carrying the class 2 integron and in 33%, 8%, and 8%, respectively, of A. nosocomialis isolates carrying the class 2 integron. At least one Tn7 transposition gene was present in 88% of A. baumannii and 18% of A. nosocomialis isolates (Table 1).
Among the 153 MDR A. baumannii isolates, 42 (28%) carried cassettes within the class 2 integron; all were resistant to trimethoprim-sulfamethoxazole, and 83% were resistant to aminoglycosides, indicating that resistance was related to the dfrA1 and aadA1 genes. MDR A. nosocomialis isolates were less frequent, but trimethoprim-sulfamethoxazole resistance was strongly associated with class 2 integrons. Nevertheless, five A. nosocomialis isolates with class 2 arrays (In2-1) were susceptible to all the agents tested (data not shown).
Here, we describe class 1 and 2 integrons that were strongly associated with CC109/1 (IC-I) and CC113/79 A. baumannii isolates, respectively. One concern with this study is that isolates were epidemiologically related, and some were from cross-infections. Nevertheless, the cohort study design provided accurate prevalence data; the cases moved to the ICU represented unrelated sources of these clones. This result may explain the high prevalence of intI2 in Latin America (5), where CC113/79 predominates (14). Indeed, in Latin America, the low prevalence of IC-I (<20%) (14, 15) can explain the low frequency of the class 1 integron among local A. baumannii isolates. In Europe, IC-I and class 1 integrons have been frequently found (3, 16).
Both integrons were found simultaneously in a few A. baumannii isolates of CC113/79 and CC110/25 in the present study. This finding has been described by others but only in isolates of undefined MLST clones (7, 17, 18). By in silico analysis of 11 A. baumannii genomes, including three IC-I and six IC-II, only class 1 integrons were found (19).
In other non-baumannii species of Acinetobacter, there have been few reports of integrons (20, 21). In the present study, a similar distribution of class 1 and 2 integrons was observed in A. nosocomialis, including in CC260/71 isolates. A class 1 integron containing a metallo-β-lactamase cassette was described in this clone from Japan and Korea (20, 21); however, we did not find such genes in A. nosocomialis (10).
In the present study, the In2-4 array was found in 46% of CC113/79 A. baumannii isolates. In Argentina, this array was also frequently observed (5), and this finding is in line with the report of the CC113/79 spread in this country (14). Similarly, the description of dfrA1, sat2, and aadA1 cassettes in Argentina, Chile, and Uruguay may also be explained by the CC113/79 spread in Latin America (5, 7). A single Brazilian study of MDR A. baumannii found class 2 integrons in 23% of isolates (22), which also carried the main cassette array (dfrA1-sat2-aadA1) found in our study.
The present study contributes to the understanding of the distribution of class 1 and 2 integrons in A. baumannii and A. nosocomialis isolates and of how the distribution is affected by the global dispersion of international clones in different regions of the world.
ACKNOWLEDGMENTS
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Comissão Fullbright-Brasil, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), the Agência Nacional de Vigilância Sanitária (ANVISA) of Brazil, and the Fogarty International Program in Global Infectious Diseases of the National Institutes of Health (grant TW006563).
REFERENCES
- 1.Rosenthal VD, Maki DG, Jamulitrat S, Medeiros EA, Todi SK, Gomez DY, Leblebicioglu H, Abu Khader I, Miranda Novales MG, Berba R, Ramírez Wong FM, Barkat A, Pino OR, Dueñas L, Mitrev Z, Bijie H, Gurskis V, Kanj SS, Mapp T, Hidalgo RF, Ben Jaballah N, Raka L, Gikas A, Ahmed A, Thu LTA, Guzmán Siritt ME, INICC Members. 2010. International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003 to 2008, issued June 2009. Am J Infect Control 38:95–104.e2. doi: 10.1016/j.ajic.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y-C, Huang Y-T, Tan C-K, Kuo Y-W, Liao C-H, Lee P-I, Hsueh P-R. 2011. Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J Antimicrob Chemother 66:1839–1846. doi: 10.1093/jac/dkr200. [DOI] [PubMed] [Google Scholar]
- 3.Karah N, Giske CG, Sundsfjord A, Samuelsen Ø. 2011. A diversity of OXA-carbapenemases and class 1 integrons among carbapenem-resistant Acinetobacter baumannii clinical isolates from Sweden belonging to different international clonal lineages. Microb Drug Resist 17:545–549. doi: 10.1089/mdr.2011.0089. [DOI] [PubMed] [Google Scholar]
- 4.Karah N, Sundsfjord A, Towner K, Samuelsen O. 2012. Insights into the global molecular epidemiology of carbapenem nonsusceptible clones of Acinetobacter baumannii. Drug Resist Updat 15:237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Ramírez MS, Morales A, Vilacoba E, Márquez C, Centrón D. 2012. Class 2 integrons dissemination among multidrug resistance (MDR) clones of Acinetobacter baumannii. Curr Microbiol 64:290–293. doi: 10.1007/s00284-011-0068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagano M, Martins AF, Machado ABMP, Barin J, Barth AL. 2013. Carbapenem-susceptible Acinetobacter baumannii carrying the ISAba1 upstream blaOXA-51-like gene in Porto Alegre, southern Brazil. Epidemiol Infect 141:330–333. doi: 10.1017/S095026881200074X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramírez MS, Piñeiro S, Centrón D. 2010. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother 54:699–706. doi: 10.1128/AAC.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansson K, Sundstrom L, Pelletier A, Roy PH. 2002. IntI2 integron integrase in Tn7. J Bacteriol 184:1712–1721. doi: 10.1128/JB.184.6.1712-1721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girão VBDC, Martins N, Cacci LC, Coelho-Souza T, Nouer SA, Riley LW, Moreira BM. 2013. Dissemination of Acinetobacter nosocomialis clone among critically ill patients and the environment. J Clin Microbiol 51:2707–2709. doi: 10.1128/JCM.00915-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martins N, Martins IS, de Freitas WV, de Matos JA, Girão VBDC, Coelho-Souza T, Maralhães ACDG, Cacci LC, de Figueiredo MP, Dias RCS, Costa-Lourenço APR, Ferreira ALP, Dalla-Costa L, Nouér SA, Santoro-Lopes G, Riley LW, Moreira BM. 2013. Imported and intensive care unit-born Acinetobacter baumannii clonal complexes: one-year prospective cohort study in intensive care patients. Microb Drug Resist 19:216–223. doi: 10.1089/mdr.2012.0174. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Kong F, Cheng X, Yan B, Du X, Gai J, Ai H, Shi L, Iredell J. 2008. Integron gene cassettes in Acinetobacter spp. strains from South China. Int J Antimicrob Agents 32:441–445. doi: 10.1016/j.ijantimicag.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 12.White PA, McIver CJ, Rawlinson WD. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramírez MS, Quiroga C, Centrón D. 2005. Novel rearrangement of a class 2 integron in two nonepidemiologically related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 49:5179–5181. doi: 10.1128/AAC.49.12.5179-5181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stietz MS, Ramírez MS, Vilacoba E, Merkier AK, Limansky AS, Centrón D, Catalano M. 2013. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I–III. Infect Genet Evol 14:294–301. doi: 10.1016/j.meegid.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 15.Clímaco EC, de Oliveira ML, Pitondo-Silva A, Oliveira MG, Medeiros M, Lincopan N, da Costa Darini AL. 2013. Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in southeast Brazil. Infect Genet Evol 19:127–133. doi: 10.1016/j.meegid.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Turton JF, Kaufmann ME, Glover J, Coelho JM, Warner M, Pike R, Pitt TL. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol 43:3074–3082. doi: 10.1128/JCM.43.7.3074-3082.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramírez MS, Stietz MS, Vilacoba E, Jeric P, Limansky AS, Catalano M, Centrón D. 2011. Increasing frequency of class 1 and 2 integrons in multidrug-resistant clones of Acinetobacter baumannii reveals the need for continuous molecular surveillance. Int J Antimicrob Agents 37:175–177. doi: 10.1016/j.ijantimicag.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Taherikalani M, Maleki A, Sadeghifard N, Mohammadzadeh D, Soroush S, Asadollahi P, Asadollahi K, Emaneini M. 2011. Dissemination of class 1, 2 and 3 integrons among different multidrug resistant isolates of Acinetobacter baumannii in Tehran hospitals, Iran. Pol J Microbiol 60:169–174. [PubMed] [Google Scholar]
- 19.Liu C-C, Tang CY, Chang K-C, Kuo H-Y, Liou M-L. 2014. A comparative study of class 1 integrons in Acinetobacter baumannii. Gene 544:75–82. doi: 10.1016/j.gene.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, Nagao M, Matsumura Y, Matsushima A, Ito Y, Takakura S, Ichiyama S. 2011. Interspecies dissemination of a novel class 1 integron carrying blaIMP-19 among Acinetobacter species in Japan. J Antimicrob. Chemother 66:2480–2483. doi: 10.1093/jac/dkr336. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Roh KH, Lee Y, Chung H-S, Yum JH, Yong D, Lee K, Chong Y. 2013. Clonal change of blaSIM-1-carrying Acinetobacter spp. from 2003 to 2008 in the hospital where it was initially discovered. Microb Drug Resist 19:37–41. doi: 10.1089/mdr.2012.0038. [DOI] [PubMed] [Google Scholar]
- 22.Fonseca ÉL, Freitas FDS, Scheidegger ÉMD, Jacinto T, Vicente ACP. 2011. Class 2 integrons in multidrug-resistant Acinetobacter baumannii circulating in different Brazilian geographic regions. Int J Antimicrob Agents 38:95–96. doi: 10.1016/j.ijantimicag.2011.03.013. [DOI] [PubMed] [Google Scholar]


