Abstract
Recognition of extracellular peptides by plasma membrane-localized receptor proteins is commonly used in signal transduction. In plants, very little is known about how extracellular peptides are processed and activated in order to allow recognition by receptors. Here, we show that induction of cell death in planta by a secreted plant protein GRIM REAPER (GRI) is dependent on the activity of the type II metacaspase METACASPASE-9. GRI is cleaved by METACASPASE-9 in vitro resulting in the release of an 11 amino acid peptide. This peptide bound in vivo to the extracellular domain of the plasma membrane-localized, atypical leucine-rich repeat receptor-like kinase POLLEN-SPECIFIC RECEPTOR-LIKE KINASE 5 (PRK5) and was sufficient to induce oxidative stress/ROS-dependent cell death. This shows a signaling pathway in plants from processing and activation of an extracellular protein to recognition by its receptor.
Keywords: ligand, protease, receptor-like kinase, secreted protein
Introduction
Recognition of extracellular signals is central for plant development and survival. Plant-encoded extracellular peptides and proteins are important components for developmental and stress response regulators (Boller & Felix, 2009; Butenko et al, 2009). In the immune response, plants also recognize pathogen-associated molecular patterns (PAMPs) of microbial origin (Boller & Felix, 2009). The Arabidopsis thaliana genome encodes several hundred secreted proteins (Butenko et al, 2009; Murphy et al, 2012) and more than 400 membrane-spanning receptor-like protein kinases (RLKs) (Shiu & Bleecker, 2003). This suggests a large number of potential ligand–receptor interactions providing a complex network of extracellular signaling modules in plants (Boller & Felix, 2009). Nonetheless, only a few peptide–receptor interactions have so far been identified (Butenko et al, 2009). For the known plant extracellular ligand–receptor systems, the peptide ligands are either small (e.g. systemin, PEP1, CLAVATA) or only a short stretch of amino acids (aa) within the proteins (e.g. flg22, elf18) is recognized by their receptor (Altenbach & Robatzek, 2007; Boller & Felix, 2009). In animals, a number of secreted proteins are processed by proteolytic cleavage to release the active signaling peptide (Pimenta & Lebrun, 2007), and in plants, similar mechanisms are involved in peptide activation (Murphy et al, 2012).
Almost 700 proteases (Tsiatsiani et al, 2012; Rawlings et al, 2014) encoded in the Arabidopsis genome have diverse functions and specificities ranging from the processing of signal peptides required for subcellular targeting to degradation of proteins (van der Hoorn, 2008). However, plant protease substrates remain largely unexplored (Tsiatsiani et al, 2012). Metacaspases, distant relatives of animal caspases (Vercammen et al, 2007; Tsiatsiani et al, 2011), are a class of cysteine-dependent proteases in plants, fungi and protozoa. Metacaspases are important regulators of biotic and abiotic stress responses, development and cell death in plants (Hoeberichts et al, 2003; Vercammen et al, 2007; He et al, 2008; Coll et al, 2010; Tsiatsiani et al, 2011; Watanabe & Lam, 2011). To date, several substrates have been identified for plant metacaspases (Tsiatsiani et al, 2011)—a Tudor staphylococcal nuclease (Sundström et al, 2009) in Picea abies and a number of substrates for A. thaliana METACASPASE-9 (AtMC9; Tsiatsiani et al, 2013; Vercammen et al, 2006). However, there are no instances reported for plants where a protease processes a secreted (pre)protein, thereby producing a ligand for a known receptor in a specific biological process.
We have previously described an ozone (O3)-sensitive Arabidopsis mutant named grim reaper (gri; Wrzaczek et al, 2009b). The cause for the O3 sensitivity of the mutant was the presence of a truncated fragment of GRI in the gri insertion mutant. A 66-aa fragment of the secreted GRI protein, that is present in the mutant, induced cell death, as measured by elevated ion leakage, upon infiltration into plant leaves. Cell death induction by GRI-peptide was dependent on the plant hormone salicylic acid but also on production of extracellular superoxide. The gri mutant displayed enhanced resistance to a virulent bacterial pathogen.
Here, we show that a subfragment of Arabidopsis GRI contains sufficient information to induce elevated ion leakage. A metacaspase, AtMC9 (Bollhöner et al, 2013), is required in vivo for the activation of GRI in the extracellular space and is able to directly cleave GRI in vitro. AtMC9-processed peptide-mediated signaling is dependent upon binding to POLLEN-SPECIFIC RECEPTOR-LIKE KINASE 5 (PRK5), an atypical, enzymatically inactive RLK, which serves as a receptor for the peptide. Our results are an important step in understanding the processing of extracellular peptide ligands and their recognition through receptors.
Results
A 20-aa GRI-peptide contains information sufficient to induce elevated ion leakage
The extracellular Arabidopsis protein GRIM REAPER (GRI) is involved in reactive oxygen species (ROS)-mediated cell death (Wrzaczek et al, 2009b). Under superoxide-producing conditions infiltration of a 66-aa part of GRI, GRIp31–96, that is, present in the gri mutant, into Arabidopsis leaves induced cell death, as measured by elevated ion leakage (Fig 1A). Background ion leakage in the control infiltration (with GST) is caused by the wounding due to mechanical stress of infiltration (Fig 1A). When testing four shorter and overlapping peptides (Supplementary Fig S1A) in the leaf infiltration assay, only the 20-aa peptide GRIp65–84 induced ion leakage similarly to bacterially produced GST-GRIp31–96 and biochemically pure GRIp31–96 (Fig 1A; Supplementary Fig S1B shows dead cells visualized by Trypan blue staining). The three other peptides were inactive. Notably, the 20-aa-long peptide GRIp65–84 induced elevated ion leakage in a dose-responsive manner (Fig 1B).
Figure 1. The LRR RLK PRK5 is required for GRI-peptide-induced ion leakage.
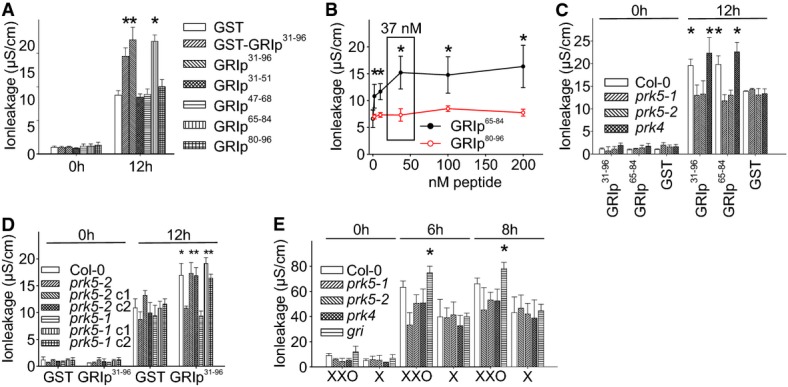
- Infiltration of GRIp65–84 induced cell death similar to GRIp31–96. Bacterially produced 37 nM GST, GST-GRIp31–96 or biochemically pure GRI-peptides (GRIp31–96, GRIp31–51, GRIp47–68, GRIp65–84, GRIp80–96) were infiltrated into leaves of Col-0 plants.
- Infiltration of increasing concentrations of GRIp65–84 into Col-0 leaves, electrolyte leakage was measured after 12 h. Background (red line) shows ion leakage from infiltration of leaves with increasing concentrations of (inactive) GRIp80–96.
- Infiltration of leaves with 37 nM GRIp31–96 induced elevated ion leakage in Col-0 and prk4, but not in prk5-1 or prk5-2. Infiltration with GST caused the same background effect for all lines.
- Genomic complementation of prk5 rescues the insensitivity to induction of elevated ion leakage by GRIp31–96.
- Enzymatic superoxide production from xanthine/xanthine oxidase (XXO) induced more electrolyte leakage in gri compared to Col-0 or prk5-1,prk5-2 and prk4 after infiltration into leaves. Infiltration with xanthine buffer (X) was used as a control.
Data information: All panels show average ± standard deviation (SD) of four replicates consisting of four leaf disks each. Asterisks indicate statistically significant differences from GST infiltration (A, C, D), from infiltration with (inactive) GRIp80–96 (B) or from Col-0 (E) according to Sidak’s test (P < 0.05). All experiments were repeated at least four times with similar results.
A leucine-rich repeat RLK mediates GRI-peptide-induced ion leakage
GRI is related to the Solanaceae stigma-specific protein STIG1 (Goldman et al, 1994). Tomato LeSTIG1 interacted in vitro with the ectodomains of two RLKs, the pollen receptor kinases LePRK1 and LePRK2 (Tang et al, 2004; Löcke et al, 2010; Huang et al, 2014). Therefore, we tested the potential interaction of GRI with RLKs. Leaves from Arabidopsis T-DNA insertion lines for leucine-rich repeat (LRR) RLKs homologous to the two tomato RLKs were infiltrated with the 66-aa GRIp31–96 and 20-aa GRIp65–84 peptides and scored for cell death. Two T-DNA insertion alleles in At1g50610 (SALK_016815 and SALK_101260) in the last exon and in the 5′ UTR region, respectively, displayed reduced ion leakage levels in response to peptide infiltration (Fig 1C). This gene has recently been named PRK5 (Chang et al, 2013). Thus, the mutants are referred to as prk5-1 (SALK_016815; Chang et al, 2013) and prk5-2 (SALK_101260), respectively. Complementation of prk5 with a genomic clone consisting of a 1,500 base pair promoter region and the coding region of PRK5 restored the wild-type phenotype (Fig 1D). PRK5 has previously been described as a pollen-specific RLK (Chang et al, 2013), but RT–PCR analysis demonstrated the presence of low levels of PRK5 transcript in leaves (Supplementary Fig S2). While PRK5 transcript levels are low in plant organs other than pollen tubes under normal growth conditions (Supplementary Fig S3), analysis of publicly available expression data suggests that transcript abundance is increased in response to biotic and abiotic stresses (Supplementary Figs S4, S5, S6, S7 and S8). Similarly, GRI transcript abundance is lower in leaves than in flowers (Wrzaczek et al, 2009b). Our previous results suggested that GRI-induced cell death as evidenced by increased ion leakage was dependent on superoxide production. Therefore, we used infiltration of an enzymatic system, xanthine with xanthine oxidase (XXO), to produce superoxide in the extracellular space and analyze the response of prk5. Compared to wild-type plants, the loss-of-function mutants prk5 and prk4 showed slightly less, statistically not significant (but reproducible), ion leakage as induced by extracellular superoxide while the gain-of-function mutant gri (Wrzaczek et al, 2009b) exhibited increased sensitivity (Fig 1E). This indicates that in leaves, PRK5 could act as a downstream element for ROS-dependent cell death induced by GRI or a smaller subdomain of it.
PRK5 is a plasma membrane-localized, enzymatically atypical protein kinase
PRK5 belongs to the LRR RLK subtype III (Shiu & Bleecker, 2003) and consists of 686 aa (calculated MW: 76.86 kDa; pI 7.84). A transmembrane domain separates the extracellular region with a signal peptide from the intracellular kinase domain, which contains a Y-based sorting/endocytosis motif in the C-terminus (Fig 2A). Structural prediction suggests that the extracellular domain (Supplementary Fig S9A) is similar to other LRR RLKs and the intracellular domain (Supplementary Fig S9B) has the overall typical sequence and structural conservation of protein kinases. The few known plant receptors for extracellular proteins are plasma membrane-localized (Aker & de Vries, 2008). We analyzed the subcellular localization of PRK5 tagged with cyan fluorescent protein (PRK5-CFP; Fig 2B) using transient expression in Arabidopsis mesophyll protoplasts. PRK5-CFP co-localization with the plasma membrane marker CAAX-yellow fluorescent protein (CAAX-YFP; Kwaaitaal et al, 2011) (Fig 2C–F) was markedly distinct from the cytoplasmic YFP (Supplementary Fig S10A). PRK5-YFP also localized to the cell periphery in Nicotiana benthamiana epidermal cells (Supplementary Fig S10B–G). After plasmolysis, Hechtian strands (Vahisalu et al, 2008), which connect the plasma membrane to the cell wall, were visible verifying that PRK5-YFP was localized to the plasma membrane.
Figure 2. PRK5 is an atypical, enzymatically inactive RLK.
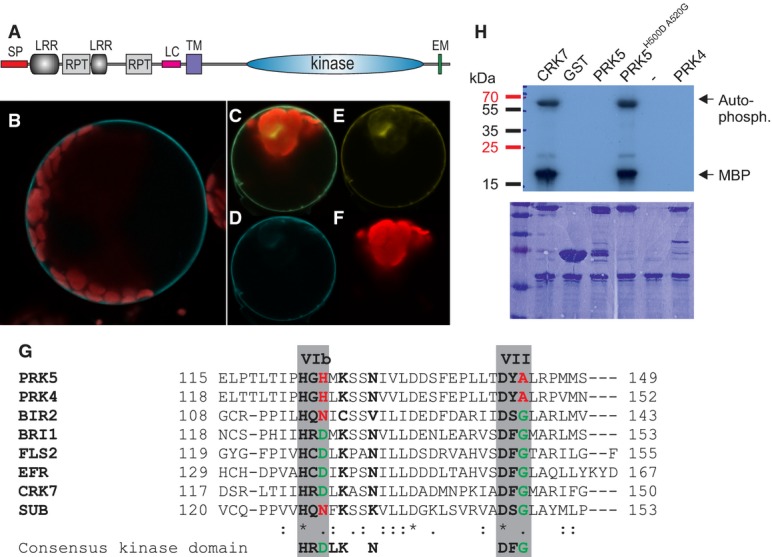
- A PRK5 domain structure: SP signal peptide (aa 1–39), LRR leucine-rich repeat, RPT internal repeat, LC region of low complexity, TM transmembrane domain (aa 282–304), EM Y-based sorting/endocytosis motif (YSSM; aa 670–673).
- B PRK5-CFP localized to the cell periphery in Col-0 mesophyll protoplasts.
- C–F Co-localization of PRK5-CFP and PM localized CAAX-YFP in Col-0 mesophyll protoplasts. (C) overlay; (D) PRK5-CFP (465–510 nm); (E) CAAX-YFP (521–587 nm); (F) chloroplast (636–711 nm).
- G Alignment of subdomains VIb and VII of the catalytic core of the kinase domains of active (BRI1, FLS2, EFR, CRK7) and inactive RLKs (PRK4, PRK5, BIR2, SUB). Residues marked in green highlight conservation of the consensus of active protein kinases while residues highlighted in red indicate deviations from the consensus sequence. An alignment of the full kinase domains for the RLKs used in this figure can be found in Supplementary Fig S11A.
- H Kinase activity of PRK5 in in vitro phosphorylation assays using γ32P-ATP and myelin-basic protein (MBP) as a substrate in the presence of 10 mM MnCl2. GST-PRK5 and GST-PRK4 did not show kinase activity. Mutation of conserved residues in kinase subdomains VIb and VII to reconstitute the consensus kinase domain motif restored GST-PRK5H500DA520G kinase activity. GST-CRK7 was used as a positive control. Upper panel shows autoradiograph, and lower panel shows the Coomassie-stained 15% SDS–polyacrylamide gel.
Data information: Experiments in (B–F and H) were repeated three times with similar results. Source data are available online for this figure.
While the overall kinase domain structure is preserved in PRK5, critical amino acids in the kinase subdomains VIb and VII (Stone & Walker, 1995) are not conserved in PRK5 (Fig 2G, Supplementary Fig S11A and B). The aspartic acid (D) residue, which is conserved in active RLKs (e.g. in FLS2, EFR, BRI1, CRK7) is altered to histidine (H) in PRK5. In the inactive RLKs STRUBBELIG (SUB; Vaddepalli et al, 2011) and BAK1-INTERACTING RECEPTOR-LIKE KINASE 2 (BIR2; Halter et al, 2014b), this residue is changed to asparagine (N). This suggested that PRK5 could be enzymatically inactive and accordingly, recombinant GLUTATHIONE-S-TRANSFERASE (GST)-tagged PRK5 (GST-PRK5) did not have kinase activity toward the artificial substrate myelin-basic protein. Intriguingly, mutations restoring the consensus kinase sequence in the catalytic core (H500D A520G; Supplementary Fig S11C) turned GST-PRK5 into an active kinase in the presence of MnCl2 (Fig 2H) and MgCl2 (Supplementary Fig S11D). Taken together, PRK5 closely resembles an active kinase based on sequence and modeling, but, at least in vitro, is enzymatically inactive.
PRK5 binds GRIp31–96 in vitro
Since PRK5 was required for cell death induction by GRIp31–96 and GRIp65–84, we investigated the interaction between GRI and the ectodomain of PRK5 in vitro. Recombinant full-length GRI (minus signal peptide; GRI31–168) or GRI31–96 fused to GST was incubated with 35S-methionine-labeled in vitro-produced extracellular domains of PRK540–281 or PRK440–279. GRI31–96 interacted directly with PRK540–281, while GRI31–168 showed slightly weaker interaction (Fig 3A; Supplementary Fig S12 shows Western analysis with α-GST, α-GRI and α-GRI-peptide antibodies). Binding of GRI31–96 and GRI31–168 to the PRK4 extracellular domain was weaker compared to binding of GRI31–96 to PRK540–281 (Fig 3A). Given the high sequence similarity of PRK4 and PRK5 (70.15% sequence identity; 77.76% sequence similarity; Supplementary Fig S12E), it is not surprising that GRI31–96 and GRI31–168 were still able to interact at least to some extent with both receptors. No interaction was, however, detected with the ectodomain of a different RLK, FLAGELLIN-SENSITIVE 2 (FLS2; Supplementary Fig S12F). These results suggest that GRI and the 66-aa peptide GRI31–96 can directly interact with the extracellular domains of receptors, preferentially with the ectodomain of PRK5.
Figure 3. A 20-aa peptide binds to the extracellular domain of PRK5.
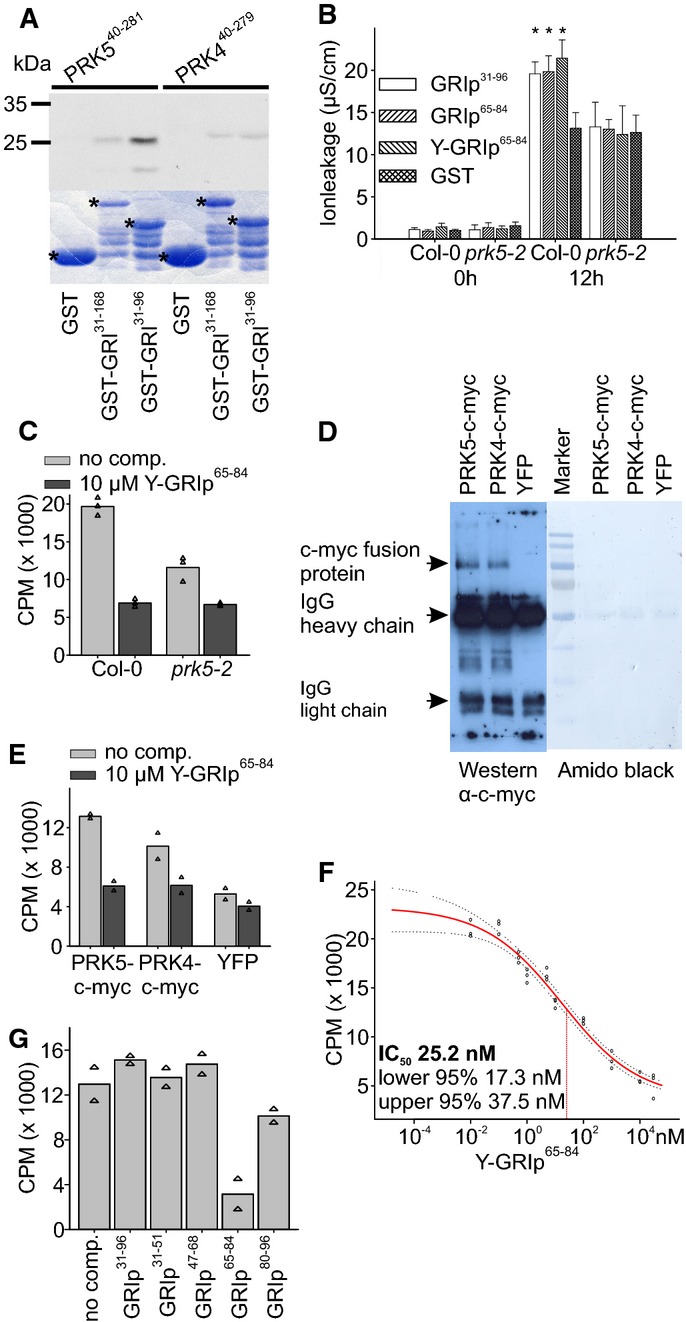
- In vitro-produced 35S-labeled ectodomains of PRK5 (PRK540–281) or PRK4 (PRK440–279) were incubated with bacterially produced GST, GST-GRI or GST-GRI31–96 and purified. GST-GRI31–96 but not GST directly bound to the ectodomain of PRK5. Binding of GST-GRI to PRK5 ectodomain and binding of GST-GRI31–96 and GST-GRI to the ectodomain of PRK4 were strongly reduced. Upper part: autoradiograph, lower part: Coomassie-stained 12% SDS–polyacrylamide gel. Asterisks in the Coomassie-stained gel indicate GST, GST-GRI and GST-GRI31–96, respectively. Supplementary Fig S12 shows a Western blot of the GST-tagged proteins.
- Infiltration of 37 nM GST, GRIp31–96, GRIp65–84 or Y-GRIp65–84 into Col-0 or prk5-2 leaves. Tyrosine-labeled GRIp65–84 still induced cell death in Col-0 but not in prk5-2 plants.
- 125I-labeled Y-GRIp65–84 (0.46 nM) bound specifically to Col-0 membrane fractions (light gray bars), the binding was significantly reduced in prk5 plants. Excess of non-radioactive Y-GRIp65–84 (10 μM) reduced binding to background levels (dark gray bars; all bars show the average of three samples, triangles show individual data points).
- Immunoprecipitation of PRK5-c-myc, PRK4-c-myc or YFP expressed in protoplasts with rabbit polyclonal anti-c-myc antibody, blotted with mouse monoclonal anti-c-myc antibody.
- Binding of 125I-Y-GRIp65–84 (0.46 nM) to immunoprecipitates (using anti-c-myc antibodies) from prk5-2 protoplasts transfected with PRK5-c-myc, PRK4-c-myc or YFP, respectively. Binding was competed out with 10 μM unlabeled Y-GRIp65–84. Bars show the average of two samples, and triangles show individual data points. Western blot is shown in panel (D).
- Analysis of 125I-Y-GRIp65–84 (0.46 nM) binding competed out with increasing amounts of unlabeled Y-GRIp65–84 to Col-0 membrane extracts. Fifty percent inhibition (IC50) occurred at 25.2 nM. Red line shows binding average competition according to a sigmoid curve, dotted lines show 95% confidence intervals, and circles show data points.
- Excess of GRIp65–84 (10 μM) but not of GRIp31–96 or other peptides (GRIp31–51, GRIp47–68, GRIp80–96) competed the binding of 0.46 nM 125I-Y-GRIp65–84 to membrane extracts from Col-0 (all bars show the average of two samples, triangles show individual data points).
Data information: Data in (B) are shown as average ± SD of four replicates consisting of four leaf disks each. Asterisks in (B) mark statistically significant differences from infiltration with GST according to Sidak’s test (P < 0.05). All experiments were repeated three times with similar results. Source data are available online for this figure.
Native PRK5 binds GRIp65–84
GRIp65–84 is a subfragment of GRIp31–96 sufficient to induce elevated ion leakage upon infiltration into Arabidopsis leaves. To investigate whether the interaction of GRI and PRK5 can take place in vivo, an additional tyrosine (Y) was added to the N-terminus of GRIp65–84 to allow radiolabeling with iodine125 (125I). The Y-GRIp65–84 peptide showed similar activity in inducing elevated ion leakage compared to GRIp65–84 and GRIp31–96 (Fig 3B). In radioligand binding assays, 125I-Y-GRIp65–84 bound to membrane fractions from wild-type plants, whereas binding was strongly reduced in prk5-2 extracts, and excess of 10 μM non-radiolabeled Y-GRIp65–84 reduced binding to background levels (Fig 3C). 125I-Y-GRIp65–84 bound to microsomal fractions of protoplasts overexpressing PRK5-c-myc and also, albeit with lower affinity, PRK4-c-myc (Fig 3D and E). Specific binding of 125I-Y-GRIp65–84 was competed out by non-radioactive Y-GRIp65–84 with an IC50 of 25.2 nM (Fig 3F). Of the four short peptides, only GRIp65–84 competed for binding (Fig 3G). Interestingly, the 66-aa-long GRIp31–96 did not compete for binding of 125I-Y-GRIp65–84 (Fig 3G), indicating that, even though it interacted with the ectodomain of PRK5 in vitro, binding activity to membrane fractions would require further processing. Reasons might be that the receptor used in the in vitro assay was without co-receptors or other interacting proteins which in vivo might set additional constraints for ligand binding. In addition, as shown in other similar systems (Löcke et al, 2010), other extracellular proteins interacting with GRI might add further constraints for the ligand–receptor interaction in vivo. Together, the results suggest that the extracellular domain of PRK5 serves as a sensor for peptides derived from GRI through direct protein–protein interaction.
A metacaspase is required for activation of GRI-peptide
Although infiltration of both the 66-aa GRIp31–96 and the 20-aa GRIp65–84 induced cell death in planta (Fig 1A), the 66-aa peptide did not compete for binding of 125I-Y-GRIp65–84 in vivo (Fig 3G). Furthermore, Western blot analysis on leaf extracts of epitope-tagged GRI overexpressing plants displayed two distinct bands (Wrzaczek et al, 2009b). Together, these data suggested that GRI might be processed by proteolytic cleavage (Wrzaczek et al, 2009a) and analysis of GRI protein sequence suggested that the metacaspase AtMC9 might be able to cleave GRI. The in vitro and in vivo substrate specificity for AtMC9 has been described in detail (Tsiatsiani et al, 2013). The sequence SKTR64–67 in the 66-aa cell death-inducing peptide GRIp31–96 holds the characteristic of the AtMC9 preference for basic residues at substrate positions P3 and P1 (K and R; Vercammen et al, 2006, 2004).
In accordance with this, recombinant AtMC9 (rAtMC9; Vercammen et al, 2004) directly cleaved bacterially produced maltose-binding protein (MBP)-GRI fusion protein in vitro (Fig 4A). The inactive mutant rAtMC9mut (Belenghi et al, 2007) showed no proteolytic activity toward MBP-GRI (Fig 4A). The shift in the molecular weight of MBP-GRI25–168 in Western blot analysis with α-MBP antibody suggested cleavage of GRI at the SKTR motif, which is located at the N-terminus of GRIp65–84. The position of the AtMC9 cleavage site(s) in the MBP-GRI25–168 protein was determined by LC-MS/MS following in-solution labeling with a trideutero-acetyl group (<AcD3>) of primary alpha-amines of newly formed N-termini generated by AtMC9 cleavage. Analysis led to the identification of the peptides <AcD3>-68LLVSHYK74 and <AcD3>-98GTSLLHCCK107, thus showing that in vitro AtMC9 cleaves MBP-GRI25–168 not only after arginine 67 (R67 of the SKTR motif) but also after lysine 97 (K97, the second K of the KANK sequence; Supplementary Fig S13A–C). To assess the in vivo relevance of GRI-peptide cleavage by AtMC9, we used the atmc9 mutant (Bollhöner et al, 2013). The short 20-aa peptide GRIp65–84 was able to induce cell death in atmc9 (Fig 4B) but not the longer 66-aa GRIp31–96. In addition, atmc9 displayed slight, statistically not significant but reproducible, reduction in the ion leakage induced by XXO compared to wild-type plants (Supplementary Fig S14). This suggests that AtMC9 activity is required to modify the 66-aa GRIp31–96 for the induction of elevated ion leakage.
Figure 4. Processing of GRI-peptide by METACASPASE-9 is required for induction of elevated ion leakage.
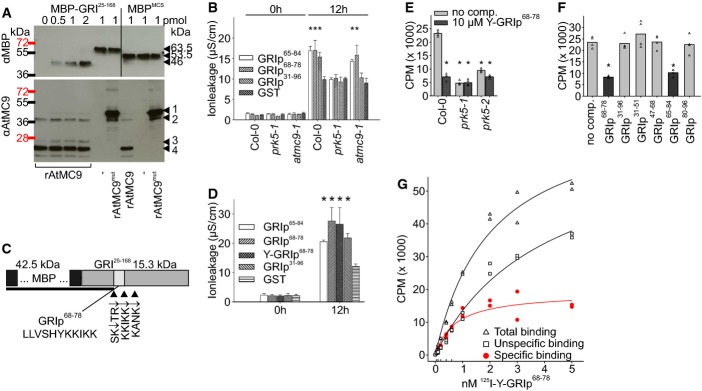
- Recombinant AtMC9 (rAtMC9) cleaved GRI25–168 in vitro. Bacterially produced MBP-GRI25–168 (from 0 to 2 pmol left to right) was incubated with 1 pmol rAtMC9 or inactive rAtMC9mut (rAtMC9C147AC29A), and cleavage products were analyzed by Western blot with anti-MBP and anti-AtMC9 antibodies. Arrowheads in the anti-MBP blot (from top to bottom) indicate MBP-GRI25–168 (63.5 kDa), MBPMCS (53.5 kDa), rAtMC9-cleaved MBP-GRI25–168 (46 kDa). Arrowheads in the anti-AtMC9 blot indicate: 1: N-terminal domain + p20 + p10 subunits of AtMC9, 2: p20 + p10 subunits of AtMC9, 3: N-terminal domain + p20 subunit of AtMC9, 4: p20 subunit of AtMC9.
- Infiltration of wild-type, prk5-1 and atmc9-1 leaves with 37 nM GRIp31–96, GRIp65–84, GRIp68–78 or GST. GRIp65–84 and GRIp68–78 but not GRIp31–96 were able to induce elevated ion leakage in the atmc9-1 mutant.
- Schematic representation of the GRI and the cleavage sites for rAtMC9. The cleavage product detected in Western blot analysis with anti-MBP antibody is shown as black bar. Mass spectrometric analysis of MBP-GRI25–168 cleavage with rAtMC9 provided evidence for cleavage after SKTR and KANK; further analysis of GRIp31–96 cleavage by rAtMC9 provided evidence for cleavage after SK, SKTR and after KKIKK. The position of the resulting 11-aa-long peptide (68LLVSHYKKIKK78) is indicated by a white inset.
- Infiltration of Col-0 leaves with 37 nM of GRIp65–84, GRIp68–78, Y-GRIp68–78, GRIp31–96 or GST. Y-GRIp68–78 showed similar activity in cell death induction compared to the other GRI-derived peptides.
- 125I-labeled Y-GRIp68–78 (0.46 nM) bound specifically to Col-0 membrane fractions (light gray bars), the binding was significantly reduced in prk5-1 and prk5-2 plants. Excess of non-radioactive Y-GRIp65–84 (10 μM) reduced binding to background levels (dark gray bars; all bars show the average of four samples, triangles show individual data points).
- Excess of GRIp68–78 and GRIp65–84 (10 μM) but not of GRIp31–96 or other peptides (GRIp31–51, GRIp47–68, GRIp80–96) competed the binding of 0.46 nM 125I-Y-GRIp68–78 to membrane extracts from Col-0 (all bars show average of four samples, triangles show individual data points).
- Saturation binding curve for 125I-Y-GRIp68–78 to Col-0 membrane extracts. Specific binding was calculated by subtracting non-specific binding from the total binding. The affinity of 125I-Y-GRIp68–78 to the receptor (Kd = 1.9 nM) was calculated by non-linear regression analysis. Scatchard plot is shown in Supplementary Fig S17.
Data information: Data in (B) and (D) are shown as average ± SD of four replicates consisting of four leaf disks each. Asterisks in (B) and (D) mark statistically significant differences from infiltration with GST according to Sidak’s test (P < 0.05). Asterisks in (E) and (F) mark statistically significant differences from peptide binding to Col-0 membrane fractions without competitor according to Sidak’s test (P < 0.01). All experiments were repeated three times with similar results. Source data are available online for this figure.
To investigate the cleavage of GRI in more detail, we analyzed the fragments generated by incubation of GRIp31–96 (the 66-aa subfragment of GRI, which is produced in the gri mutant; Wrzaczek et al, 2009b) with rAtMC9. Reverse-phase-HPLC and mass spectrometric analysis revealed cleavage of the peptide after lysine 65 (K65) and arginine 67 (R67) in the SKTR motif and an additional site after lysine 78 (K78) in the KKIKK pattern (Fig C; Supplementary Fig S15A–C). In combination with the results from cleavage of MBP-GRI, this suggested that an 11-aa-long peptide, GRIp68–78, could be produced by cleavage with AtMC9.
To address the relevance of this cleavage for GRI-peptide activity, we tested an 11-aa peptide GRIp68–78 (Fig 4C). GRIp68–78 induced cell death in Col-0 but not in prk5-1 (Fig 4B) or prk5-2 (Supplementary Fig S16A). GRIp68–78 but not GRIp31–96 induced ion leakage in atmc9-1 (Fig 4B). As cell death induction by GRI-peptide is dependent on salicylic acid and extracellular superoxide (Wrzaczek et al, 2009b), we tested induction of elevated ion leakage by GRIp68–78 in mutants deficient in salicylic acid (salicylic acid deficient 2 [sid2]) and extracellular superoxide production (respiratory burst oxidase homolog D [rbohD]). Neither GRIp31–96 nor GRIp68–78 induced elevated ion leakage in sid2 and rbohD (Supplementary Fig S16B). These results suggest that salicylic acid and extracellular superoxide are still required for cell death induction by the processed GRIp68–78. A tyrosine-labeled version of the 11-aa peptide, Y-GRIp68–78, also induced elevated ion leakage (Fig 4D) and 125I-labeled Y-GRIp68–78 bound to membrane extracts from wild-type plants (Fig 4E). However, binding was reduced to background levels in prk5-1 and prk5-2 (Fig 4E). Binding of 125I-Y-GRIp68–78 was competed out by addition of non-radioactive GRIp68–78 and GRIp65–84 but not by other peptides (Fig 4F). The high binding background could result from anionic interactions be due to the strong basic nature of the 11-aa peptide (Fig 4G, Supplementary Fig S17), which may affect calculation of the dissociation constant (Kd) of 1.9 nM for GRIp68–78. The results suggest that an 11-aa peptide derived from GRI based on identified AtMC9-cleavage sites is sufficient to induce cell death in Arabidopsis leaves and binds with high specificity to the extracellular domain of the receptor PRK5. The gri mutant has previously been found to be more resistant to the virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Wrzaczek et al, 2009b). However, prk5 and atmc9 did not display altered pathogen resistance (Supplementary Fig S18). A reason for this might be that the infiltration with the pathogen into gri—where a ‘pre-activated’ GRI-derived peptide could already be present—leads to a number of cells undergoing cell death, which would lead to the initiation of a hypersensitive response (HR). In the prk5 and atmc9 mutants this pre-activated GRI-derived peptide is not recognized (in prk5) or produced (in atmc9). Thus, prk5 and atmc9 did not exhibit altered sensitivity to the virulent pathogen. However, more detailed analysis of this aspect will be required in the future using prk5 gri and atmc9 gri double mutants as well as overexpression of prk5. Also, a complete loss-of-function allele for gri will be crucial for further dissection of the roles of GRI and derived peptides.
Our hypothesis that GRI is an in vivo target of AtMC9 is supported by the fact that both proteins have been reported to be present in the extracellular space (Vercammen et al, 2006; Wrzaczek et al, 2009b). Additional lines of evidence suggest that proteolytic processing is required for the biological activity of GRI. The 66-aa GRIp31–96 was unable to compete out the binding of radioactively labeled 125I-Y-GRIp65–84 in plant membrane fractions (Figs 3G and 4F) and was unable to induce elevated ion leakage in the atmc9 mutant. While metacaspases are involved in stress adaptation and cell death regulation in plants (Tsiatsiani et al, 2011; Lam & Zhang, 2012), the role of AtMC9 in these processes has previously been unknown. Our results show that AtMC9 directly cleaves GRI and is thus involved in the processing and activation of the extracellular peptide.
Discussion
In this study, we describe how the secreted protein GRI is cleaved by a protease and a resulting peptide is subsequently recognized by a transmembrane receptor. GRI is a secreted Arabidopsis protein with similarity to STIG1 from tobacco and tomato. A 66-aa N-terminal fragment of GRI, which is produced in the gain-of-function mutant gri, was previously shown to induce cell death when infiltrated into Arabidopsis leaves (Wrzaczek et al, 2009b). We show that recombinant GRI and GRIp31–96 were cleaved by the metacaspase AtMC9 in vitro, releasing an 11-aa peptide (Fig 4) that was sufficient for the induction of cell death as inferred from ion leakage measurements. Physiological evidence indicated that this metacaspase-dependent processing of GRI was required for cell death induction in planta.
We identified PRK5, a receptor-like kinase, as the receptor for GRI-derived peptides generated through cleavage by AtMC9. PRK5 is required for cell death induction by GRI. PRK5 is present in leaves at low levels similar to GRI itself. While STIG1 and PRK5 have been described to function in floral organs (Goldman et al, 1994; Chang et al, 2013), our results show that GRI and its receptor also have functions in leaves. Involvement of a signaling component in several different processes is common for plant signaling. For example BRI1-ASSOCIATED KINASE (BAK1) has originally been described to function in brassinosteroid signaling but also has important roles in PAMP and DAMP signaling (Liebrand et al, 2014).
Our in silico and in vitro analyses suggest that PRK5 is enzymatically inactive (Fig 2G and H). Estimates indicate that 10 to 20% of all RLKs in Arabidopsis might be catalytically inactive (Castells & Casacuberta, 2007; Blaum et al, 2014). Only a few—to our knowledge—have been linked to a biological process based on mutant analysis; examples include BIR2 (Halter et al, 2014b), SUB (Vaddepalli et al, 2011) and SHORT SUSPENSOR (SSR; Bayer et al, 2009). While SUB has been suggested to bind a ligand (Vaddepalli et al, 2011), no ligands have so far been identified for SSR. BIR2 is suggested to control BAK1-receptor complex assembly in the absence of peptide ligands and thus might not recognize a ligand either (Halter et al, 2014a,b). PRK5 is the first atypical, kinase-inactive, plant RLK that acts as a primary receptor for a peptide ligand.
We have previously shown that cell death induction by a 66-aa GRI-peptide occurs under conditions that lead to superoxide production (Wrzaczek et al, 2009b), for example, through mechanical stress and wounding caused by infiltration of the peptide into Arabidopsis leaves. Removal of superoxide production, either by co-infiltration of the peptide with superoxide dismutase or by peptide infiltration into a mutant deficient in the NADPH oxidase RBOHD, reduced cell death to background levels. Extracellular superoxide was still required for cell death induction by GRIp68–78 (Supplementary Fig S16B). Wounding induces ROS production and is one of the signals that have been shown to initiate the so-called ‘ROS wave’ (Mittler et al, 2011). Our results suggest that ROS can act as a parallel signal, perhaps together with other wound-induced cues, to sensitize cells to cell death induction by GRI-derived peptides (Supplementary Fig S19).
It is currently unknown how extracellular ROS are sensed by cells. Accurate ROS sensing most likely relies on a broad range of independent mechanisms (Wrzaczek et al, 2013), and it is unlikely that ROS participate in classical ligand–receptor interactions. ROS might rather react with extracellular components including cell walls, lipids and also proteins/peptides. While ROS may act as a cue in parallel to GRIp68–78, the full-length GRI itself could be subject to redox regulation (Supplementary Fig S19) through two cysteine motifs (C-9X-C-2X-C) in its C-terminal region. The cysteine motifs in GRI are similar to the pattern found in DUF26 (domain of unknown function 26) proteins, C-8X-C-2X-C (Wrzaczek et al, 2010). In both cases, the cysteine motifs are suggested to be a target for redox regulation. The C-2X-C part of DUF26 and GRI also is a classical target for thioredoxins (Zhang et al, 2011). Recently, an extracellular thioredoxin has been shown to regulate stress responses through ROS (Zhang et al, 2011). Thereby, GRI could be involved in the apoplastic redox sensory mechanisms regulated through interaction with thioredoxins and reorganization of thiol bonds. This regulation could affect cleavage of GRI by modulation of the three-dimensional structure of the protein and changing accessibility of the AtMC9 cleavage sites through redox regulation of thiol bonds. Thus, GRI may be controlled dually by conformational change and proteolytic cleavage (Wrzaczek et al, 2009a). GRI was not identified in a recent analysis of the AtMC9 degradome (Tsiatsiani et al, 2013). However, that study was performed on young seedlings as compared to the older plants used in the work described here. It is interesting to note that AtMC9 activity can be regulated through S-nitrosylation (Belenghi et al, 2007). Evidence suggests that there is a significant cross talk between reactive nitrogen species (RNS) and ROS, but the clear relationship between the two remains elusive (Wang et al, 2013). The functional unit of GRI and metacaspase could be regulated by ROS/RNS on multiple levels and be part of the apoplastic redox sensing machinery in plants.
GRI is a member of a small protein family with six members in Arabidopsis. The C-terminus of GRI containing the cysteine repeats is highly similar to STIG1 and the Arabidopsis orthologs, but the N-terminal part after the signal peptide, which contains the cell death-inducing peptide motif, shows strikingly lower levels of conservation. Future research should address the question whether the conservation of the C-terminus of GRI is linked to cell death regulation or whether it is involved in other processes. No functions have been described for any of the GRI orthologs in Arabidopsis, but the conservation of the C-terminal cysteine motifs might point toward a common mode of regulation among GRI, STIG1 and related proteins. Interestingly, for CLAVATA3/ESR-RELATED 18 (CLE18), two different peptides derived from the precursor protein have been shown to have individual and possibly antagonistic functions (Murphy et al, 2012). This suggests that more than one biologically active peptide could be generated from a single precursor. It remains to be determined whether this also applies to GRI and related proteins. It also remains to be seen whether GRI interacts with other extracellular proteins as has been shown for tomato LeSTIG1 (Löcke et al, 2010).
The combination of GRI, AtMC9 and PRK5 provides a functional unit comprising secreted protein, protease and the receptor for the cleavage product. This scheme is likely to be employed in the regulation of many other ligand–receptor interactions in plants. Future research on the functions of GRI, AtMC9 and PRK5 in plant development including stigma–pollen interactions and cell death regulation, and the roles of the other proteins similar to GRI will increase our understanding of extracellular signaling in plants.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as wild-type. The gri (Wrzaczek et al, 2009b) and atmc9 (Bollhöner et al, 2013) lines were previously described. The prk5-1, prk5-2 and prk4 T-DNA insertion lines were obtained from the European Arabidopsis Stock Centre (http://www.arabidopsis.info).
Seeds were sown on 1:1 peat–vermiculite mixture, stratified for 2 days and grown under controlled conditions (Vahisalu et al, 2008) under 12-h/12-h day/night cycle (temperature 23/18°C, relative humidity 70/90%). For in vivo radio-ligand binding assays, A. thaliana seedlings were grown on agar plates at 12-h/12-h day/night cycle, temperature 23/18°C for 7 days.
Peptide and XXO infiltration experiments as well as pathogen infection assays were performed as previously described (Wrzaczek et al, 2009b). Protoplast preparation, transfection and extracts for ligand binding assays and Western analysis were prepared as described (Lee et al, 2011). For transient gene expression studies, Arabidopsis protoplasts were transfected with PRK5-CFP, PRK4-CFP, or controls (cytoplasmic YFP, plasma membrane-localized CAAX-YFP (Kwaaitaal et al, 2011) under the control of 35S promoter and incubated for 48 h at room temperature in darkness prior to confocal microscopy.
Plasmid constructs
Constructs were created by PCR, and restriction or Gateway sites were introduced via the PCR primers. For expression of GRI as a MBP fusion, the coding sequence of GRI minus the signal peptide was cloned EcoRI/PstI into the pMAL-c2x vector (New England Biolabs). For expression of GRI as a GST-fusion protein, the coding sequence of GRI or GRIp31–66 minus the signal peptide was cloned EcoRI/NotI or EcoRI/SalI, respectively, into the pGEX-4T-1 vector (GE Healthcare Life Science). The kinase domains of PRK5 and PRK4 were cloned EcoRI/NotI into pGEX-4T-1. For coupled in vitro transcription and translation, the ectodomains of PRK5, PRK4 and FLS2 were cloned into pZeRO-2.1 (Invitrogen). For genomic complementation lines, the coding region of PRK5 including 1,500-bp promoter region was amplified by PCR and cloned into pGreenII0179 (Hellens et al, 2000). Plants were transformed by Agrobacterium tumefaciens-mediated gene transfer. Homozygous single-insert plants carrying the transgene were selected based on antibiotic resistance. The coding regions of PRK5 and PRK4 were cloned PacI/EcoRI into a modified version of the pGWR8 (Rozhon et al, 2010) vector containing a 6× c-myc or YFP tag under the control of the UBQ10 promoter. For confocal microscopy of protoplasts, the coding regions of PRK5 and PRK4 were Gateway-cloned into the CZN575 vector containing the sCFP3a tag.
Trypan blue staining
Trypan blue staining was performed as described (Dat et al, 2003).
GRI-derived peptides and radioiodine-labeling
Peptides were synthesized and purified to > 95% purity on a reverse-phase high-pressure liquid chromatography by GenScript (USA), Proteogenix (France; Y-GRI65–84 and Y-flg22) or in house on an Applied Biosystems 433A Peptide Synthesizer at VIB Ghent (GRIp68–97). Peptides were dissolved in H2O (stock solution 10 mg/ml) and diluted to the required concentrations just before experiments. Y-GRI65–84 and Y-GRI68–78 were radiolabeled with [125I] iodine using chloramine-T to yield 125I-Y-GRI65–84 and Y-GRI68–78, respectively, with a specific radioactivity > 2,000 Ci/mmol by BIOTREND Chemikalien (Germany).
Radio-ligand binding assays
Binding assays were performed as described (Bauer et al, 2001) with modifications. Plant material (0.1 g) was homogenized in 200 μl binding buffer (25 mM MES [2-(N-morpholino)ethanesulfonic acid] pH 6.0, 3 mM MgCl2, 10 mM NaCl) containing protease inhibitor cocktail (1:100; Fermentas/Thermo Fisher Scientific). Lysates were centrifuged at 4°C for 15 min at 10,000 g. The pellet was resuspended in 100 μl binding buffer and filtered through Miracloth. Binding assays were incubated for 20 min on ice in a total volume of 100 μl with 46 fmol 125I-Y-GRIp65–84 either alone (total binding) or in the presence of 10 μM Y-GRIp65–84 or GRIp65–84 (non-specific binding) for standard assays (concentrations for radiolabeled or cold peptides in saturation and competition assays are indicated in the figures).
Protoplasts were lysed in binding buffer containing 1% (w/v) octylphenoxypolyethoxyethanol (Nonidet NP-40), 0.1% SDS and 0.5% (w/v) sodium deoxycholate as detergents. After 2 h shaking at 4°C, the lysates were centrifuged at 4°C for 15 min at 10,000 g, and the supernatant was used directly for binding assays or for immunoprecipitation with subsequent binding assays.
Membrane fractions or immunoprecipitates were collected by vacuum filtration on glass fiber filters (Macherey–Nagel MN GF-2; preincubated in binding buffer containing 1% bovine serum albumin, 1% bactopeptone, 1% bactotryptone, 1% polyethylenimine). Prior to peptide binding filters were rinsed with 1 ml binding buffer. For 125I-Y-GRIp65–84 after filtration, filters were washed under constant vacuum with 1 ml binding buffer, 10 ml wash buffer I (20 mM Tris pH 7.5, 5 mM EDTA, 100 mM NaCl, 1% Triton X-100) and 5 ml wash buffer II (20 mM Tris pH 7.5, 5 mM EDTA, 1 M NaCl, 1% Triton X-100) (Jonak et al, 2000). For 125I-Y-GRIp68–78, filters were washed under constant vacuum with 1 ml binding buffer and twice with 5 ml wash buffer I (20 mM Tris pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100). Radioactivity retained on the filters was measured using a Wallac Wizard3 Gamma Counter. Specific binding was calculated by subtracting non-specific from total binding.
Western analysis of protoplasts transfected with PRK5-c-myc, PRK4-c-myc or YFP used anti-c-myc A-14 rabbit polyclonal and 9E10 mouse monoclonal antibodies (Santa Cruz), respectively.
In vitro interaction analysis and protein kinase assays
Recombinant glutathione-S-transferase (GST) or maltose-binding protein (MBP) fusion proteins of GRI, GRIp31–96 or the intracellular kinase domains of PRK5 and PRK4 were produced in Escherichia coli BL21 cells and purified according to manufacturer’s instructions. In vitro interaction tests of the extracellular domain of PRK5, PRK4 and FLS2 with GRI were performed as described (Nakagami et al, 2004). Western analysis for GST-fusion proteins was done using anti-GST antibody (mouse monoclonal antibody; Sigma Aldrich), anti-GRI antibody (rabbit polyclonal antibody raised against the epitope TRKCASGVKCEYGYC; Inbiolabs) or anti-GRI-pep (rabbit polyclonal antibody raised against the epitope VVDQEDDPEYYIL; Inbiolabs), respectively. Radioactive in vitro protein kinase assays using myelin-basic protein as an artificial substrate were performed as described (Idänheimo et al, 2014) in 10 mM HEPES pH 7.4, 1 mM dithiothreitol with either 10 mM MgCl2 or MnCl2.
Microscopy
PRK5 subcellular localization was analyzed by confocal microscopy on a Leica SP5 II HCS A inverted confocal microscope using a solid state blue laser for CFP, YFP and chloroplast autofluorescence (detection with 465–510, 521–587 and 636–674 nm range, respectively).
In vitro AtMC9 cleavage assay
Recombinant His-6 purified AtMC9 protease (Vercammen et al, 2004) and the inactive AtMC9C147AC29A mutant (Belenghi et al, 2007) were pre-activated in assay buffer containing 50 mM MES (pH 5.5), 150 mM NaCl, 10% (w/v) sucrose, 0.1% (w/v) CHAPS and 10 mM DTT for 15 min at room temperature. The proteases were then mixed with the substrate in several pmol quantity ratios and incubated at 30°C for 30 min. The reaction was terminated by addition of Laemmli–SDS–PAGE loading buffer and heating to 95°C for 5 min. Proteins were separated on 12% SDS–PAGE gels and transferred to Immobilon P (Millipore) membrane for Western blot analysis. AtMC9 was probed with a rabbit polyclonal antibody (Vercammen et al, 2004) and MBP-tagged substrates with a MBP-tag rabbit polyclonal antibody (Santa Cruz) and visualized by chemiluminescence (Western Lightning Plus-ECL; PerkinElmer). In-solution acetyl-2H(3) labeling and subsequent MS/MS analysis of neo N-termini was performed as described (Melzer et al, 2012).
Mass spectrometric analysis
In-solution trideutero-acetyl (<AcD3>)-labeling and subsequent MS/MS analysis of neo N-termini was performed as previously described (Helsens et al, 2008) with minor modifications. Approximately 5 μg of AtMC9 and MBP-GRI25–168 (total amount in the protein mix) were reacted for 90 min at 30°C in AtMC9 assay buffer supplemented with 2 mM TCEP. The reaction was stopped by raising the pH to 8.0 with NaOH and alkylation of cysteines with 10 mM iodoacetamide for 1 h at 30°C in the dark. The buffer was exchanged to 50 mM triethylammonium bicarbonate buffer. Labeling was performed in solution with a threefold molar excess of N-hydroxysuccinimide ester of trideutero-acetate (produced in-house), twice for 1 h at 30°C. To quench remaining NHS-ester, a fourfold molar excess (over NHS-ester) of glycine was added for 10 min at 30°C. Trypsin digestion was performed overnight, after which the digest was incubated with a fourfold molar excess of hydroxylamine for 10 min at 30°C and was subsequently acidified with trifluoroacetic acid (TFA). Peptides were analyzed by LC-MS/MS using a Thermo LTQ Orbitrap XL mass spectrometer. Spectra were identified with the Mascot search algorithm in the TAIR10 database (concatenated with MBP-GRI25–168 sequence).
The Mascot search parameters and all identified spectra matching the MBP-GRI25–168 sequence were grouped in Supplementary Dataset S1.
In vitro GRIp31–96 peptide cleavage by AtMC9 and separation by RP-HPLC
The GRIp31–96 peptide was dissolved to a concentration of 1 mM in 5% formic acid and adjusted to pH 3.8 with NaOH. 200 μM GRIp31–96 peptide was incubated per 40 μl reaction mixture (pH of the reaction mixtures was adjusted to pH 5.5 with NaOH) containing increasing concentration of recombinant AtMC9 (0, 31, 125 or 500 nM rAtMC9) in AtMC9 assay buffer (supplemented with 40 mM DTT and 40 mM MES buffer). After incubation for 30 min at 30°C, the reaction was stopped by addition of 5 μl 10% TFA. Cleavage products in the samples were separated by reverse-phase high-performance liquid chromatography (RP-HPLC, Agilent Technologies 1200 series) on a 2.1 mm (internal diameter) C18 column in solvent A (2/98% ACN/H20, 0.1% TFA) with a gradient increase of 1% solvent B (70/30% ACN/H2O, 0.1% TFA) per minute. Buffer controls were run with the same parameters. Peptide elution was monitored by measuring UV absorbance at 280 nm, and 1 min fractions were collected from 24 to 120 min in a 96-well plate. Fractions corresponding to each peak in the 500 nM rAtMC9 sample were measured by MALDI-TOF MS (Ultraflex, Bruker), and the measured masses were linked to GRIp31–96 peptide fragments. The fractions of the 0 nM rAtMC9 sample corresponding to the suspected full-length GRIp31–96 peptide were analyzed by LC-MS/MS using a Thermo LTQ Orbitrap XL mass spectrometer, and the correct GRIp31–96 mass was deduced from the MS precursor masses. Detected peptides are listed in Supplementary Dataset S1.
Primer sequences
Primers used for cloning are listed in Supplementary Table S1.
Sequence analysis
Sequences were aligned using PSI-Coffee (Di Tommaso et al, 2011) at the T-Coffee web server (http://tcoffee.crg.cat/apps/tcoffee/index.html).
Protein structure predictions
Protein structure predictions were done using I-TASSER (Roy et al, 2010) (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) or Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id==index) (Kelley & Sternberg, 2009).
Statistical analysis
Statistical analysis was performed in IBM SPSS Statistics (version 22). Ion leakage and binding data were analyzed using one-way ANOVA using Sidak’s post hoc test. Bacterial growth was analyzed using two-way ANOVA using Tukey’s HSD post hoc test. Plots were created in Sigmaplot and in R.
Acknowledgments
We thank Tuomas Puukko and Leena Grönholm for technical help. We thank Dr. Silke Robatzek and Dr. Sarah K. Coleman for help with radio-ligand binding, Dr. Claudia Jonak for suggestions for analysis of protein kinase activity and Dr. Jan Willem Borst, Prof. Theodorus Galdella, Dr. Silke Robatzek and Dr. Claudia Jonak for materials. We thank Dr. Mikko Frilander for access to the isotope laboratory at the Institute of Biotechnology, University of Helsinki, and Dr. Jarkko Salojärvi for help with statistics. We thank Drs Sarah K. Coleman, Pinja Jaspers, Julia Krasensky, Johanna Leppälä, Maija Sierla and Jorma Vahala for critical comments on the manuscript. This research was supported by the Academy of Finland Centre of Excellence program and Helsinki University Biocentrum Helsinki program (to JK), Research Foundation-Flanders (grant number G.0038.09N to FVB) and Ghent University (Special Research Fund and Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy to FB), the Swedish Energy agency and Research councils VR, Vinnova and Formas (to HT). AG is supported by a post-doctoral grant from the Academy of Finland (decision #140187); MW is supported by the University of Helsinki (post-doctoral grant and 3-year fund allocation) and the Academy of Finland (decision #275632). JPV was supported by the Finnish Cultural Foundation (grant number 00111000). LT was supported by VIB International PhD Program and EMBO (LTF-776-2013). Financial support for the laboratory of YH was provided by the Academy of Finland. Centre of Excellence programme, the University of Helsinki, the European Research Council Advanced Investigator Grant Symdev, and the Gatsby Foundation.
Author contributions
MW and JPV contributed equally to this work. SS, LT and HHRR contributed equally to this work. MW, JPV, SS, LT, HHRR, HT, KG, FVB, YH and JK designed research, and MW, JPV, HHRR, SS, LT, AG, DK, AL, AS and BB carried out experiments. MW, JPV and JK wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figures
Supplementary Table S1
Supplementary Dataset S1
Review Process File
Source Data for Figure 2H
Source Data for Figure 3A
Source Data for Figure 3F
Source Data for Figure 4A
References
- Aker J, de Vries SC. Plasma membrane receptor complexes. Plant Physiol. 2008;147:1560–1564. doi: 10.1104/pp.108.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach D, Robatzek S. Pattern recognition receptors: from the cell surface to intracellular dynamics. Mol Plant Microbe Interact. 2007;20:1031–1039. doi: 10.1094/MPMI-20-9-1031. [DOI] [PubMed] [Google Scholar]
- Bauer Z, Gómez-Gómez L, Boller T, Felix G. Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J Biol Chem. 2001;276:45669–45676. doi: 10.1074/jbc.M102390200. [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. Paternal control of embryonic patterning in Arabidopsis thaliana. Science. 2009;323:1485–1488. doi: 10.1126/science.1167784. [DOI] [PubMed] [Google Scholar]
- Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inzé D, Delledonne M, Van Breusegem F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem. 2007;282:1352–1358. doi: 10.1074/jbc.M608931200. [DOI] [PubMed] [Google Scholar]
- Blaum BS, Mazzotta S, Nöldeke ER, Halter T, Madlung J, Kemmerling B, Stehle T. Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J Struct Biol. 2014;186:112–121. doi: 10.1016/j.jsb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Zhang B, Stael S, Denancé N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H. Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. Plant peptides in signalling: looking for new partners. Trends Plant Sci. 2009;14:255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J Exp Bot. 2007;58:3503–3511. doi: 10.1093/jxb/erm226. [DOI] [PubMed] [Google Scholar]
- Chang F, Gu Y, Ma H, Yang Z. AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol Plant. 2013;6:1187–1201. doi: 10.1093/mp/sss103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- Dat JF, Pellinen R, Beeckman T, van de Cotte B, Langebartels C, Kangasjärvi J, Inzé D, Van Breusegem F. Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 2003;33:621–632. doi: 10.1046/j.1365-313x.2003.01655.x. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MH, Goldberg RB, Mariani C. Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J. 1994;13:2976–2984. doi: 10.1002/j.1460-2075.1994.tb06596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Blaum BS, Stehle T, Kemmerling B. BIR2 affects complex formation of BAK1 with ligand binding receptors in plant defense. Plant Signal Behav. 2014a;9:e28944. doi: 10.4161/psb.28944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, Nürnberger T, Zipfel C, Clouse S, Borst JW, Boeren S, de Vries SC, Tax F, Kemmerling B. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol. 2014b;24:134–143. doi: 10.1016/j.cub.2013.11.047. [DOI] [PubMed] [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem. 2008;283:774–783. doi: 10.1074/jbc.M704185200. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Helsens K, Timmerman E, Vandekerckhove J, Gevaert K, Martens L. Peptizer, a tool for assessing false positive peptide identifications and manually validating selected results. Mol Cell Proteom. 2008;7:2364–2372. doi: 10.1074/mcp.M800082-MCP200. [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, ten Have A, Woltering EJ. A tomato metacaspase gene is upregulated during programmed cell death in Botrytis cinerea-infected leaves. Planta. 2003;217:517–522. doi: 10.1007/s00425-003-1049-9. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- Huang WJ, Liu HK, McCormick S, Tang WH. Tomato pistil factor STIG1 promotes in vivo pollen tube growth by binding to phosphatidylinositol 3-phosphate and the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell. 2014;26:3538–3555. doi: 10.1105/tpc.114.123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idänheimo N, Gauthier A, Salojärvi J, Siligato R, Brosché M, Kollist H, Mähönen AP, Kangasjärvi J, Wrzaczek M. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem Biophys Res Commun. 2014;445:457–462. doi: 10.1016/j.bbrc.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Jonak C, Beisteiner D, Beyerly J, Hirt H. Wound-induced expression and activation of WIG, a novel glycogen synthase kinase 3. Plant Cell. 2000;12:1467–1475. doi: 10.1105/tpc.12.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kwaaitaal M, Schor M, Hink MA, Visser AJ, de Vries SC. Fluorescence correlation spectroscopy and fluorescence recovery after photobleaching to study receptor kinase mobility in planta. Methods Mol Biol. 2011;779:225–242. doi: 10.1007/978-1-61779-264-9_13. [DOI] [PubMed] [Google Scholar]
- Lam E, Zhang Y. Regulating the reapers: activating metacaspases for programmed cell death. Trends Plant Sci. 2012;17:487–494. doi: 10.1016/j.tplants.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Lee H, Chah OK, Sheen J. Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature. 2011;473:376–379. doi: 10.1038/nature09958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TW, van den Burg HA, Joosten MH. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014;19:123–132. doi: 10.1016/j.tplants.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Löcke S, Fricke I, Mucha E, Humpert ML, Berken A. Interactions in the pollen-specific receptor-like kinases-containing signaling network. Eur J Cell Biol. 2010;89:917–923. doi: 10.1016/j.ejcb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Melzer IM, Fernández SB, Bösser S, Lohrig K, Lewandrowski U, Wolters D, Kehrloesser S, Brezniceanu ML, Theos AC, Irusta PM, Impens F, Gevaert K, Zörnig M. The Apaf-1-binding protein Aven is cleaved by Cathepsin D to unleash its anti-apoptotic potential. Cell Death Differ. 2012;19:1435–1445. doi: 10.1038/cdd.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave? Trends Plant Sci. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell. 2012;24:3198–3217. doi: 10.1105/tpc.112.099010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kiegerl S, Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J Biol Chem. 2004;279:26959–26966. doi: 10.1074/jbc.M312662200. [DOI] [PubMed] [Google Scholar]
- Pimenta DC, Lebrun I. Cryptides: buried secrets in proteins. Peptides. 2007;28:2403–2410. doi: 10.1016/j.peptides.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon W, Mayerhofer J, Petutschnig E, Fujioka S, Jonak C. ASKΘ, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 2010;62:215–223. doi: 10.1111/j.1365-313X.2010.04145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA, Jr, Välineva T, Saarikettu J, Frilander MJ, Suarez MF, Zavialov A, Ståhl U, Hussey PJ, Silvennoinen O, Sundberg E, Zhivotovsky B, Bozhkov PV. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol. 2009;11:1347–1354. doi: 10.1038/ncb1979. [DOI] [PubMed] [Google Scholar]
- Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S. LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J. 2004;39:343–353. doi: 10.1111/j.1365-313X.2004.02139.x. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV. Metacaspases. Cell Death Differ. 2011;18:1279–1288. doi: 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L, Gevaert K, Van Breusegem F. Natural substrates of plant proteases: how can protease degradomics extend our knowledge? Physiol Plant. 2012;145:28–40. doi: 10.1111/j.1399-3054.2011.01534.x. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, Van Damme P, Gevaert K, Van Breusegem F. The Arabidopsis METACASPASE9 degradome. Plant Cell. 2013;25:2831–2847. doi: 10.1105/tpc.113.115287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P, Fulton L, Batoux M, Yadav RK, Schneitz K. Structure-function analysis of STRUBBELIG, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS One. 2011;6:e19730. doi: 10.1371/journal.pone.0019730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inzé D, van Breusegem F. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem. 2004;279:45329–45336. doi: 10.1074/jbc.M406329200. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan JA, De Rycke R, Brackenier A, Inzé D, Harris JL, Van Breusegem F. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J Mol Biol. 2006;364:625–636. doi: 10.1016/j.jmb.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases? J Cell Biol. 2007;179:375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Loake GJ, Chu C. Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front Plant Sci. 2013;4:314. doi: 10.3389/fpls.2013.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011;66:969–982. doi: 10.1111/j.1365-313X.2011.04554.x. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Kangasjärvi J. Scorched earth strategy: Grim Reaper saves the plant. Plant Signal Behav. 2009a;4:631–633. doi: 10.4161/psb.4.7.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Kollist H, Kangasjärvi J. Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proc Natl Acad Sci USA. 2009b;106:5412–5417. doi: 10.1073/pnas.0808980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpinski S, Karpinska B, Kangasjärvi J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010;10:95. doi: 10.1186/1471-2229-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Kangasjärvi J. ROS signaling loops—production, perception, regulation. Curr Opin Plant Biol. 2013;16:575–582. doi: 10.1016/j.pbi.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Zhao BC, Ge WN, Zhang YF, Song Y, Sun DY, Guo Y. An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol. 2011;157:1884–1899. doi: 10.1104/pp.111.182808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table S1
Supplementary Dataset S1
Review Process File
Source Data for Figure 2H
Source Data for Figure 3A
Source Data for Figure 3F
Source Data for Figure 4A


