Abstract
Genetic factors underlying aging are remarkably conserved from yeast to human. The fission yeast Schizosaccharomyces pombe is an emerging genetic model to analyze cellular aging. Chronological lifespan (CLS) has been studied in stationary-phase yeast cells depleted for glucose, which only survive for a few days. Here, we analyzed CLS in quiescent S. pombe cells deprived of nitrogen, which arrest in a differentiated, G0-like state and survive for more than 2 months. We applied parallel mutant phenotyping by barcode sequencing (Bar-seq) to assay pooled haploid deletion mutants as they aged together during long-term quiescence. As expected, mutants with defects in autophagy or quiescence were under-represented or not detected. Lifespan scores could be calculated for 1199 mutants. We focus the discussion on the 48 most long-lived mutants, including both known aging genes in other model systems and genes not previously implicated in aging. Genes encoding membrane proteins were particularly prominent as pro-aging factors. We independently verified the extended CLS in individual assays for 30 selected mutants, showing the efficacy of the screen. We also applied Bar-seq to profile all pooled deletion mutants for proliferation under a standard growth condition. Unlike for stationary-phase cells, no inverse correlation between growth and CLS of quiescent cells was evident. These screens provide a rich resource for further studies, and they suggest that the quiescence model can provide unique, complementary insights into cellular aging.
Keywords: cellular aging, longevity, quiescence, G0 phase, competitive growth
Aging is the major risk factor for complex pathologies such as cancer, cardiovascular disease, diabetes, and neurodegeneration. Model organisms have been used extensively to study the genetic basis of aging, and several aging-related processes are remarkably conserved from yeast to human (Lopez-Otin et al. 2013). Since the first discovery of lifespan-extending mutations in worms (Kenyon et al. 1993), numerous genes have been uncovered that positively or negatively affect longevity in various model systems (Fontana et al. 2010; Gems and Partridge 2013)
Chronological lifespan (CLS), defined as the time cells survive in a nondividing state, has been useful to study cellular aging in the budding yeast, Saccharomyces cerevisiae (Longo et al. 2012). To this end, researchers typically assay survival during stationary phase after exhaustion of glucose, and several genome-wide screens for S. cerevisiae CLS mutants have been performed (Powers et al. 2006; Fabrizio et al. 2010; Matecic et al. 2010). The distantly related fission yeast, Schizosaccharomyces pombe, provides an emerging, complementary model for cellular aging. In S. pombe, chronological aging has also been studied in stationary-phase cultures limited by glucose, a condition in which cells mostly arrest in G2 phase and die within a few days; several genetic and environmental factors affecting CLS during stationary phase have been reported (Roux et al. 2006; Roux et al. 2009). Three aging screens have been performed in S. pombe: a chemical screen has uncovered compounds that extend CLS (Stephan et al. 2013); a genetic screen has identified four genes whose overexpression results in extension of CLS (Roux et al. 2010); and another genetic screen has identified deletion mutants resistant to TORC1-dependent growth inhibition, which included 26 mutants with altered CLS (Rallis et al. 2014).
When S. pombe cells are deprived of nitrogen in the absence of any mating partner, they reversibly arrest in a differentiated G0-like state, called quiescence (Yanagida 2009; Marguerat et al. 2012; Sajiki et al. 2009; Takeda et al. 2010). The Yanagida laboratory has pioneered studies of quiescent S. pombe cells, including genetic analyses of quiescence entry, short-term maintenance, and exit; unlike stationary-phase cells limited for glucose, quiescent cells remain metabolically active by recycling nitrogen and can survive for several weeks if glucose remains available (Shimanuki et al. 2013; Yanagida 2009). Such quiescent cells are thus physiologically adapted for long-term survival and may therefore provide a distinct, complementary model system to study chronological aging.
Here, we apply Barcode sequencing (Bar-seq) (Smith et al. 2009; Han et al. 2010) to analyze the lifespans of 2847 haploid prototroph gene deletion mutants in S. pombe (77.7% of all nonessential deletion mutants) (Kim et al. 2010), because they age together in a pool in a quiescent state without nitrogen. We provide CLS data for both wild-type and mutant strains during long-term quiescence. We focus on mutants with longer CLS than wild-type and independently verify 30 of those mutants. Using Bar-seq, we also profile the proliferation of the deletion mutants growing competitively in a pool and explore the relationship between growth and lifespan.
Materials and Methods
Construction of prototroph deletion strain library
The auxotrophic ade6-M216 (or ade6-M210) ura4-D18 leu1-32 markers of the Bioneer deletion library (Kim et al. 2010) rendered it unsuitable to screen for CLS under nitrogen-depleted conditions. We therefore applied the principle of S. pombe SGA (Baryshnikova et al. 2010) to cross out all auxotrophic markers from the Bioneer v2.0 library; thus, we obtained a prototroph deletion library. To this end, the haploid v2.0 deletion mutants were crossed with the 972 h− strain on SPA plates and left to sporulate at 25° for 2 d. The plates were transferred to 42° for 3 d to eliminate vegetative cells. Spores were then transferred to yeast extract with supplements (YES) medium and left to germinate for 2 d. The library was then successively spotted on Edinburgh minimal medium (EMM; Formedium) to select for prototrophs and on YES medium with G418 to select for the kanMX4 cassette used for generating deletions. Altogether, we performed three rounds of EMM and YES+G418 selection. Because leu1-32 is strongly linked to the mating-type locus, the selection for prototrophs also led to a selection for h− strains. We estimated that <1% of the segregants were h+ cells, based on quantitations of mating frequency in the mutant pool. In the process of converting the library to prototrophs, some mutants have been lost due to genetic linkage. We therefore ended up with 2847 mutants in the prototroph pool instead of 3004 in the original library
Generation of pooled deletion strain library
Using the RoToR robot (Singer Instruments), we compacted the prototroph deletion library into 9 plates at 384 colonies per plate. Strains were grown for 2 d on YES plates containing ampicillin, kanamycin and G418. Colonies were then washed off the plates with 5 ml of 20% (v/v) glycerol in YES per plate, pooled together, aliquoted, and stored at −80°. Pooled library cultures grown at 32° to OD600 0.6 in EMM without nitrogen were tested for spores. To this end, 5% (w/v) SDS was added to the cells following incubation at 30° for 30 min before plating onto YES agar plates; growth was not observed after 3 d of incubation at 32°. This result indicated absence of spores, reflecting that the pool almost exclusively consists of h− cells (see above).
We examined the prototroph mutant pool using FACS and visual analyses of proliferating mutant cells: in both EMM and YES media the cells were mainly in G2 phase, and there was no indication of diploid cells (data not shown). Some auxotroph mutants were included in the supposedly prototroph library. We therefore examined the original (not prototroph) deletion mutants of lys7, his1, ura1, ade9 whose barcodes were detected in the prototroph pool. These deletion strains did not grow when streaked on corresponding selective plates, suggesting that these deletion mutants are auxotrophic. However, when using the RoToR for plating (as used when making the prototroph library), some residual growth of the prototroph mutants was evident on selective media. This result suggests that when a cell mass is printed onto selective media (without corresponding supplements, but not in the case for antibiotics), residual growth can lead to a slight increase in biomass. This phenomenon suggests a community effect with nutrient sharing or colony compartmentalization as seen for budding yeast (Cap et al. 2012). Alternatively, or in addition, some auxotrophic mutants might be leaky and allow some growth, especially when plated using the RoToR.
Chronological lifespan assays during long-term quiescence
We thawed 250 μl of the prototroph library pool, inoculated in 250 ml YES medium, and left it at 25° for 16 hr without shaking. We then centrifuged 50 ml from the culture at 420×g for 2 min, washed the cell pellet twice with 50 ml EMM, resuspended the cells in 250 ml EMM, and grew them overnight at 30° with shaking (130 rpm) up to OD600 ∼0.5. The cells were then collected by centrifugation at 420×g for 2 min, washed twice in an equivalent volume of pre-warmed EMM without nitrogen (EMM-N), and resuspended in 800 ml EMM-N. The OD600 was adjusted to 0.15, and the cells were grown for 2 d at 32° with shaking (130 rpm) to a final OD600 ∼0.6. This condition was used as the reference timepoint (t = 0) after cells stopped to proliferate. We then collected 50–100 ml of cell culture once per week (until viability was <10%) by centrifugation at 960×g for 3 min, one wash in 0.5 ml EMM-N, and resuspension in 0.5 ml EMM-N+0.5 ml 50% (v/v) glycerol. Cells were then stored at −80° until use for DNA extraction and generation of the Bar-seq libraries.
The medium of the quiescent cultures was replaced twice per week for the first 4 wk, and once per week thereafter to avoid depletion of glucose and other nutrients as well as accumulation of toxic by-products from metabolism. When initially replacing the medium less than twice per week, cell viability did decrease to ∼40–50% within 4 wk. To determine cell viability during long-term quiescence, cells were counted using a Beckman Coulter counter, diluted to ∼200 cells per 100 μl of YES, and 100 μl of diluted cells were plated on 3 YES plates each (300 μl total). After 3 d of incubation at 32°, the colonies on the plates were counted and viability was calculated for each timepoint relative to the reference timepoint. Two independent biological replicates of the deletion pool and individual strains were assayed.
To determine chronological lifespans of individual mutants compared with wild-type, 40 ml cultures were prepared in EMM-N as described. Samples were taken at weekly intervals, serially diluted (OD600 0.1, 0.01, 0.001), and 5 µl of diluted cell suspensions were spotted onto YES plates. Cells were left to grow at 32° for 3 d before photos were taken.
Re-growth of quiescent cells
For the re-growth CLS screen, 100 µl aliquots from quiescent cells stored at −80° in glycerol were spread on YES plates and left to grow at 32° for 1 d (0, 4, 8 wk in quiescence) or 2 d (12, 14 wk in quiescence) at 32°. Cells were scraped from the agar with 1 ml YES and the suspension was transferred to a 1.5-ml tube and centrifuged at 960×g for 1 min. These cell pellets were used for DNA extraction and barcode sequencing.
Genomic DNA extraction and Bar-seq library preparation
The MasterPure Yeast DNA purification kit (Epicentre) was used for DNA extraction. Barcodes were amplified with Ex Taq DNA polymerase (TaKaRa) using two rounds of PCR as previously described (Han et al. 2010). The following primers were used for the first round of PCR. For amplification of uptags, forward primer 5′-CACGACGCTCTTCCGATCTxxxxGAGGCAAGCTAAGATATC-3′ and reverse 5′-AGCAGAAGACGGCATACGAGATATTGGCGTGACTAGTTCAGACGTGTGCTCTTCCGATCT GCCTTACTTCGCATTTA-3′ reverse. For amplification of downtags, forward 5′- CACGACGCTCTTCCGATCTxxxxCCAGTGTCGAAAAGTATC-3′ and reverse primer 5′-AGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTTGCGTTGCGTAGG-3′. The xxxx within the sequence represents four unique nucleotide indexes that allowed us to multiplex for sequencing (index sequences available on request). We used 10 pmol per primer for each reaction and the following program for the thermocycler: 94°/4 min, (94°/20 sec, 53°/20 sec, 72°/20 sec) × 30 cycles, 72°/20 sec. For the second round of PCR, the forward primer was 5′- AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and the reverse primers was 5′-AGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′. The program used for the thermocycler for the second round of PCR was as follows: 94°/4 min, (94°/20 sec, 56°/20 sec, 72°/20 sec) × 20 cycles, 72°/20 sec. The PCR products were run on 1.5% (w/v) agarose gels, and the bands were excised and purified using the QIAquick gel extraction kit (Qiagen). The amplified and purified barcodes were quantified using the Qubit Fluorometer (Invitrogen) and Bioanalyzer (Agilent). The samples were then mixed together in equal molar ratios and used for Illumina sequencing using either MiSeq (MiSeq v2 Reagent kit) or HiSequation 2000 (for re-growth screen) instruments. Fifty cycles of single-end sequencing were used, with 20 multiplexed samples for each library.
Barcode decoding
The barcode sequences of the Bioneer v2 library were experimentally characterized by high-throughput sequencing. Using a primer extension procedure used previously for paired-end-sequencing-based decoding (Han et al. 2010), we amplified the barcode sequences together with the flanking genomic DNA sequences from the genomic DNA of pooled mutants. The PCR products were single-end sequenced on a MiSeq instrument for 101 cycles. Each MiSeq sequencing read contains a 4-bp multiplexing index, 18-bp PCR primer sequence, 20-bp barcode, 26-bp universal spacer, and 33-bp flanking genomic sequence. After grouping reads with the same barcode sequence together, their flanking genomic sequences were compared with the expected flanking sequences obtained from the file http://pombe.kaist.ac.kr/nbtsupp/download/All_primers%20used_in_gene%20deletion.xls (Kim et al. 2010). Our analysis identified barcodes for 2473 genes. Among them, 1871 genes have both uptag and dntag decoded, 254 genes have only uptag decoded, and 348 genes have only dntag decoded. The decoded barcodes are listed in Supporting Information, Table S1 and used in the data analysis.
Data analysis
The decoded barcodes for Bioneer v2 were combined with the barcodes for Bioneer v1 (Han et al. 2010) to maximize the set of mutants that can be interrogated. Uptags and dntags, together with universal primers and 20 multiplex index sequences, were used to construct a FASTA database, in which each gene had 20 sequence entries of exactly 51 bp (multiplex index, 4 bp; universal primer, 18 bp; up/downtag, 20 bp; universal primer, 9 bp). This database contains 98,280 sequence entries and is available on request. To identify and quantify sequence reads per mutant in a given sample, raw sequence data were aligned against the database using Bowtie 0.12.7 (Langmead et al. 2009); only a single mismatch within the first 42 bp were tolerated, and only unique matches across the entire database were retained. Two independent biological replicates were performed, exploiting the variation to assess statistical significance (Robinson et al. 2014). Timepoints for which we obtained less than 10 reads were excluded from further analysis. The number of reads for each mutant in a timepoint “i” was normalized for the sequencing depth of the same timepoint (reads per mutanti/depthi). Fold changes relative to the reference timepoint (reads per mutant0/depth0) were determined. To identify CLS mutants, a lifespan score was calculated. The average of weighted up-tags and down-tags for each timepoint was calculated as previously described (Han et al. 2010; Sun et al. 2013). To weight a barcode at a specific timepoint i, we determined the sum of reads for that barcode at timepoint i and reference (timepoint 0) divided by the sum of reads of both barcodes at timepoints 0 and i. For example, for the uptag at timepoint 8 wk, the weighted value was: (Up8+Up0)/(Up8+Dn8+Up0+Dn0). Then, the median of all timepoints (except t = 0, reference) was determined and the average for the two biological replicates was defined as the lifespan score. The lifespan score represents the median of all fold-changes throughout the time course for the two replicates. Growth scores were calculated as follows. The number of reads for each mutant in a timepoint “i” was normalized for the sequencing depth of the same sample (reads per mutanti/depthi). Fold changes relative to the reference timepoint at 120 min (reads per mutant120/depth120) were then determined. The average of weighted up-tags and down-tags for each timepoint was calculated as described (Han et al. 2010; Sun et al. 2013), and the median of all timepoints, except t = 120 min (reference timepoint), was determined. Then, the average of both biological repeats was determined and defined as the growth score, which thus represents the median of all fold-changes throughout the time course.
Classification and verification of mutants
Genetic interactions were analyzed using the S. pombe genetic interactome tool (Ryan et al. 2012). Diagnostic PCRs for both deletion junctions were performed to confirm deletion of the correct gene for 30 random deletion mutants that showed CLS phenotypes. PCR primer sequences were as reported (Kim et al. 2010). The MasterPure Yeast DNA purification kit (Epicentre) was used for DNA extraction and the following program for the thermocycler: 94°/2 min, (94°/30 sec, 53°/30 sec, 72°/90 sec) × 30 cycles, 72°/7 min.
Results and Discussion
Lifespan of wild-type cells during quiescence in absence of nitrogen
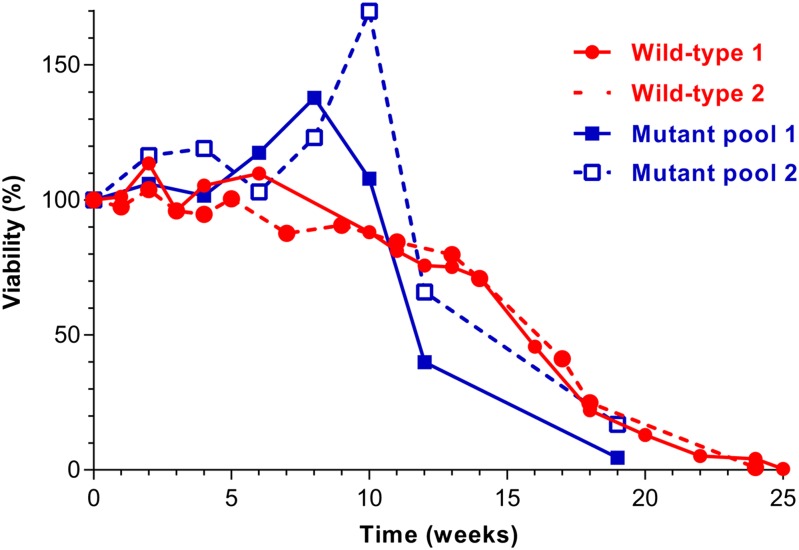
We first determined the CLS of wild-type fission yeast during long-term quiescence in the absence of nitrogen by determining cell viability in weekly intervals (Figure 1, red curves). Cell viability remained above 70% for approximately 14 wk, followed by a stronger decrease over the next month, but only after 24–25 weeks did viability decrease below 1%. The median CLS was approximately 16.6 wk (polynomial regression/order 2). This long CLS is in stark contrast to stationary-phase fission yeast cells limited by glucose, which survive for less than 1 wk under standard conditions and up to ∼18 days with low initial glucose concentration (Roux et al. 2009). This difference indicates that fission yeast can cope much better with nitrogen deprivation than with glucose deprivation, which might suggest that the ecological conditions encountered by wild strains are not normally limited for glucose. It has been proposed that the natural environment of both fission and budding yeasts contains abundant carbon sources but variable nitrogen sources (Weisman 2010; Backhus et al. 2001). Moreover, the CLS determined here is much longer than for quiescent budding yeast cells in the absence of nitrogen, which show greatly reduced viability after 12 d (Dziedzic and Caplan 2012). We therefore propose that long-term quiescence of fission yeast cells can serve as a complementary condition to study chronological aging. This condition could provide insight into aging of terminally differentiated postmitotic cells or for stem cells that alternate between proliferating and quiescent states.
Figure 1.
CLS assays during long-term quiescence under nitrogen deprivation. Two independent biological repeats each for wild-type and for deletion mutant pool were performed. Viability was determined from colony-forming units (CFUs) on plated cells. The CFUs determined at timepoint 0 was defined as 100% survival, and survival at the other timepoints was determined relative to this reference timepoint. For the Bar-seq screen, the mutant-pool experiments shown here were used. The timepoints at which pooled mutants were harvested for Bar-seq analysis are indicated in blue on the X-axis.
Lifespan of prototrophic deletion pool during quiescence
The Bioneer v2.0 haploid deletion library contains 3004 nonessential mutants in an auxotroph ade6-M216 leu1-32 ura4-D18 background (Kim et al. 2010). These mutants therefore need to be grown in medium supplemented with adenine, leucine, and uracil, which would provide a nitrogen source during long-term quiescence. To screen for deletion mutants with altered CLS in the absence of nitrogen, we therefore needed to cross out the auxotrophic markers from all deletion mutants to generate a prototroph deletion library (see Materials and Methods). This back-crossing of all mutants to a wild-type strain also tested for co-segregation of any phenotypes with the deletion marker. All prototroph mutants were then pooled together in preparation for the parallel screening.
We next monitored overall CLS of the prototrophic deletion library pool during quiescence (Figure 1, blue curves). Cell viability remained at higher levels than for wild-type cells for approximately 10 wk, transiently even surpassing the 100% initial viability of the reference timepoint, followed by a stronger decrease in viability than for wild-type over the next 9 wk. These differences compared with the CLS profiles of wild-type cells could be expected as different mutants will lose viability at different rates in the pool. The transient increase in viability of the pool could reflect that nitrogen sources released from dead mutants allow other mutants to temporarily proliferate again.
Bar-seq screen for deletion mutants with altered lifespans during quiescence
We next screened the 2847 prototrophic deletion mutants in parallel for altered CLS during quiescence in the absence of nitrogen. Using the two independent biological repeats of the mutant pool quiescence experiments shown in Figure 1, we collected samples at timepoints 0, 4, 8, 12, and 14 wk. These samples were then further processed for Bar-seq analysis (see Materials and Methods). This approach takes advantage of the two unique barcode sequences associated with each deletion mutant, called uptag and downtag (Kim et al. 2010). Changes in relative abundance of the mutants as a function of chronological aging can thus be detected by next-generation sequencing of these barcodes. Mutants with shorter CLS than the pool average will become under-enriched, whereas those with longer CLS will become enriched with time relative to the reference timepoint at the beginning of the time course. Such parallel quantitative phenotyping is highly sensitive and efficient (Han et al. 2010; Sun et al. 2013). Barcode decoding for v2 Bioneer was performed in this study (see Materials and Methods), and the barcodes are provided in Table S1.
In studies reporting CLS screens for budding yeast mutants, aging cells have been re-grown before DNA extraction to avoid any noise from dead cells (Fabrizio et al. 2010; Matecic et al. 2010). Here, as our main approach, we chose to directly extract DNA from quiescent cells rather than re-growing them beforehand. This approach has the advantage that the measured proportion of the mutants represents their proportion in the pool at time of sample collection, without any bias being introduced by differences in quiescence exit or growth rate among the mutants. DNA is expected to become rapidly degraded on cell death and therefore should not substantially affect the assay. Moreover, it is less likely that DNA from nonlysed dead cells was present in our samples, because the time interval between timepoints was 2–4 wk rather than a few days as in the budding yeast studies. Nevertheless, we also performed one CLS screen (two biological replicates) with cells that were re-grown before DNA extraction and Bar-seq analysis. The two complementary approaches showed good agreement (see below).
Of the 2847 prototroph mutants, we detected 75% and 65% with ≥1 or ≥10 sequence reads, respectively, at timepoint zero for each of the replicates. Seventy percent of sequencing reads could be mapped to the decoded barcodes (10,555,488 reads out of a total of 15,087,758 processed reads over all 20 samples, including 5 timepoints, 2 tags, and 2 repeats). The deletion junction information was downloaded from the KAIST website. The barcodes that failed to be detected may have deletion junctions different from those provided by KAIST for the following reasons: the sequences could have been altered due to oligonucleotide synthesis errors or in vivo mutations, or some strains (especially those that have been upgraded) could have been created with primers different from the original design. Such discrepancies have also been observed when decoding the v1.0 haploid deletion library (Han et al. 2010).
The number of sequencing reads for the uptags always correlated with those of the downtags for all samples (rPearson ∼0.64–0.95). The sequencing counts for each mutant are provided in Table S2. We obtained at least one barcode read for 2563 deletion mutants and could determine reliable CLS data for 1199 of these mutants (Table S3).
Reassuringly, barcode reads for deletion mutants in genes reported to be necessary for entry into or maintenance of quiescence (Sajiki et al. 2009) were either absent or present at very low numbers (<10) throughout the time course (Table S2). A similar pattern was evident for all autophagy mutants; although some of these mutants were detected at the reference timepoint and in the pools of proliferating cells, the counts decreased dramatically from 4 wk onwards (Table S2). This finding was expected because autophagy plays important roles for lifespan during quiescence in S. pombe (Kohda et al. 2007) and other organisms (Markaki and Tavernarakis 2013).
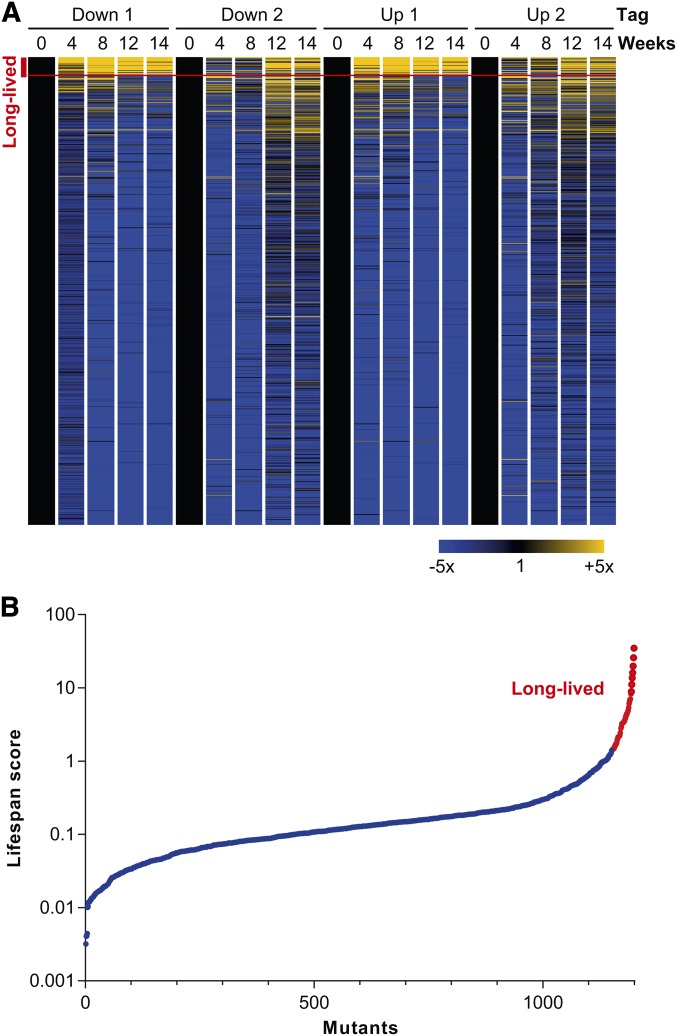
Many mutants decreased in abundance over the time course relative to the reference timepoint (Figure 2A). The 10% of mutants with the lowest lifespan scores are provided in Table S4. Several different deficiencies and indirect effects, not necessarily related to chronological aging, could underlie these phenotypes. However, the few mutants that increased over the time course relative to the reference timepoint (Figure 2A), and therefore have longer lifespans than the pool average, are more likely to directly interfere with the aging process. We therefore focused the analysis and discussion below on these long-lived mutants (Table 1). The data for the alternate CLS screen with cells re-grown before Bar-seq analysis are provided in Table S5 (sequence counts per mutant in each sample) and in Table S6 (long-lived mutants unique for re-growth screen).
Figure 2.
CLS profiling and lifespan scores of deletion mutants. (A) CLS profiles showing fold-changes for the two barcode tags relative to the reference timepoint (t = 0). The columns show the data for the two biological repeats for the downtags (Down 1 and 2) and uptags (Up 1 and 2) at the five timepoints analyzed (0–14 wk). The rows correspond to the 1199 deletion mutants for which lifespan scores could be determined. The fold changes in tag abundance relative to timepoint 0 are color-coded as indicated at bottom right. The mutants are ordered by lifespan scores, with highest scores at top. The 48 mutants defined as "long-lived" (Table 1) are indicated with a red bracket at top left. The heat map was generated with GeneSpring (Agilent). (B) Plot of ordered lifespan scores for 1199 deletion mutants (see Materials and Methods). The 48 long-lived mutants are indicated in red.
Table 1. Long-lived mutants.
| Mutant | Product Description in PomBase (Wood et al. 2012) | Verified Single Mutants | Long-Lived in Re-growth Screen | GO Cellular Component (Blake et al. 2013) |
|---|---|---|---|---|
| SPBC1198.07c | Mannan endo-1,6-alpha-mannosidase (predicted) | Yes | Yes | Integral component of membrane, endoplasmic reticulum |
| SPBC14C8.15 | Triglyceride lipase-cholesterol esterase (predicted) | Not tested | Yes | Integral component of membrane, Golgi apparatus |
| SPBC18H10.18c | Sequence orphan | Yes | Yes | Integral component of membrane |
| SPCC594.02c | Conserved fungal protein | Yes | Yes | Integral component of membrane, endoplasmic reticulum |
| SPBC30D10.09c | HVA22/TB2/DP1 family protein | Not tested | Yes | Integral component of membrane, endoplasmic reticulum |
| gyp3 | GTPase activating protein Gyp3 (predicted) | Yes | Yes | Integral component of membrane |
| vps66 | Acyltransferase (predicted) | Yes | Yes | Integral component of membrane, Golgi apparatus, endoplasmic reticulum |
| frp2 | Ferric chelate reductase (predicted) | Yes | Yes | Plasma membrane, endoplasmic reticulum |
| ncs1 | Neuronal calcium sensor-related protein Ncs1 | Yes | Yes | Plasma membrane |
| rho2 | Rho family GTPase Rho2 | Yes | Yes | Plasma membrane |
| SPCC1020.08 | Wybutosine biosynthesis protein Tyw1 (predicted) | Not tested | Yes | Endoplasmic reticulum |
| SPAC23D3.03c | GTPase-activating protein (predicted) | Yes | Yes | Golgi apparatus |
| fta5 | Cell surface glycoprotein | Not tested | No | Golgi apparatus, endoplasmic reticulum |
| vta1 | Vps20-associated protein Vts1 (predicted) | Yes | Yes | |
| SPAC8E11.05c | Conserved fungal protein | Yes | Yes | |
| SPACUNK4.16c | Trehalose-phosphate synthase (predicted) | Yes | Yes | |
| SPBC1921.04c | Dubious | Yes | Yes | |
| SPCC320.03 | Transcription factor (predicted) | Yes | Yes | |
| SPCC594.01 | DUF1769 family protein | Yes | Yes | |
| abp2 | ARS binding protein Abp2 | Not tested | Yes | |
| apl1 | AP-2 adaptor complex subunit Apl1 (predicted) | Yes | Yes | |
| aps2 | AP-2 adaptor complex subunit Aps2 (predicted) | Yes | Yes | |
| clr4 | Histone H3 lysine methyltransferase Clr4 | Not tested | No | |
| cmb1 | Cytosine-mismatch binding protein 1 | Not tested | No | |
| cmk2 | MAPK-activated protein kinase Cmk2 | Not tested | Yes | |
| apl3 | AP-2 adaptor complex subunit Alp3 (predicted) | Yes | Yes | |
| SPBC947.09 | ThiJ domain protein | Not tested | No | |
| sds23 | Inducer of sexual development Sds23/Moc1 | Yes | Yes | |
| SPCC306.11 | Sequence orphan | Not tested | No | |
| gpx1 | Glutathione peroxidase Gpx1 | Not tested | No | |
| hht1 | Histone H3 h3.1 | Yes | Yes | |
| mni1 | Mago Nashi interacting protein Mni1 (predicted) | No | Yes | |
| mrc1 | Mediator of replication checkpoint 1 | Yes | Yes | |
| msh3 | MutS protein homolog 3 | Yes | Yes | |
| mug161 | CwfJ family protein, splicing factor (predicted) | Not tested | Yes | |
| osr1 | Short chain dehydrogenase (predicted) | Not tested | No | |
| SPBC4B4.12c | Sequence orphan | Not tested | No | |
| pek1 | MAP kinase kinase Pek1 | Not tested | Yes | |
| plb1 | Phospholipase B homolog Plb1 | Yes | Yes | |
| dad2 | DASH complex subunit | Not tested | No | |
| mug80 | Cyclin Clg1 (predicted) | Not tested | Yes | |
| set9 | Histone lysine methyltransferase Set9 | Yes | Yes | |
| spo4 | Serine/threonine protein kinase Spo4 | Yes | Yes | |
| tfs1 | Transcription elongation factor TFIIS | Yes | Yes | |
| tif213 | Translation initiation factor eIF2 gamma subunit (predicted) | Yes | Yes | |
| gsk3 | Serine/threonine protein kinase | Yes | Yes | |
| SPCC794.03 | Amino acid permease (predicted) | Yes | Yes | |
| SPBC8E4.02c | Sequence orphan | Yes | Yes |
We computed a lifespan score for each mutant that represents the proportion of each mutant in the pool across the time course relative to the reference timepoint (see Materials and Methods). Lifespan scores could be computed for 1199 mutants; these scores are plotted in Figure 2B, with values provided in Table S3. We identified 48 long-lived mutants, defined as those with lifespan scores >1.44 (Figure 2B; Table 1). Using the same method to determine long-lived mutants, the re-growth CLS screen also identified 39 (81%) of the 48 mutants in Table 1, in addition to another 55 long-lived mutants (Table S6). The two approaches therefore showed generally good agreement. The long-lived mutants were not enriched for any Gene Ontology (GO) categories. We exploited a genetic interactome study for fission yeast (Ryan et al. 2012) to examine the functional relationships among the long-lived mutants. For 35 out of 48 long-lived mutants, genetic interaction data were available. Notably, 25 out of these 35 genes showed at least one genetic interaction with other genes in our list of long-lived mutants. This result indicates that the genes identified by the screen show substantial functional coherence.
Verification of CLS mutants
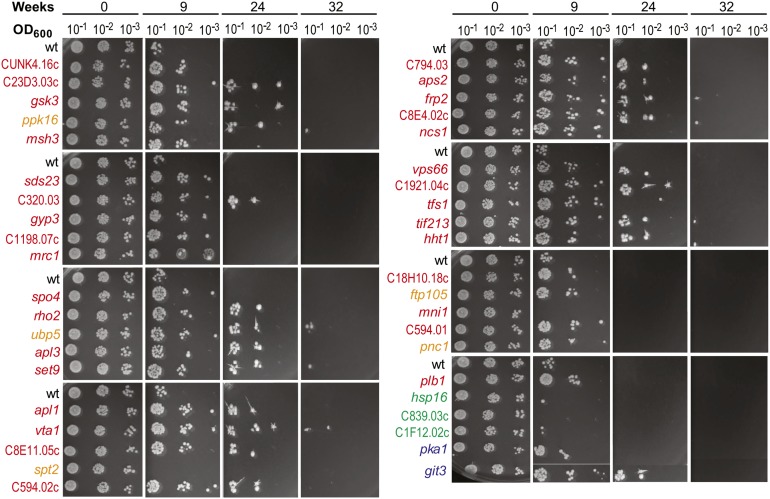
We performed independent lifespan assays for selected mutants when aged as individual cultures rather than in a competitive pool. The individual mutant cells, along with wild-type reference cells, were shifted to a nitrogen-depleted minimal medium, and survival was monitored every week by spotting serial dilutions onto agar plates. We randomly selected and tested 31 out of the 48 long-lived mutants for extended CLS compared with wild-type (Figure 3, red). All except 1 (mni1) of these 31 mutants were also long-lived when assayed as individual cultures: growth was detected in the higher dilutions at 9 wk (where wild-type cells did not grow), and even after 24 and 32 wk in some cases. Note that in the quantitative assay of Figure 1, wild-type cells largely remained viable for 9 wk, whereas in the qualitative assay of Figure 3 wild-type cells were largely dead at 9 wk. In the quantitative assay, cells were plated singly on rich media, whereas in the qualitative assay cells were spotted at high density. We speculate that long-term quiescent cells can revive better without competition by neighboring cells.
Figure 3.
Independent verification of CLS for selected mutants in individual cultures. Mutant and wild-type (wt) control cells were assayed at 0, 9, 24, and 32 wk after entering quiescence in the absence of nitrogen. As indicated on top, 10-fold serial dilutions from each timepoint were spotted on agar plates for each culture. The mutant strains are color-coded as follows: red, long-lived mutants (Table 1); orange, mutants with high lifespan scores ∼0.4-1.2; green, lifespan score <0.14; blue, two mutants known to be long-lived during quiescence but for which no lifespan score could be computed.
We verified that the correct genes were deleted for the 30 confirmed long-lived mutants by PCR testing for the predicted genomic-marker junctions created by gene deletion. Deletion for three mutants (SPBC8E4.02c, rho2, gsk3) could not be confirmed at both the 5′ and 3′ junctions, and for another three mutants (vsp66, aps2, SPACUNK4.16c) we could only confirm one of the two junctions. Although not conclusive, these results raise some doubts about the nature of these deletion mutants.
We also independently tested three random mutants (hsp16, SPBC839.03c, SPAC1F12.02c) with lifespan scores of <0.14, indicative of short-lived or sick mutants (Figure 3, green). All these mutants had largely lost viability at 9 wk and thus were somewhat shorter-lived than wild-type. In addition, five mutants (ppk16, ubp5, ftp105, spt2, pnc1) with lifespan scores between approximately 0.4 and 1.2 were randomly selected to test for any long-lived mutants, relative to wild-type, among mutants that did not make it onto the long-lived list but were among the top 15% with respect to lifespan score (Figure 3, orange). Notably, four of these five mutants (ppk16, ubp5, ftp105, pnc1) were longer-lived than wild-type in individual cultures. This result suggests that many mutants with lower lifespan scores can feature longer CLS compared with wild-type. Finally, we also tested the two mutants pka1 and git3, for which no lifespan scores could be computed but that exhibit extended lifespan with limiting glucose during stationary phase (Roux et al. 2006). Both of these mutants also exhibited extended lifespan in our CLS assay (Figure 3, blue).
Comparison of CLS mutants to published data
In budding yeast, three CLS screens of either individual or pooled stationary-phase cultures after glucose exhaustion have been reported (using microarrays instead of sequencing to quantify barcodes) (Powers et al. 2006; Fabrizio et al. 2010; Matecic et al. 2010). None of the orthologs of our 48 long-lived mutants have been identified in these budding yeast screens, which might indicate differences between the two chronological aging models and/or the two organisms. Notably, however, only five mutants (far7, mds3, dcw1, puf3, trk1) have been identified in two of the budding yeast screens, and no mutant has been identified in all three screens (Fabrizio et al. 2010; Matecic et al. 2010; Powers et al. 2006). However, several types of genes identified here also exhibit pro-aging effects in other systems. Deletion of tif213, encoding a subunit of the translation initiation factor eIF2, results in extended lifespan in our screen, similar to budding yeast mutants deficient in translation initiation activities (Bitterman et al. 2002). Two other regulatory genes corresponding to long-lived mutants in our screen, tfs1 and spo4, encode a transcription elongation factor and serine/threonine kinase, respectively, as do the long-lived worm mutants SPT4 and AKT1 (Hamilton et al. 2005). Tfs1 affects chromatin patterns via global RNA polymerase II transcription (Reyes-Turcu et al. 2011). The histone methyltransferase Set9 exhibits pro-aging effects both in our system and in worms (Hamilton et al. 2005), as does the histone methyltransferase Set2 in budding yeast (Ryu et al. 2014). The predicted product of SPAC23D3.03c, a GTPase-activating protein whose absence extends lifespan in our system, is involved in vesicle-mediated transport. The PP2A-type phosphatase inhibitor Sds23 exhibits pro-aging effects both in our system and in budding yeast (Breslow et al. 2008). In contrast, Sds23 has also been reported to have anti-aging effects in fission yeast, because its overexpression extends lifespan of glucose-depleted cells during stationary phase (Roux et al. 2010). The gsk3 mutant, whose gene encodes glycogen-synthase kinase 3, was among the most long-lived in our study, and inhibition of the orthologous GSK-3 gene ameliorates a fly model of Alzheimer’s disease (Sofola et al. 2010). Gsk3 kinase promotes protein translation (Shin et al. 2011). There is a rich literature on protein translation and aging (Delaney et al. 2011) showing that reduced translation increases lifespan (Rallis et al. 2013; Selman et al. 2009).
Many of our long-lived mutants correspond to proteins associated with membranes. The following proteins are integral membrane proteins: Vps66, Gyp3, SPBC1198.07c, SPBC14C8.15, SPBC18H10.18c, SPCC594.02c, and SPBC30D10.09c. Three other proteins localize to the plasma membrane (Rho2, Ncs1 and Frp2), four proteins localize to the Golgi complex (Vps66, Fta5, SPAC23D3.03c and SPBC14C8.15), and eight proteins localize to the endoplasmic reticulum (Vps66, Fta5, SPBC1198.07C, SPCC1020.08, Frp2, SPCC594.02c, SPBC30D10.09c) (Matsuyama et al. 2006). Mutations in genes involved in the secretory pathway of the endoplasmic reticulum extend replicative lifespan in budding yeast (Labunskyy et al. 2014). SPACUNK4.16c is a predicted trehalose phosphate synthase, and the budding yeast tsl1 mutant also exhibits extended chronological lifespan because of diminished trehalose synthesis (Kyryakov et al. 2012).
Several genes seem to have opposite effects on lifespan in budding and fission yeast. In budding yeast, mutants of the adenine biosynthesis pathway are long-lived (Matecic et al. 2010), whereas the corresponding mutants either decrease in abundance or are not detected in our screen (Table S2). The MutS mutants msh2 and msh6 are short-lived in budding yeast (Breslow et al. 2008), whereas the related mismatch-repair-defective msh3 mutant was long-lived in our screen. The gyp1 mutant, lacking a GTPase-activating protein, is short-lived in budding yeast (Giaever et al. 2002), whereas the gyp3 and SPAC23D3.03c GTPase-activating protein mutants are long-lived in our screen. The mkk1 MAP kinase kinase mutant is short-lived in budding yeast (Fabrizio et al. 2010), whereas the orthologous pek1 mutant is long-lived in our screen. The clg1 cyclin mutant is short-lived in budding yeast (Fabrizio et al. 2010), whereas the orthologous mug80 cyclin mutant is long-lived in our screen. Finally, the gpx2 glutathione peroxidase mutant is short-lived in budding yeast (Breslow et al. 2008), whereas the corresponding gpx1 mutant is long-lived in our screen. These opposite effects might reflect differences between the two distantly related yeasts and/or between the two distinct chronological aging models involving glucose depletion in budding yeast and long-term quiescence on nitrogen depletion in fission yeast.
We also identified the following proteins not previously implicated in chronological aging: the Hht1 histone, the Abp2 DNA replication origin-binding protein, the Mrc1 replication-checkpoint mediator, the Cmk2 MAPK-activated protein kinase, the Vps66 acyltransferase (predicted), the Ncs1 calcium sensor, the Tfs1 transcription factor, and the Plb1 phospholipase, along with several orphans.
Notably, our screen did not identify any mutants that are known to be long-lived during stationary phase under glucose limitation (Rallis et al. 2014; Roux et al. 2010; Roux et al. 2009; Roux et al. 2006). The following fission yeast mutants, which are long-lived during stationary phase, were not detected in our screen due to low numbers of sequencing counts: pka1 (cAMP-dependent protein kinase catalytic subunit) (Roux et al. 2006); sck2 (S6 protein kinase) (Roux et al. 2006); and git3 (glucose receptor) (Roux et al. 2009). We therefore independently tested pka1 and git3 mutants for longevity during quiescence. Notably, both mutants showed extended lifespan in individual cultures under nitrogen limitation (Figure 3). These results suggest that there is at least some similarity in the genetic basis underlying extended CLS in the two chronological aging models. The Pka1 and TOR pathways mainly control cellular responses to glucose and nitrogen, respectively, and are known to have common targets. For example, in budding yeast these two pathways converge on a single kinase to control entry to quiescence (Pedruzzi et al. 2003) or independently target processes vital for quiescence maintenance and survival (Stephan et al. 2009).
Growth profiling of deletion mutants by Bar-seq
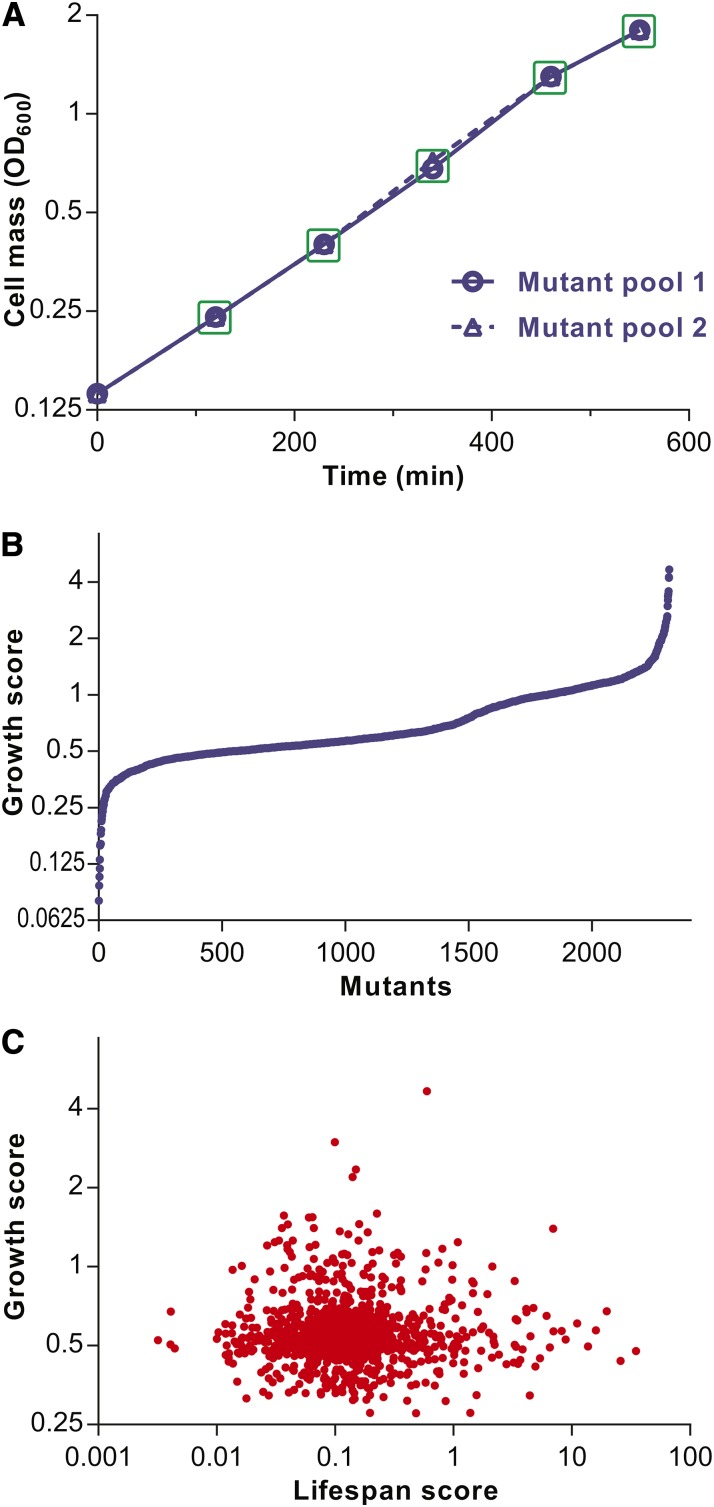
To our knowledge, the S. pombe deletion library has not been specifically analyzed for mutants with differences in proliferation under a standard, nonstress condition. We exploited the sensitive and efficient approach of parallel phenotyping to also compare the proliferation of the deletion mutants grown competitively in the same culture, followed by quantitative Bar-seq analyses. We performed two independent biological repeats of the mutant pool grown exponentially in EMM for 9 hr, with samples for Bar-seq analysis harvested at 5 timepoints between 120 and 550 min (Figure 4A). The sequence counts for each mutant in each sample are provided in Table S7. The sequence counts were normalized to the sequencing depth for each sample, and a growth score was determined for each mutant (see Materials and Methods). Growth scores could be computed for 2314 mutants; these scores are plotted in Figure 4B, with values provided in Table S8. The 231 most rapidly proliferating mutants are listed in Table S9, and the 231 most slowly proliferating mutants are listed in Table S10.
Figure 4.
Growth profiling of deletion mutants and comparison to CLS. (A) Growth curves for two independent biological repeats of deletion mutant pool. The mutant pools were grown in EMM medium to exponential phase and diluted to OD600 0.14 (timepoint 0). The OD600 was measured at 120, 230, 340, 460, and 550 min, and the pooled mutants were harvested at the same timepoints for Bar-seq analysis (green squares). (B) Plot of ordered growth scores for 2314 deletion mutants (see Materials and Methods). (C) Plot of growth scores vs. corresponding lifespan scores of 1193 mutants for which both scores could be determined. Growth and lifespan scores show no significant correlation (rPearson = −0.009; P = ∼0.75).
We looked for functional enrichments among the top 10% and 20% (231 and 463 genes, respectively) most slowly or most rapidly proliferating deletion mutants. The most slowly growing mutants were significantly enriched for the following categories (P < 0.02): conserved genes (Wood et al. 2002); core environmental stress response (CESR) genes that are downregulated during stress (Chen et al. 2003); ribosomal protein genes; top 500 most highly expressed genes (Wilhelm et al. 2008); and the GO category "macromolecule metabolism" (Blake et al. 2013), among others. These enrichments are expected for genes with important roles in cell growth. In budding yeast, mutants lacking ribosomal proteins have been identified as slow growers in a screen of individually growing mutants (Yoshikawa et al. 2011), and the expression of genes encoding ribosomal and macromolecule metabolic functions are induced during rapid growth (Castrillo et al. 2007; Brauer et al. 2008) . Also, in fission yeast ribosomal proteins are required for normal growth rate (Kim et al. 2010).
The most rapidly growing mutants were significantly enriched for the following categories (P < 0.02): genes without orthologs (Wood et al. 2012); CESR genes that are upregulated during stress (Chen et al. 2003); top 500 most lowly expressed genes (Wilhelm et al. 2008); and nitrogen-starvation response genes (Mata et al. 2002), among others. These categories are often the opposite of the categories for slowly growing mutants. Cells coordinate gene regulation with the growth rate to accommodate their physiological needs; under good conditions, cells grow rapidly but are stress-sensitive, whereas under poor conditions cells grow slowly or not at all (quiescence) but become stress-resistant (Lopez-Maury et al. 2008). This balance between cellular growth and maintenance is reflected in two large, antagonistically regulated gene expression programs in fission yeast, the CESR upregulated and downregulated genes (stress-related and growth-related genes, respectively) (Chen et al. 2003; Lopez-Maury et al. 2008; Pancaldi et al. 2010). It is interesting that deleting genes that are upregulated during stress or starvation leads to higher growth rates, which might reflect an evolutionary tradeoff between proliferation and stress protection. This finding also suggests that some of these stress-related genes are directly involved in inhibiting cell growth.
There are some indications that growth and lifespan are inversely correlated. For example, slow growth achieved by temperature manipulations results in extended lifespan of fish (Lee et al. 2013), and slow growth before entry into stationary phase is associated with longevity during stationary phase in fission yeast (Rallis et al. 2013). We therefore looked into the relationship between lifespan and growth in our data by plotting the growth scores and the corresponding lifespan scores for all mutants where both scores were available (Figure 4C). No significant inverse correlation was evident between lifespan and growth. Because many short-lived mutants might not be directly involved in chronological aging, we repeated the correlation analysis only for the 152 mutants with lifespan scores >0.4. Again, no significant inverse correlation between lifespan and growth was evident (rPearson = −0.03; P = ∼0.73). We conclude that slow growth is not associated with extended lifespan in our quiescence model, in contrast to the stationary-phase model. This difference could be rationalized by the extended time interval between the growth and quiescence phases (weeks or months), whereas the interval between the growth and stationary phases is much smaller (days). Quiescence might therefore represent a differentiated state that is not affected by the physiology during exponential growth before entry into quiescence. This finding further argues for the distinctiveness of the quiescence model, which might provide unique insights into chronological aging.
Supplementary Material
Acknowledgments
We thank Sandra Codlin for help with the MiSeq sequencing and Antonia Lock for critical reading of the manuscript. This research was funded by a BBSRC Research Grant (grant number BB/I012451/1) and a Wellcome Trust Senior Investigator Award (grant number 095598/Z/11/Z) to J.B., and by grants to L.-L.D from the Chinese Ministry of Science and Technology and the Beijing Municipal Government.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014415/-/DC1
Data deposited to European Nucleotide Archive (ENA), accession number PRJEB6977 (secondary accession ERP006656).
Communicating editor: C. S. Hoffman
Literature Cited
- Backhus L. E., DeRisi J., Bisson L. F., 2001. Functional genomic analysis of a commercial wine strain of Saccharomyces cerevisiae under differing nitrogen conditions. FEMS Yeast Res. 1: 111–125. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Dixon S., Vizeacoumar F. J., Myers C. L., et al. , 2010. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 470: 145–179. [DOI] [PubMed] [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A., 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277: 45099–45107. [DOI] [PubMed] [Google Scholar]
- Blake J. A., Dolan M., Drabkin H., Hill D. P., Li N., et al. , 2013. Gene Ontology annotations and resources. Nucleic Acids Res. 41: D530–D535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Huttenhower C., Airoldi E. M., Rosenstein R., Matese J. C., et al. , 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., et al. , 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cap M., Stepanek L., Harant K., Vachova L., Palkova Z., 2012. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell 46: 436–448. [DOI] [PubMed] [Google Scholar]
- Castrillo J. I., Zeef L. A., Hoyle D. C., Zhang N., Hayes A., et al. , 2007. Growth control of the eukaryote cell: a systems biology study in yeast. J. Biol. 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Toone W. M., Mata J., Lyne R., Burns G., et al. , 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14: 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J. R., Murakami C. J., Olsen B., Kennedy B. K., Kaeberlein M., 2011. Quantitative evidence for early life fitness defects from 32 longevity-associated alleles in yeast. Cell Cycle 10: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziedzic S. A., Caplan A. B., 2012. Autophagy proteins play cytoprotective and cytocidal roles in leucine starvation-induced cell death in Saccharomyces cerevisiae. Autophagy 8: 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Hoon S., Shamalnasab M., Galbani A., Wei M., et al. , 2010. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 6: e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L., Partridge L., Longo V. D., 2010. Extending healthy life span–from yeast to humans. Science 328: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D., Partridge L., 2013. Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 75: 621–644. [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Hamilton B., Dong Y., Shindo M., Liu W., Odell I., et al. , 2005. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19: 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. X., Xu X. Y., Zhang M. J., Peng X., Du L. L., 2010. Global fitness profiling of fission yeast deletion strains by barcode sequencing. Genome Biol. 11: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kim D. U., Hayles J., Kim D., Wood V., Park H. O., et al. , 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda T. A., Tanaka K., Konomi M., Sato M., Osumi M., et al. , 2007. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 12: 155–170. [DOI] [PubMed] [Google Scholar]
- Kyryakov P., Beach A., Richard V. R., Burstein M. T., Leonov A., et al. , 2012. Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front Physiol 3: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labunskyy V. M., Gerashchenko M. V., Delaney J. R., Kaya A., Kennedy B. K., et al. , 2014. Lifespan extension conferred by endoplasmic reticulum secretory pathway deficiency requires induction of the unfolded protein response. PLoS Genet. 10: e1004019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. S., Monaghan P., Metcalfe N. B., 2013. Experimental demonstration of the growth rate–lifespan trade-off. Proc. Biol. Sci. 280: 20122370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V. D., Shadel G. S., Kaeberlein M., Kennedy B., 2012. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 16: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maury L., Marguerat S., Bähler J., 2008. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 9: 583–593. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., 2013. The hallmarks of aging. Cell 153: 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., et al. , 2012. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markaki M., Tavernarakis N., 2013. Metabolic control by target of rapamycin and autophagy during ageing - a mini-review. Gerontology 59: 340–348. [DOI] [PubMed] [Google Scholar]
- Mata J., Lyne R., Burns G., Bähler J., 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Matecic M., Smith D. L., Pan X., Maqani N., Bekiranov S., et al. , 2010. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 6: e1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., et al. , 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24: 841–847. [DOI] [PubMed] [Google Scholar]
- Pancaldi V., Schubert F., Bähler J., 2010. Meta-analysis of genome regulation and expression variability across hundreds of environmental and genetic perturbations in fission yeast. Mol. Biosyst. 6: 543–552. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., et al. , 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12: 1607–1613. [DOI] [PubMed] [Google Scholar]
- Powers R. W., 3rd, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S., 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20: 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C., Codlin S., Bähler J., 2013. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 12: 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C., Lopez-Maury L., Georgescu T., Pancaldi V., Bähler J., 2014. Systematic screen for mutants resistant to TORC1 inhibition in fission yeast reveals genes involved in cellular ageing and growth. Biol. Open 3: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F. E., Zhang K., Zofall M., Chen E., Grewal S. I., 2011. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol. 18: 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D. G., W. Chen, J. D. Storey, and D. Gresham, 2014 Design and analysis of Bar-seq experiments. G3 (Bethesda) 4: 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A. E., Arseneault G., Chartrand P., Ferbeyre G., Rokeach L. A., 2010. A screen for genes involved in respiration control and longevity in Schizosaccharomyces pombe. Ann. N. Y. Acad. Sci. 1197: 19–27. [DOI] [PubMed] [Google Scholar]
- Roux A. E., Leroux A., Alaamery M. A., Hoffman C. S., Chartrand P., et al. , 2009. Pro-aging effects of glucose signaling through a G protein-coupled glucose receptor in fission yeast. PLoS Genet. 5: e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A. E., Quissac A., Chartrand P., Ferbeyre G., Rokeach L. A., 2006. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell 5: 345–357. [DOI] [PubMed] [Google Scholar]
- Ryan C. J., Roguev A., Patrick K., Xu J., Jahari H., et al. , 2012. Hierarchical modularity and the evolution of genetic interactomes across species. Mol. Cell 46: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H. Y., Rhie B. H., Ahn S. H., 2014. Loss of the Set2 histone methyltransferase increases cellular lifespan in yeast cells. Biochem. Biophys. Res. Commun. 446: 113–118. [DOI] [PubMed] [Google Scholar]
- Sajiki K., Hatanaka M., Nakamura T., Takeda K., Shimanuki M., et al. , 2009. Genetic control of cellular quiescence in S. pombe. J. Cell Sci. 122: 1418–1429. [DOI] [PubMed] [Google Scholar]
- Selman C., Tullet J. M. A., Wieser D., Irvine E., Lingard S. J., et al. , 2009. Ribosomal Protein S6 Kinase 1 Signaling Regulates Mammalian Life Span. Science 326: 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M., Uehara L., Pluskal T., Yoshida T., Kokubu A., et al. , 2013. Klf1, a C2H2 zinc finger-transcription factor, is required for cell wall maintenance during long-term quiescence in differentiated G0 phase. PLoS ONE 8: e78545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Wolgamott L., Yu Y., Blenis J., Yoon S. O., 2011. Glycogen synthase kinase (GSK)-3 promotes p70 ribosomal protein S6 kinase (p70S6K) activity and cell proliferation. Proc. Natl. Acad. Sci. USA 108: E1204–E1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M., Heisler L. E., Mellor J., Kaper F., Thompson M. J., et al. , 2009. Quantitative phenotyping via deep barcode sequencing. Genome Res. 19: 1836–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola O., Kerr F., Rogers I., Killick R., Augustin H., et al. , 2010. Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet. 6: e1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J., Franke J., Ehrenhofer-Murray A. E., 2013. Chemical genetic screen in fission yeast reveals roles for vacuolar acidification, mitochondrial fission, and cellular GMP levels in lifespan extension. Aging Cell 12: 574–583. [DOI] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K., 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 106: 17049–17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. L., Li M., Suo F., Liu X. M., Shen E. Z., et al. , 2013. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 9: e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Yoshida T., Kikuchi S., Nagao K., Kokubu A., et al. , 2010. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc. Natl. Acad. Sci. USA 107: 3540–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., 2010. Fission yeast TOR and Rapamycin, Vol. 27, pp. 251-269 in The Enzymes, edited by Michael N. Hall and Fuyuhiko Tamanoi. [Google Scholar]
- Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., et al. , 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., et al. , 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Wood V., Harris M. A., McDowall M. D., Rutherford K., Vaughan B. W., et al. , 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40: D695–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M., 2009. Cellular quiescence: are controlling genes conserved? Trends Cell Biol. 19: 705–715. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K., Tanaka T., Ida Y., Furusawa C., Hirasawa T., et al. , 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28: 349–361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.