Abstract
The telomeric transcriptome comprises multiple long non-coding RNAs generated by transcription of linear chromosome ends. In a screening performed in Schizosaccharomyces pombe, we identified factors modulating the cellular levels of the telomeric transcriptome. Among these factors, Cay1 is the fission yeast member of the conserved family of Cactins, uncharacterized proteins crucial for cell growth and survival. In cay1Δ mutants, the cellular levels of the telomeric factor Rap1 are drastically diminished due to defects in rap1+ pre-mRNA splicing and Rap1 protein stability. cay1Δ cells accumulate histone H3 acetylated at lysine 9 at telomeres, which become transcriptionally desilenced, are over-elongated by telomerase and cause chromosomal aberrations in the cold. Overexpressing Rap1 in cay1+ deleted cells significantly reverts all telomeric defects. Additionally, cay1Δ mutants accumulate unprocessed Tf2 retrotransposon RNA through Rap1-independent mechanisms. Thus, Cay1 plays crucial roles in cells by ultimately harmonizing expression of transcripts originating from seemingly unrelated genomic loci.
Keywords: Cactin, fission yeast, heterochromatin, telomeres, TERRA
Introduction
Telomeres are protective nucleoprotein structures located at the ends of linear eukaryotic chromosomes. Telomeres prevent chromosome ends from being recognized as DNA lesions and handled by DNA repair machineries (Jain & Cooper, 2010; O'Sullivan & Karlseder, 2010). Aberrant DNA repair at telomeres results in attrition, recombination or fusions, and consequent genome rearrangements, which can promote cell transformation (Jain & Cooper, 2010; O'Sullivan & Karlseder, 2010). The structural organization and regulation of telomeres in the fission yeast Schizosaccharomyces pombe share a high degree of conservation with several multicellular organisms including mammals and this, together with the ease of genetic manipulation, makes S. pombe an ideal model organism for telomere studies (Jain & Cooper, 2010).
The core structure of telomeres comprises arrays of G-rich DNA tandem repeats protruding over the complementary C-rich strand to form a single-stranded 3′ overhang (the G-overhang), and the multiprotein complex shelterin (Jain & Cooper, 2010; O'Sullivan & Karlseder, 2010). Two central building blocks of shelterin are the double-stranded telomeric DNA-binding protein Taz1 (TRF1 and TRF2 in mammals) and Rap1 (Jain & Cooper, 2010; O'Sullivan & Karlseder, 2010). Rap1 and Taz1 exert multiple crucial functions at telomeres together or independently, including regulation of telomerase, protection from aberrant DNA processing, and promoting telomeric DNA replication (Jain & Cooper, 2010; O'Sullivan & Karlseder, 2010). Rap1 is recruited to telomeres mostly through interaction with Taz1 (or TRF2), and this interaction stabilizes Rap1 cellular levels (Kanoh & Ishikawa, 2001; Celli & de Lange, 2005; Chen et al, 2011).
Telomeres, as other constitutive heterochromatin centers, are refractory to transcription and are enriched in molecular marks typical of repressive chromatin including histone H3 methylated at lysine 9 (H3K9me), and they are poor in acetylated H3K9 and H4 (Cam et al, 2005; Gomez et al, 2005). Both Taz1 and Rap1 support telomeric silencing, and their ablation leads to transcriptional activation of reporter genes inserted at subtelomeres, as well as to loss of repression of natural subtelomeric genes including the telomere-linked RecQ-type DNA helicases tlh1+ and tlh2+ (Cooper et al, 1997; Kanoh & Ishikawa, 2001; Fujita et al, 2012). Mammalian Rap1 also regulates expression of subtelomeric and intrachromosomal genes (Martinez et al, 2010). Despite their repressive state, telomeres are actively transcribed into different long non-coding RNA (lncRNA) species, including TERRA (telomeric repeat-containing RNA) first discovered in mammalian cells (Azzalin et al, 2007; Schoeftner & Blasco, 2008). TERRA is mainly synthesized by DNA-dependent RNA polymerase II (RNAPII), which uses the C-rich telomeric strand to produce TERRA molecules comprising a subtelomeric tract and a variably long tract of G-rich telomeric RNA repeats (Azzalin et al, 2007; Luke et al, 2008; Schoeftner & Blasco, 2008; Nergadze et al, 2009). The ensemble of lncRNA species transcribed from chromosomes ends—the telomeric transcriptome—also comprises molecules structurally different from TERRA. In fission yeast, we have documented the existence of ARIA, a TERRA antisense transcript composed entirely of C-rich telomeric RNA repeats (Bah et al, 2012; Greenwood & Cooper, 2012). Additionally, the S. pombe telomeric transcriptome comprises the two antiparallel subtelomeric lncRNAs ARRET and αARRET (Bah et al, 2012; Greenwood & Cooper, 2012). ARRET is also found in budding yeasts and plants (Luke et al, 2008; Vrbsky et al, 2010), while ARIA and αARRET have been reported only in fission yeast.
The telomeric transcriptome is regulated at both transcriptional and post-transcriptional levels. Rap1 limits association of RNAPII with TERRA transcription start sites, and rap1Δ and taz1Δ cells accumulate all telomeric lncRNAs (Bah et al, 2012; Greenwood & Cooper, 2012). Silencing of ARRET and αARRET requires the H3K9 methyltransferase Clr4 and Swi6, the fission yeast orthologue of heterochromatin protein HP1 (Greenwood & Cooper, 2012). Also, all telomeric transcriptome species accumulate in mutants of the non-canonical poly(A) polymerases Cid12 and Cid14 (Bah et al, 2012), suggesting that polyadenylation promotes telomeric RNA degradation. In fission yeast, expression of the telomeric transcriptome is also affected by the genetic background. For example, in two independent strain backgrounds, deletion of rap1+ stabilized primarily ARIA or both TERRA and ARIA to similar extents (Bah et al, 2012; Greenwood & Cooper, 2012). In addition, chromosome end transcription in mammals readily responds to changes in environmental conditions such as changes in temperature (Schoeftner & Blasco, 2008).
We have screened a fission yeast gene deletion collection and identified new factors modulating the cellular levels of telomeric lncRNAs. One such factor is the fission yeast member of the eukaryotic family of Cactins (herein referred to as Cay1 for ‘Cactin in fission yeast protein 1’), which are evolutionarily conserved proteins of unclear molecular function involved in different fundamental processes such as control of gene expression, cell cycle progression, inflammation response, development, and embryogenesis (Lin et al, 2000; Atzei et al, 2010a,b; Tannoury et al, 2010; Szatanek et al, 2012; Baldwin et al, 2013). We show that cay1+ deletion impairs splicing of rap1+ pre-mRNA and Rap1 protein stability. Consequently, Rap1 levels are dramatically decreased in cay1Δ cells, leading to aberrant telomere elongation, increased acetylation of subtelomeric H3K9 and desilencing of telomeric RNA. Re-establishing physiological levels of Rap1 protein in cay1Δ cells largely reverts all telomeric aberrations. We also show that cay1Δ cells accumulate transcripts from Tf2 LTR retrotransposons through Rap1-independent mechanisms. Our studies offer the first molecular characterization of a member of the Cactin family and reveal its centrality in pre-mRNA splicing, protein stability, telomere maintenance, and retrotransposon expression.
Results
A screening of a complete S. pombe deletion library for telomeric RNA regulators identifies Cay1
To identify factors regulating TERRA cellular levels, we screened a collection of fission yeast strains individually deleted for 3,004 non-essential genes corresponding to ∼60% of the fission yeast genes. We prepared total RNA in a 96-well format and dot blot hybridized it using a radioactive double-stranded telomeric probe. Although this probe does not discriminate between TERRA and ARIA, its high specific activity allows detection of low amounts of transcripts, contrary to 5′ end-labeled oligonucleotides specific for the two telomeric strands. Telomeric signals were normalized through 18S rRNA signals for the same dot blot membranes (Supplementary Fig S1A). 31 and 20 deletion strains showed TERRA/ARIA levels at least twofold higher or threefold lower than wild-type cells (wt), respectively (Supplementary Table S1). We then focused on strains with up-regulated telomeric signal (UP-TERRA strains) and performed new dot blot analysis using U6 snRNA as a normalizer. This second normalizer was used to confirm that the increased telomeric signal in the identified UP-TERRA strains was not an artifact deriving from down-regulation of 18S rRNA rather than up-regulation in TERRA/ARIA. This second screening confirmed a two-to fourfold increase in telomeric RNAs in 8 UP-TERRA strains (Fig 1A; Supplementary Fig S1B), while in the other 23 strains, TERRA/ARIA signal was more modestly increased over wt. The 8 remaining strains are deleted for the following genes: mpn1+(U6 snRNA-specific RNA exonuclease Mpn1), vip1+(RNA-binding protein Vip1), cay1+ (spbc2f12.12c+, conserved eukaryotic protein Cay1 similar to human C19orf29/Cactin), pof3+(F-box protein Pof3), poz1+(telomeric protein Poz1), spbc2a9.02+(NAD-dependent epimerase/dehydratase family protein), spbp8b7.08c+(leucin carboxyl methyltransferase), and rap1+(telomeric protein Rap1). The identification of the telomeric factors Rap1 and Poz1, which promote silencing at chromosome ends (Kanoh & Ishikawa, 2001; Bah et al, 2012; Fujita et al, 2012), validated the framework of our screening.
Figure 1. Characterization of UP-TERRA mutants.
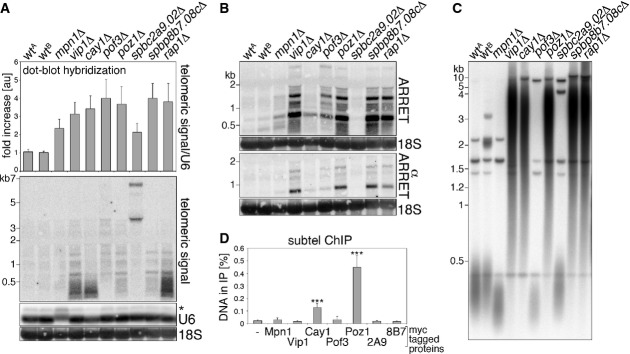
- A Telomeric lncRNA levels in the indicated deletion strains as analyzed by dot blot (top) or Northern blot hybridization (bottom) using a double-stranded telomeric probe. Telomeric dot blot signals are expressed as fold increase relative to wtA after normalization through U6 snRNA. Bars and error bars are averages and s.d. from 4 independent experiments. U6 and 18S rRNA are shown as loading controls for Northern blotting. The asterisk indicates unspliced U6. Molecular weights are on the left in kilobases.
- B ARRET and αARRET Northern blot analysis of the indicated strains using strand-specific probes.
- C Telomere length analysis of ApaI-digested DNA from the indicated strains.
- D Quantification of subtelomeric DNA from ChIP experiments using strains carrying the indicated Myc-tagged proteins (2A9 and 8B7: SPBC2A9.02-Myc and SPBP8B7.08c-Myc, respectively). Immunoprecipitated DNA is expressed as fraction of input DNA. An untagged strain (−) served as a negative control. Bars and error bars are averages and s.d. from three independent experiments. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. *P < 0.001 relative to untagged strain.
We then Northern blot hybridized total RNA from the 8 UP-TERRA mutants grown to exponential phase. vip1Δ, cay1Δ, and rap1Δ samples showed a clear increase in telomeric hybridization signal as compared to wt, while a more modest increase was observed for mpn1Δ, pof3Δ, poz1Δ, and spbp8b7.08cΔ samples (Fig 1A). In spbc2a9.02Δ cells, telomeric transcripts accumulated in two distinct bands of more than 3 kb rather than in a smear (Fig 1A), suggesting changes in the transcriptional landscape of chromosome ends. The differences between the results obtained in dot blot and Northern blot experiments possibly derive from different growth conditions; for dot blot experiments, strains were grown collectively in 96-well plates, thus not necessarily allowing the same number of generations and cell density, while for Northern blots, strains were grown separately to exponential phase. Moreover, while in dot blot experiments RNA is concentrated in one spot, in Northern blot experiments, RNA is electrophoresed prior to hybridization, and TERRA/ARIA signals are therefore spread throughout the gel lanes. ARRET RNA was also up-regulated, albeit to different extents, in all mutants except for spbc2a9.02Δ, while αARRET was clearly stabilized in vip1Δ, poz1Δ, spbp8b7.08cΔ, and rap1Δ (Fig 1B). Moreover, all UP-TERRA mutants except for spbc2a9.02Δ had altered telomere length (Fig 1C). In vip1Δ, cay1Δ, poz1Δ, spbp8b7.08cΔ, and rap1Δ cells telomeres were elongated, spanning in size between 0.5 and 10 kb, while in mpn1Δ and pof3Δ, telomeres were shorter than in wt. Finally, we Myc epitope tagged all eight TERRA regulators at their endogenous loci and performed chromatin immunoprecipitations (ChIPs) using anti-Myc antibodies followed by quantitative real-time PCR (qPCR) detecting subtelomeric DNA sequences adjacent to the first telomeric repeat. As expected, Poz1-Myc and Rap1-Myc co-immunoprecipitated significant amounts of subtelomeric DNA (Fig 1D and below). Among the other factors, only Cay1-Myc tested positive in our ChIP experiments, suggesting a direct role at telomeres and prompting us to analyze Cay1 functions in greater detail.
Cay1 represses Tf2 LTR retrotransposon transcripts
To probe a more global function for Cay1 in gene silencing, we hybridized whole-genome tiling arrays with cDNA prepared from cay1Δ and wt cells. As expected, we observed accumulation of subtelomeric transcripts, while transcripts from centromeres were not affected (Fig 2A; Supplementary Fig S2A). 47 gene transcripts were enriched more than twofold, while 62 were at least 0.7-fold less abundant than in wt (Supplementary Table S2). The most up-regulated transcripts corresponded to subtelomeric genes, such as the telomere-linked DNA helicases tlh1+ and tlh2+, and to the intrachromosomal genes ecl1+, aes1+, spbc1271.07/8c+, gdh2+, ppr1+, and spbc19c7.04c+ (Fig 2A). Strikingly, transcripts from all 13 Tf2 retrotransposons and many solo LTRs dispersed throughout the three fission yeast chromosomes (Bowen et al, 2003) strongly accumulated in cay1Δ cells (Fig 2A; Supplementary Fig S2A and B). Tf2 transcript hybridization signals were increased most robustly at the flanking LTR sequences (Supplementary Fig S2A). Northern blot analysis confirmed the stabilization of Tf2 and tlh1/2+ transcripts upon cay1+ deletion (Fig 2B). In cay1Δ cells, Tf2 transcripts mostly accumulated as unprocessed precursors running more slowly than processed Tf2s detected in wt cells (Fig 2B). Processed Tf2 transcripts, as detected in wt cells, lack a 5′-end sequence of 198 nucleotides that is removed through mechanisms that remain to be fully elucidated (Durand-Dubief et al, 2007). Although at lower levels than for subtelomeres, Cay1-Myc immunoprecipitated together with Tf2, Tf2 LTR, and tlh1/2+ DNA (Fig 2C).
Figure 2. cay1Δ cells accumulate Tf2 LTR retrotransposon transcripts.
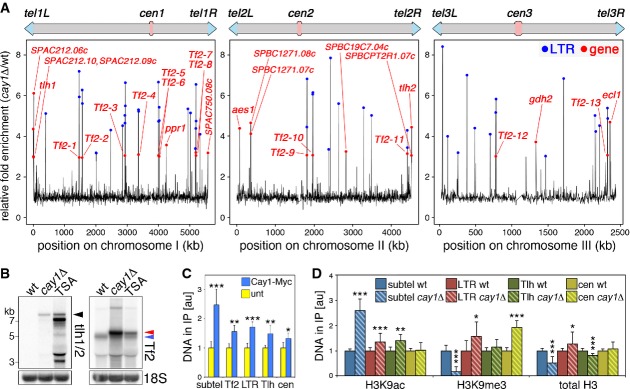
- A Chromosome maps of differentially expressed genes in cay1Δ cells shown as fold enrichment over wt. Red dots are overexpressed genes and Tf2 retrotransposons, blue dots are overexpressed LTR sequences.
- B Northern blot analysis of tlh1/2+, Tf2 retrotransposons, and 18S rRNA (loading control) in wt, cay1Δ, and trichostatin A (TSA)-treated wt cells. Black arrowhead indicates tlh1/2+ mRNA; red arrowhead unprocessed Tf2 transcripts; and blue arrowhead processed Tf2 transcripts.
- C ChIP analysis of Cay1-Myc binding to the indicated genomic loci. subtel: subtelomeres; Tf2: Tf2 retrotransposon genes; LTR: Tf2 LTR sequences; Tlh: tlh1/2+ genes; cen: centromeres. Immunoprecipitated DNA is normalized to input DNA and expressed as fold increase over untagged strains (unt).
- D ChIP analysis of H3K9ac, H3K9me3, and total H3 at the indicated genomic loci. Immunoprecipitated DNA is normalized to input DNA and expressed as fold increase over wt after subtraction of values obtained for negative control immunoprecipitations performed with beads only.
Data information: Bars and error bars are averages and s.d. from at least 4 (C) or 8 (D) independent experiments. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. *P < 0.05, **P < 0.01, *P < 0.001 relative to wt.
We next performed ChIP experiments using antibodies against acetylated H3K9 (H3K9ac), trimethylated H3K9 (H3K9me3), and total histone H3. H3K9ac increased by two- to threefold at subtelomeres in cay1Δ cells as compared to wt, while it was only marginally increased at Tf2 LTRs and tlh1/2+ genes and completely unaffected at centromeres (Fig 2D). H3K9me3 diminished by fivefold at subtelomeres, it increased by twofold at centromeres, and it remained largely unaffected at Tf2 LTRs and tlh1/2+ genes (Fig 2D). Altogether, these data suggest that Cay1 promotes telomeric heterochromatinization by restricting the levels of H3K9ac and promoting accumulation of H3K9me3, while it is not required for heterochromatin establishment at centromeres. Consistently, cay1+ deletion in a previously established reporter strain (Nimmo et al, 1998) desilenced a gene inserted subtelomerically, but not a centromeric reporter gene (Supplementary Fig S3). Moreover, it appears that Tf2 and tlh1/2+ transcript accumulation in cay1Δ cells does not derive from major changes in H3K9 methylation and acetylation. Finally, total H3 diminished by twofold at subtelomeres, while it was not substantially changed at the other loci (Fig 2D). Similarly, cellular H3 levels were diminished by twofold in cay1Δ as compared to wt cells (Supplementary Fig S4A), implying that reduced H3 density at chromosome ends as well as its diminished cellular levels could contribute to impaired telomere silencing. Nevertheless, telomeric transcripts were not stabilized to cay1Δ levels in histone H3/H4 gene deletion strains with different H3 cellular levels (Mellone et al, 2003) (Supplementary Fig S4B and C). Likewise, we did not observe major changes in telomere length in the same H3/H4 deletion strains (Supplementary Fig S4D). Because reduced cellular H3 levels are known to stabilize Tf2 transcripts (Zhou et al, 2013), we reasoned that Tf2 transcript increase in cay1Δ cells could be linked to reduced total H3 levels. Nonetheless, Tf2 RNA accumulated to extents similar to the ones observed in cay1Δ cells only in H3/H4 mutants with total H3 levels significantly lower than cay1Δ cells (Supplementary Fig S4B and C), suggesting that Cay1 does not repress Tf2 transcripts through regulation of total H3 levels.
Rap1 pre-mRNA splicing and protein stability are impaired in cay1Δ cells
Because the telomere elongation observed in cay1Δ is reminiscent of what was observed in rap1Δ and taz1Δ cells (Cooper et al, 1997; Kanoh & Ishikawa, 2001), we examined cellular Taz1 and Rap1 levels using antibodies against endogenous proteins and found that total Rap1 was reduced to approximately 10% of wt levels, while Taz1 was unaffected (Fig 3A). Similarly, Rap1-Myc but not Taz1-Myc cellular levels were severely diminished in cells deleted for cay1+ (Supplementary Fig S5A). Consistently, the density of telomere-bound Rap1-Myc was much lower in cay1Δ than in wt cells, while Taz1-Myc telomeric association was not affected (Supplementary Fig S5B). Cay1-Myc cellular levels and telomeric association were not substantially altered in cells deleted for rap1+, taz1+, or poz1+ (Supplementary Fig S5A and B).
Figure 3. Rap1 pre-mRNA splicing and protein stability are impaired in cay1Δ cells.
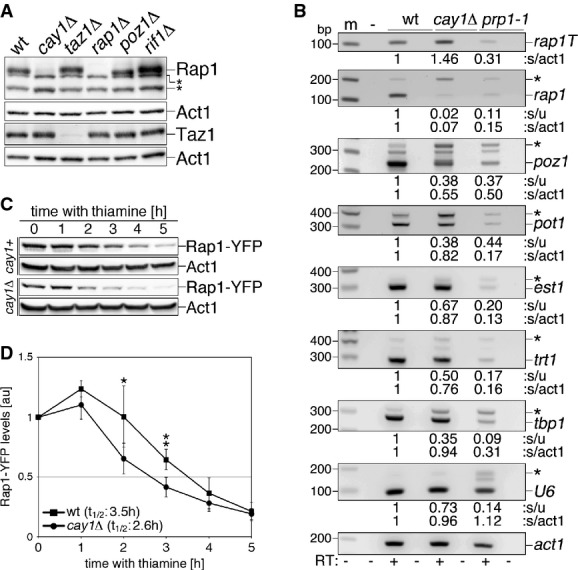
- A Western blot analysis of endogenous Rap1 and Taz1 in the indicated strains. Act1 was used as a loading control. Asterisks indicate cross-reacting bands.
- B RT–PCR analysis of splicing efficiencies of the indicated pre-mRNAs in wt and cay1Δ cells. The prp1-1 strain carries a thermosensitive allele of the splicing factor Prp1 and was used as a positive control for splicing impairment. prp1-1 cells were grown at 36°C for 4 h before harvesting. Amplification products corresponding to unspliced and spliced mRNAs are indicated by asterisks and gene names, respectively. rap1T indicates amplification products obtained with oligonucleotides spanning rap1+ exon 3 and thereby amplifying both spliced and unspliced mRNA. Intronless act1+ was used as a loading control. Numbers at the bottom of each panel are ratios between spliced and unspliced forms (s/u) and spliced and act1+ (s/act1). Values are expressed as fold increase over wt.
- C Western blot analysis of Rap1-YFP protein levels in the indicated strains upon nmt1 promoter shutoff by thiamine addition. Membranes were probed with antibodies against GFP and Act1 (loading control).
- D Quantification of Rap1-YFP protein levels in experiments as in (C). Rap1-YFP levels are expressed as fold increase over time 0 after normalization through Act1. Data points and error bars are averages and s.d. from at least four independent experiments. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. *P < 0.05, **P < 0.01 for cay1+ versus cay1Δ samples.
To elucidate the mechanisms leading to Rap1 insufficiency in cay1Δ cells, we first tested the state of rap1+ mRNA by PCR and observed a 1.5-fold increase in total levels in cay1Δ cells when we used oligonucleotide pairs amplifying a region within exon 3 and therefore not discriminating between spliced and unspliced forms (Fig 3B). This suggests that cay1+ deletion does not compromise rap1+ transcription. We then employed oligonucleotides spanning the junction between rap1+ exons 2 and 3. In cay1Δ samples, we observed a dramatic decrease in the ratio between the amplification products corresponding to spliced and unspliced mRNA, with the spliced form being down-regulated to 7% of wt (Fig 3B). Although to lower extents, pre-mRNA splicing of poz1+ was also affected and the mature form was down to 55% as compared to wt. Finally, splicing of the telomeric factors pot1+, est1+, and trt1+ and of the non-telomeric factors tbp1+ and snu6+ (U6) were also less efficient in cay1Δ cells; yet, the levels of spliced pot1+, est1+, trt1+, tbp1+, and U6 RNAs were only modestly diminished as compared to wt cells (Fig 3B). Consistently, Northern blot analysis did not reveal any major accumulation of unspliced U6 in cay1Δ cells (Fig 1A). These data show that cay1+ deletion affects splicing of different RNA substrates to different extents. Consistently, analysis of global splicing efficiencies using our tiling array data did not detect a major alteration in exon/intron abundance (Supplementary Fig S2C) following analysis pipelines that previously revealed pre-mRNA splicing defects in the mpn1Δ mutant (Shchepachev et al, 2012).
We also tested whether Cay1 regulates Rap1 protein stability. We overexpressed a Rap1-YFP fusion protein from a heterologous nmt1 promoter in wt and cay1Δ cells. The Rap1-YFP fusion gene contains all intronic and exonic sequences of rap1+. We measured Rap1-YFP protein stability by nmt1 promoter shutoff experiments and found that Rap1-YFP had a half-life of 2.6 h in cay1Δ cells and 3.5 h in cay1+ cells (Fig 3C and D). nmt1 promoter repression occurred with similar kinetics in both wt and cay1Δ cells as determined by quantitative reverse transcription PCR (qRT–PCR) measurements of rap1-yfp mRNA at different time points (unpublished observations). In conclusion, the diminished levels of Rap1 in cay1Δ cells are largely due to impaired rap1+ pre-mRNA splicing and, apparently to a lower extent, to destabilization of Rap1 protein. Cellular Rap1 protein levels were largely unaffected in several H3/H4 deletion strains (Supplementary Fig S4B) arguing against the possibility that diminished H3 levels in cay1Δ cells contribute to Rap1 insufficiency.
Cay1 and Rap1 genetically interact to regulate telomere length, transcription, and H3K9 acetylation
The diminished Rap1 levels are likely to explain the telomeric phenotypes observed in cay1Δ cells. Indeed, telomeres in cay1Δ and rap1Δ cells share several common features. First, as for rap1Δ (Miller et al, 2006), cay1Δ telomeres gradually shortened over successive generations when trt1+, the gene encoding the catalytic subunit of telomerase, was deleted (Fig 4A). Distinct bands positive to telomeric probe hybridizations appeared between 3 and 10 kb at late generations after trt1+ deletion most likely reflecting subtelomeric recombination events (Fig 4A). Moreover, similar to rap1Δ strains, we detected longer G-overhangs in cay1Δ cells as compared to wt (Supplementary Fig S6A). We then deleted cay1+ in rap1Δ cells and analyzed telomere length and integrity. Telomeres were longer in rap1Δ than in cay1Δ cells and remained at cay1Δ length in the doubly deleted strain (Fig 4B). Moreover, we tested whether cay1Δ telomeres, as it is the case for rap1Δ (Miller et al, 2005), are unprotected and therefore substrate for NHEJ. Contrary to rap1Δ cells, G1-arrested cay1Δ cells did not show any overt telomere fusion, while cay1Δrap1Δ cells fused their telomeres similar to rap1Δ cells (Fig 4C). Thus, while insufficient levels of Rap1 in cay1Δ cells can largely account for the telomere over-elongation, the ∼10% of Rap1 protein remaining in cay1Δ cells is sufficient to maintain telomere end-protection. We also deleted cay1+ in the other telomeric mutants taz1Δ, poz1Δ, and rif1Δ. Deletion of cay1+ in taz1Δ resulted in extensive loss of telomeric signal (Fig 4B), indicating that Cay1 is required to maintain telomeres in taz1Δ and vice versa. In cay1Δpoz1Δ cells, telomeres were slightly longer than in cay1Δ cells (Fig 4B), and deleting rif1+ in cay1Δ led to further lengthening of telomeres as compared to single deletion mutants (Supplementary Fig S6B), similar to what was previously seen for rap1Δrif1Δ mutants (Kanoh & Ishikawa, 2001).
Figure 4. Cay1 and Rap1 genetically interact to maintain telomere homeostasis.
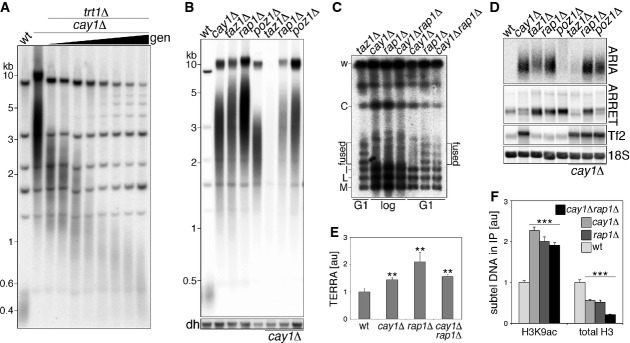
- A Telomere length analysis of ApaI-digested DNA from cay1Δtrt1Δ cells harvested at increasing generation doublings (gen).
- B Telomere length analysis of ApaI-digested DNA from the indicated strains. dh: centromeric dh repeats shown as loading control.
- C PFGE analysis of telomeric fusions in strains grown to logarithmic phase (log) or G1-arrested by nitrogen starvation (G1). Genomic DNA was digested with NotI and hybridized to C, I, L, and M probes detecting terminal fragments of chromosomes I and II. Bands corresponding to chromosome end fusions are indicated (fused).
- D Northern blot analysis of ARIA, ARRET, Tf2 retrotransposons, and 18S rRNA (loading control) in the indicated strains.
- E qRT–PCR quantification of TERRA levels expressed as fold increase over wt after normalization through act1+ mRNA. Bars and error bars are averages and s.d. from 3 independent experiments. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. **P < 0.01 relative to wt.
- F ChIP analysis of H3K9ac and total H3 for the indicated strains. Immunoprecipitated DNA is normalized to input DNA and expressed as fold increase over wt after subtraction of values obtained for negative control immunoprecipitations performed with beads only. Bars and error bars are averages and s.d. from at least four independent experiments. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. *P < 0.001 relative to wt.
As for the telomeric transcriptome, single deletion of cay1+ or rap1+ led to similar accumulation of ARIA as detected by Northern blot using strand-specific telomeric oligonucleotides (Supplementary Fig S6C) or random primer labeled double-stranded telomeric probes (Fig 4D). As TERRA was essentially undetectable in all strains, we performed qRT–PCR using telomeric C-rich oligonucleotides for reverse transcription and found that TERRA increased by about 1.5- to 2-fold in both cay1+ or rap1+ deleted cells (Fig 4E). ARRET also accumulated in the two strains to similar extents as measured by Northern blotting using strand-specific probes (Fig 4D; Supplementary Fig S6C). ARIA, TERRA, and ARRET did not further accumulate in cay1Δrap1Δ cells as compared to single mutants (Fig 4D and E; Supplementary Fig S6C). Thus, similar to what was observed for telomere length regulation, Cay1 and Rap1 genetically interact to silence chromosome ends. Telomeric transcripts were also stabilized in taz1+ deleted cells, while they were severely diminished in cay1Δtaz1Δ cells (Fig 4D; Supplementary Fig S6C), possibly due to the observed loss of telomeric sequences (Fig 4B). As observed above (Fig 1A and B), poz1+ single deletion strongly stabilized ARRET, while telomeric repeat RNA was less increased than that in cay1Δ, rap1Δ and taz1Δ cells (Fig 4D), indicating a major role for Poz1 in silencing subtelomeres. Double deletion of poz1+ and cay1+ stabilized ARIA and ARRET to levels similar to the ones observed in cay1Δ cells (Fig 4D). Finally, rif1+ deletion did not visibly affect the telomeric transcriptome nor did it show genetic interaction with cay1+ deletion (Supplementary Fig S6D). We also examined Tf2 transcript levels and did not detect accumulation or processing defects in any of the single telomeric mutants. Moreover, in all double mutant strains, Tf2 transcripts remained at levels comparable to the ones in the cay1Δ single mutant (Fig 4D; Supplementary Fig S6D).
We next analyzed H3K9 acetylation and H3 density at cay1Δ and rap1Δ chromosome ends. Subtelomeric H3K9ac accumulated to similar levels in cay1Δ and rap1Δ cells, and no further increase was observed in the double mutant (Fig 4F). Subtelomeric H3 density diminished by approximately twofold in both cay1Δ and rap1Δ cells and by approximately fourfold in the double mutant (Fig 4F). H3 cellular levels were lower in cay1Δrap1Δ double mutants than in cay1Δ cells, while no change in cellular H3 levels was observed in rap1Δ cells (Supplementary Fig S4A). These results establish that Cay1 and Rap1 restrict subtelomeric H3K9ac through the same pathway.
cay1Δ cells suffer from growth retardation and chromosomal aberrations in the cold
Colony spotting assays revealed that cay1+ deletion affects cell proliferation. cay1Δ cells grew slower than wt already at standard temperatures (30°C), and the slow growth was severely exacerbated in the cold (20°C; Fig 5A). Cold sensitivity was accompanied by cell elongation (Fig 5B and C), which is a hallmark of cell cycle checkpoint activation, and by appearance of chromosome bridges between daughter cell nuclei (Fig 5D). Consistent with published reports, rap1+ and poz1+ deleted cells grew normally at 30°C or 20°C (Miller et al, 2005; Fujita et al, 2012) and cay1Δrap1Δ and cay1Δpoz1Δ mutants behaved as cay1Δ cells at both temperatures (Fig 5A). As previously reported (Miller & Cooper, 2003), taz1Δ cells also grew slower and accumulated chromosome bridges in the cold, yet not as dramatically as cay1Δ (Fig 5A, C, and D). cay1Δtaz1Δ cells proliferated slower at 30°C than cay1Δ cells (Fig 4B).
Figure 5. Proliferation of cay1Δ cells.
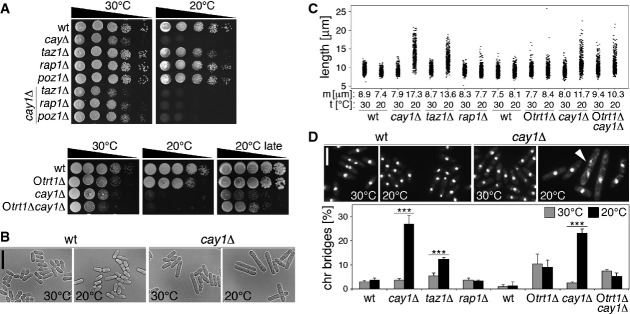
- A Serial dilutions of the indicated strains were spotted on complete medium and grown at 30°C or 20°C. Otrt1Δ and Otrt1Δcay1Δ are strains with circularized chromosomes lacking telomeric sequence (Supplementary Fig S7).
- B Representative images of wt and cay1Δ cells grown for 24 h at 30°C or 20°C. Scale bar, 20 μm.
- C Quantification of cell length for strains grown in complete medium for 24 h at 30°C or 20°C. Each dot represents a single cell and median cell length (m) is indicated for each sample. Data are from three independent experiments and at least 100 cells were scored per sample in each experiment.
- D Top: Examples of DAPI-stained wt and cay1Δ cells grown in complete medium for 24 h at 30°C or 20°C. The white arrowhead indicates a chromosome bridge. Scale bar, 10 μm. Bottom: Quantification of chromosome bridges. Bars and error bars are averages and s.d. from three independent experiments and at least 200 cells were scored per sample in each experiment. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. *P < 0.001 relative to 30°C.
We then deleted cay1+ in a trt1Δ survivor with circularized chromosomes devoid of telomeric sequences (Otrt1Δ; Supplementary Fig S7). Otrt1Δ cells grew better than cay1Δ both at 30°C and at 20°C, while Otrt1Δcay1Δ were slower than cay1Δ at 30°C (Fig 5A). However, Otrt1Δcay1Δ mutants proliferated faster and elongated lesser than cay1Δ cells in the cold and did not accumulate more chromosome bridges than Otrt1Δ cells (Fig 5A, C, and D). Thus, chromosome bridges scored in cay1Δ at 20°C largely depend on the presence of linear telomeres, while the slow growth only moderately derives from telomeric aberrations. Mutants with reduced H3 cellular levels did not recapitulate the cold sensitivity associated with cay1+ deletion (Supplementary Fig S4E).
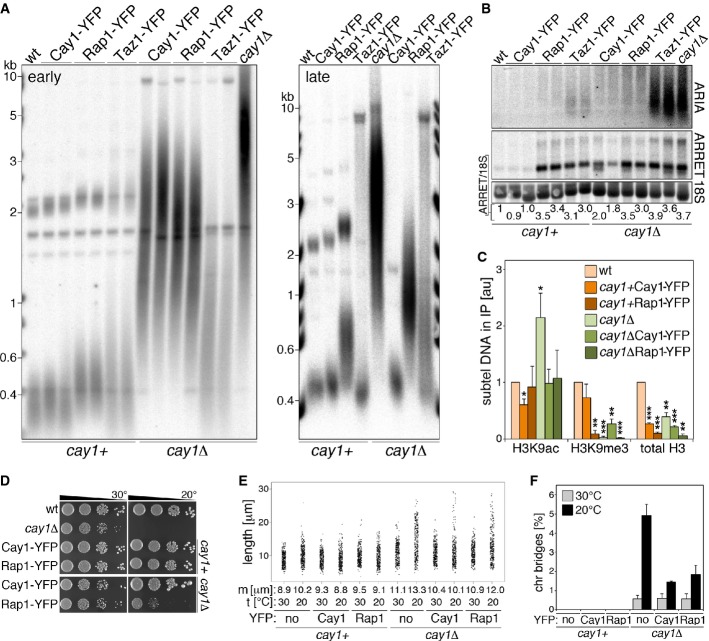
Re-establishment of Rap1 levels rescues the telomeric defects associated with cay1+ deletion
To directly test whether the telomeric defects in cay1Δ cells stem from insufficient cellular Rap1, we utilized the above-mentioned overexpression system where a rap1-yfp gene transcribed from an nmt1 promoter is stably integrated at the leu1 locus of cay1+ and cay1Δ cells. We also generated analogous strains overexpressing Cay1-YFP and Taz1-YFP. Cay1-YFP expression in cay1Δ cells reverted splicing defects of rap1+ and poz1+ mRNA and established wt-like Rap1 protein levels (Fig 6A and B). Cay1-YFP accumulated in the nucleus and formed a single dot in approximately 27% of cells both in cay1+ and in cay1Δ cells (Fig 6C). In cells simultaneously expressing Cay1-YFP and Pot1 C-terminally tagged with RFP, YFP and RFP foci never co-localized (Fig 6C); this, together with Cay1-myc ChIP results (Figs1D and 2C), indicates that Cay1 binds to telomeres at low levels or transiently, thereby impeding cytological detection of the protein at telomeric loci. rap1-yfp pre-mRNA splicing ensued efficiently in cay1+ strains, while it was impaired in cay1Δ cells. Nevertheless, the increased rap1-yfp transcripts achieved through nmt1 overexpression restored spliced rap1 levels above the ones in wt cells (Fig 6A). Consistently, Rap1-YFP in cay1Δ cells was expressed at levels comparable to endogenous Rap1 in wt strains, while it was 10 times more abundant in cay1+ cells (Fig 6B). poz1+ pre-mRNA splicing and mature mRNA levels were still impaired in cay1Δ cells overexpressing Rap1-YFP (Fig 6A). Rap1-YFP localized to the nucleus and formed 2 to 3 distinct foci, likely representing telomeres, in both cay1+ and cay1Δ cells (Fig 6D). Finally, Taz1-YFP was highly overexpressed over endogenous Taz1 in both cay1+ and cay1Δ cells and did not affect Rap1 protein levels (Fig 6B).
Figure 6. Overexpression of Rap1-YFP in cay1Δ cells.
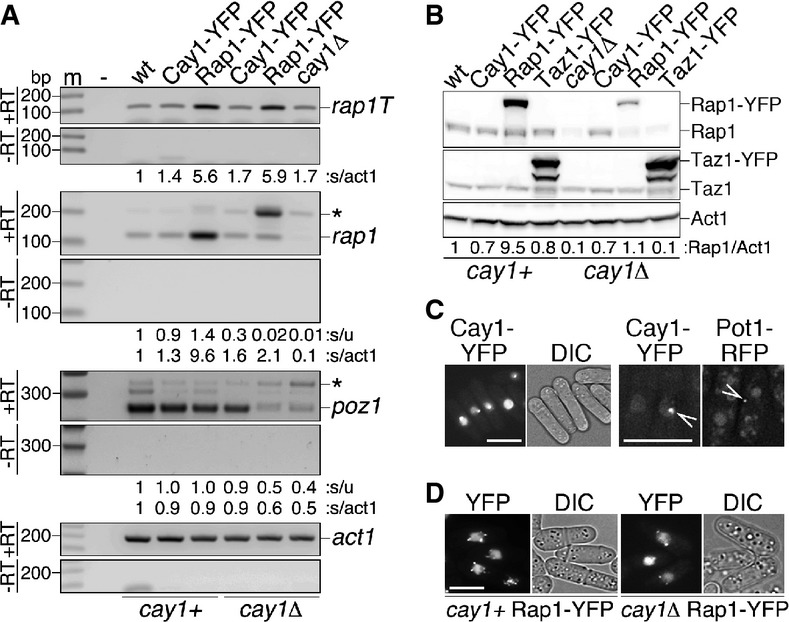
- A RT–PCR analysis of pre-mRNA splicing efficiencies of rap1+ and poz1+ pre-mRNAs in cay1+ and cay1Δ cells expressing Cay1-YFP or Rap1-YFP. Amplification products corresponding to unspliced and spliced mRNAs are indicated by asterisks and gene names, respectively. rap1T indicates amplification products obtained with oligonucleotides spanning rap1 exon 3 and thereby amplifying both spliced and unspliced mRNA. Intronless act1+ was used as a loading control. Numbers at the bottom of each panel are ratios between spliced and unspliced forms (s/u) and spliced and act1+ (s/act1). Values are expressed as fold increase over wt.
- B Western blot analysis of total proteins from early generation cay1+ and cay1Δ cells expressing Cay1-YFP, Rap1-YFP, and Taz1-YFP using antibodies against Rap1, Taz1, and Act1 (loading control). Numbers at the bottom are ratios between Rap1 proteins (endogenous plus ectopic) and Act1 (Rap1/Act1). Values are expressed as fold increase over wt.
- C Left: Representative YFP fluorescence and DIC images of cay1Δ strains expressing Cay1-YFP. Scale bar, 10 μm. Right: Representative YFP and RFP fluorescence images of Cay1-YFP- and Pot1-RFP-expressing cells. Arrows point to YFP and RFP foci within the same nucleus demonstrating lack of co-localization. Scale bar, 5 μm.
- D Representative YFP fluorescence and DIC images of cay1+ and cay1Δ strains expressing Rap1-YFP. Scale bar, 5 μm.
Continuous expression of Cay1-YFP led to a progressive shortening of telomeres, eventually re-establishing wt telomere length at late generations (Fig 7A), confirming that aberrant telomere elongation is a true outcome of cay1+ deletion. Cay1-YFP expression in cay1+ cells did not alter telomere length (Fig 7A). Rap1-YFP gradually shortened cay1Δ telomeres and re-set them to a new length that was greater than in wt cells or in cay1Δ cells expressing Cay1-YFP (Fig 7A). Rap1-YFP alone led to telomere elongation when overexpressed in cay1+ strains, although telomere length was below the one in cay1Δ strains expressing Rap1-YFP (Fig 7A). Finally, Taz1-YFP also induced telomere shortening when expressed in cay1Δ cells albeit with different kinetics than Rap1-YFP and Cay1-YFP (Fig 7A), suggesting different molecular mechanisms.
Figure 7. Rap1 ectopic expression reverts the telomeric defects of cay1Δ cells.
- A Telomere length analysis of ApaI-digested DNA from cells harvested at early (left) and late generations (right) after expression of YFP-tagged proteins.
- B Northern blot analysis of ARIA, ARRET, and 18S rRNA (loading control) at early generations after expression of YFP-tagged proteins. Numbers at the bottom are ratios between ARRET and 18S rRNA signals, expressed as fold increase over wt.
- C ChIP analysis of subtelomeric H3K9ac, H3K9me3, and total H3 in the indicated strains at early generations. Immunoprecipitated DNA is normalized to input DNA and expressed as fold increase over wt after subtraction of values obtained for negative control immunoprecipitations performed with beads only. Bars and error bars are averages and s.d. from 3 independent experiments. Statistical significance was assayed using the unpaired, two-tailed Student's t-test. *P < 0.05, **P < 0.01, *P < 0.001 relative to wt.
- D Serial dilutions of early generation cay1+ and cay1Δ cells expressing Cay1-YFP and Rap1-YFP were spotted on complete medium and grown at 30°C and 20°C.
- E Quantification of cell length in strains grown for 24 h in complete medium at 30°C or 20°C. Each dot represents a single cell and median cell length (m) is indicated for each sample. Data are from 3 independent experiments and at least 100 cells were scored per sample in each experiment.
- F Quantification of chromosome bridges of strains grown for 24 h in complete medium at 30°C or 20°C. Bars and error bars are averages and s.d. from three independent experiments and at least 200 cells were scored per sample in each experiment.
As for the telomeric transcriptome, Rap1-YFP quickly silenced ARIA in cay1Δ cells, as did Cay1-YFP but not Taz1-YFP (Fig 7B). Thus, as for telomere elongation, ARIA accumulation in cay1Δ cells is a true outcome of cay1+ deletion and primarily derives from Rap1 insufficiency and not from elongated telomeres. ARRET levels in cay1Δ were substantially reduced by Cay1-YFP although not to wt levels and were not affected by Rap1-YFP or Taz1-YFP (Fig 7B). Moreover, Rap1-YFP and Taz1-YFP expression led to accumulation of ARRET in cay1+ cells (Fig 7B). We also performed ChIP experiments using chromatin from early generation strains over-expressing Cay1-YFP and Rap1-YFP. Cay1-YFP but not Rap1-YFP slightly diminished the levels of subtelomeric H3K9 acetylation when expressed in wt cells. Most importantly, both Cay1-YFP and Rap1-YFP restored normal subtelomeric H3K9ac levels in cay1Δ cells (Fig 7C). The defects in H3K9me3 and total H3 density associated with cay1+ deletion were not resolved by either Cay1-YFP or Rap1-YFP. In addition, Cay1-YFP diminished the levels of subtelomeric H3, and Rap1-YFP of both subtelomeric H3K9me3 and H3 when expressed in wt cells (Fig 7C). Altogether, these data further confirm that accumulation of H3K9ac at subtelomeres of cay1Δ cells directly derives from cay1+ deletion and it occurs in a Rap1-dependent manner. The changes in H3K9me3 and total H3 appear to be more indirect and possibly reflecting, at least in part, a still altered telomere length in early generation cells.
Finally, as expected, Cay1-YFP averted slow growth, cell elongation, and chromosome bridges when expressed in cay1Δ cells (Fig 7D–F). Expression of Rap1-YFP in cay1Δ mutants slightly ameliorated cell growth in the cold but not at 30°C (Fig 7D), mildly reverted cell elongation (Fig 7E), but significantly diminished the frequencies of chromosome bridges in the cold (Fig 7F). Hence, because Rap1 deficiency alone does not cause chromosome bridges in the cold (Fig 5D), Rap1 itself becomes essential to deal with such aberrant structures in cay1Δ cells. On the other side, the proliferation defects of cay1Δ cells are largely independent of Rap1 deficiency, while their cold sensitivity stems to some extent from a telomeric defect.
Unprocessed Tf2 transcripts accumulate in different pre-mRNA splicing mutants
While the telomeric defects in cay1Δ cells mainly derive from Rap1 insufficiency, deletion of different telomeric factors including Rap1 did not affect Tf2 transcripts (Fig 4D; Supplementary Fig S6D). Given the here-revealed connection between Cay1 and pre-mRNA splicing, we analyzed Tf2 expression in cells where global pre-mRNA splicing was impaired. We utilized the prp1-1 strain, carrying a thermosensitive allele of the U4/U6:U5 tri-snRNP complex subunit Prp1 (Potashkin et al, 1989), and the mpn1Δ strain, which lacks the RNA exonuclease Mpn1 that specifically stabilizes the cellular levels of U6 snRNA (Shchepachev et al, 2012). Similar to what was observed for cay1Δ cells, Tf2 transcripts accumulated as unprocessed precursors both in prp1-1 cells grown at restrictive temperatures and in mpn1Δ cells (Fig 8A and B). Restoring normal pre-mRNA splicing in mpn1Δ cells through overexpression of U6 snRNA (Shchepachev et al, 2012) reverted Tf2 transcript defects (Fig 8B). Thus, the cellular levels of precursor Tf2 RNA are increased in cells suffering from inefficient pre-mRNA splicing.
Figure 8. Unprocessed Tf2 transcripts accumulate in pre-mRNA splicing mutants.
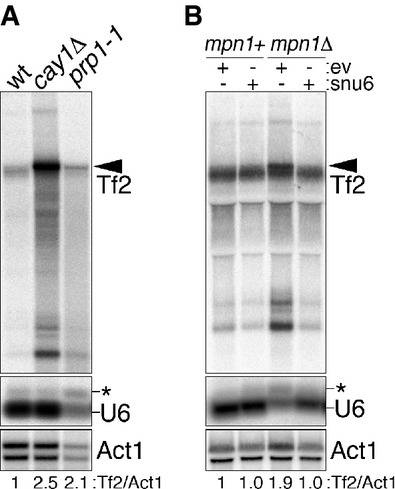
- A Northern blot analysis of Tf2 retrotransposons, U6 snRNA, and act1+ mRNA (loading control) in wt, cay1Δ, and prp1-1 cells. prp1-1 cells were grown at 36°C for 4 h before harvesting. Black arrowhead indicates unprocessed Tf2 transcripts. The asterisk indicates unspliced U6. Numbers at the bottom are the ratios between Tf2 and act1+ signals (Tf2/Act1) and expressed as fold increase over wt.
- B Northern blot analysis as in (A) in mpn1+ and mpn1Δ strains carrying a stably integrated extra copy of the snu6+ gene or empty vector control plasmids (ev).
Discussion
We present here the first comprehensive screening for regulators of telomeric lncRNAs. Given the high degree of conservation of telomere regulation and heterochromatin establishment between fission yeast and humans, we expect that the identified factors will unveil novel pathways conserved in human cells. The UP-TERRA mutants with highest levels of telomeric lncRNAs carry elongated telomeres, suggesting a possible connection between telomere transcription and lengthening. Elongated telomeres do not directly stimulate transcription of chromosome ends as expressing Cay1-YFP or Rap1-YFP in cay1Δ cells silences ARIA immediately, when telomeres are still very long. Consistently, experimentally induced telomere elongation in human cultured cells does not increase TERRA cellular levels (Arnoult et al, 2012; Farnung et al, 2012). Our data might rather suggest that increased telomeric transcription or TERRA/ARIA levels promote telomere elongation by telomerase (discussed below). Telomere elongation in UP-TERRA mutants might also be achieved through increased homologous recombination triggered by RNA:DNA hybrids involving TERRA/ARIA and telomeric DNA, as it has been suggested for budding yeast cells deficient for telomerase subunits progressing toward senescence (Balk et al, 2013; Yu et al, 2014). Yet, the fact that telomerase primarily maintains telomeres in cay1Δ, rap1Δ, and taz1Δ cells (Miller et al, 2006) argues against recombination being the major trigger of telomere elongation.
Further characterization of one UP-TERRA mutant has allowed us to dissect the molecular functions associated with Cactin in fission yeast. Cay1 supports normal telomere homeostasis largely by assuring that sufficient levels of Rap1 are maintained in cells by promoting rap1+ pre-mRNA splicing and Rap1 protein stability. Still, telomere length and ARRET in cay1Δ cells overexpressing Rap1-YFP were not ultimately restored to wt levels, and this might derive from the fact that pre-mRNAs of other telomeric factors such as poz1+ are inefficiently spliced upon cay1+ deletion. Moreover, overexpressed Rap1-YFP remains unstable in cay1Δ cells possibly affecting Rap1 turnover at telomeres and contributing to the observed telomeric defects. Similarly, aberrant splicing or protein stability of factors other than Rap1 and Poz1 could explain the proliferation defects of cay1Δ mutants. Moreover, pre-mRNA splicing inefficiency in cay1+ deleted cells is likely to explain the accumulation of unprocessed Tf2 transcripts, as these accrue also in independent splicing mutants. Again, pre-mRNA splicing of one or more factors directly promoting Tf2 expression and/or 5′ processing could be impaired in cay1Δ cells. Yet, an alternative and very intriguing hypothesis is that the spliceosome machinery could directly cleave the 5′ end of Tf2s in a process reminiscent of the spliceosome-dependent maturation of ter1+ telomerase RNA (Box et al, 2008).
It will be important to determine how Cay1 promotes splicing and protein stabilization. Protein interaction studies in different organisms have revealed physical interactions between Cactins and active spliceosomal complexes (Jurica et al, 2002; Rappsilber et al, 2002; Zhou et al, 2002; Bessonov et al, 2008; Herold et al, 2009). Moreover, in Arabidopsis thaliana, fluorescently tagged Cactin accumulates within nuclear speckles containing known splicing factors such as SR45 and RSP31 (Baldwin et al, 2013). These data strongly suggest that fission yeast Cay1 is a component of active spliceosomes and the single focus formed by Cay1-YFP might represent a nuclear center containing spliceosomal factors. Yet, Cay1 affects splicing of different introns to different extents leading to defects that remain essentially undetectable when global pre-mRNA splicing is analyzed, at least using tiling array-based approaches. Expanding our PCR-based pre-mRNA splicing analysis or resorting to RNA deep sequencing should allow collecting enough information about the pre-mRNAs that are most inefficiently spliced in cay1Δ cells. Parallel studies are also needed to unveil how Cay1 promotes Rap1 protein stabilization. It is possible that cay1Δ mutants inefficiently splice pre-mRNAs encoding regulators of Rap1 protein stability. Nonetheless, human Cactin was found to interact with the NEDD4-related ubiquitin ligase NEDL2 (Lu et al, 2013); hence, Cay1 might modulate Rap1 cellular levels through ubiquitination events. Although transiently or at low levels, Cay1-Myc localizes to telomeres, suggesting that Cay1-mediated post-translational stabilization of Rap1 might occur in the context of telomeric chromatin. Moreover, Cay1 physical association with telomeres and Tf2s suggests that Cay1 might also regulate expression of these loci in cis, for example, by participating in chromatin remodeling processes.
We have also discovered that Rap1 (and therefore Cay1) negatively regulates transcription of chromosome ends by restricting subtelomeric H3K9 acetylation, implying that Rap1 promotes histone deacetylation or suppresses histone acetylation. This finding mechanistically explains our previous observations that RNAPII occupancy at TERRA transcription start sites is diminished in rap1Δ mutants (Bah et al, 2012). In mouse embryonic fibroblasts deficient for the RNA component of telomerase, telomere shortening leads to progressive loss of heterochromatin marks including H3K9me3 and the HP1γ orthologue CBX3, as well as accumulation of H3K9ac at telomeres (Benetti et al, 2007). Consistently, TERRA expression increases in telomerase-deficient budding yeast strains carrying shortened telomeres (Cusanelli et al, 2013). Moreover, treating human cultured cells with the histone deacetylase inhibitor trichostatin A induces accumulation of TERRA (Azzalin & Lingner, 2008). On the other side, telomere elongation achieved through telomerase overexpression in human cancer cells suppresses TERRA expression through increased density of telomeric H3K9me3 and HP1α (Arnoult et al, 2012). Altogether, these observations point toward a unified model where TERRA (and possibly ARIA in fission yeast) could be part of a self-feeding molecular loop controlling telomere length homeostasis in telomerase-positive cells. When telomeres shorten, heterochromatin relaxation and TERRA transcription would stimulate telomerase accessibility to telomeres and telomere elongation. In budding yeast, TERRA transcribed from short telomeres assembles into nuclear clusters containing the telomerase RNA component TLC1. TERRA/TLC1 clusters are then redirected to the telomeres where TERRA was transcribed from (Cusanelli et al, 2013). This, together with the reported physical association of TERRA with telomerase (Redon et al, 2010), suggests a direct role for TERRA in chaperoning telomerase to telomeres that need to be elongated. Upon telomere elongation, a compact heterochromatic state would progressively be re-established, thus repressing TERRA transcription and preventing telomerase from further elongating telomeres.
Cactin proteins in different organisms share two conserved domains named Cactin mid- and C-terminal domains (Lin et al, 2000; Atzei et al, 2010b). Cay1 and human Cactin share 27% and 50% identity among their mid- and C-terminal domains, respectively. Cactin deficiency leads to embryonic lethality in A. thaliana, Danio rerio, and Caenorhabditis elegans and to cell cycle arrest in Toxoplasma gondii (Atzei et al, 2010b; Tannoury et al, 2010; Szatanek et al, 2012; Baldwin et al, 2013). Similarly, we now show that Cay1 promotes cell growth in S. pombe. We propose that these cellular defects could stem from improper splicing of specific pre-mRNAs, as well as from defective telomere length maintenance and/or expression of retroviral elements. Analyzing global pre-mRNA splicing, telomere homeostasis and retrotransposon expression upon Cactin inhibition in different model organisms will allow testing these hypotheses directly. Moreover, Cactin was first reported in Drosophila melanogaster as an interactor of the fly IκB orthologue Cactus, which regulates Dorsal (NF-κB) transciptional program (Lin et al, 2000). Human Cactin also modulates NF-κB gene expression and interacts with an IκB-related protein (Atzei et al, 2010a). Intriguingly, mammalian Rap1 interacts with inhibitors of IκB and it activates NF-κB-dependent gene expression (Teo et al, 2010). Cactin and Rap1 proteins might genetically interact to modulate NF-κB-dependent gene expression programs, similar to what they do in fission yeasts to control expression of the telomeric transcriptome. Finally, Cactin deficiency results in substantial changes in the expression profile of a multitude of genes in T. gondii (Szatanek et al, 2012) and human Cactin physically interacts with RDBP, a negative regulator of RNAPII (Lehner et al, 2004), suggesting possible functions for Cactin at the interface between gene transcription and post-transcriptional RNA processing. Overall, Cactins appear to be master regulators of multiple crucial cellular pathways; it will be fascinating to comprehensively identify those pathways and clarify to what extent they functionally overlap.
Materials and Methods
Strains and media
Original Schizosaccharomyces pombe strains were kind gifts from J. P. Cooper, M. G. Ferreira, and R. Allshire or were purchased from Bioneer Corporation (Supplementary Table S3). Strains were grown according to standard methods at the indicated temperatures in amino acid-supplemented yeast extract medium (YES) or EMM2 medium (Sabatinos & Forsburg, 2010). When required, 150 μg/ml geneticin (GIBCO), 100 μg/ml nourseothricin (Werner Bioagents), 5 μg/ml thiamine (Sigma), or 30 μg/ml trichostatin A (Selleckchem) were added. Gene deletions and C-terminal tagging were performed by single-step gene replacement using pFA6a-derived plasmids (Bahler et al, 1998; Hentges et al, 2005; Van Driessche et al, 2005) obtained from Euroscarf. Strains carrying nmt1 promoter-controlled cay1-yfp, rap1-yfp, or taz1-yfp were obtained by selecting for stable integration of NotI digested pDUAL-derived expression plasmids (RIKEN BRC) into the leu1 locus.
RNA isolation and analysis
Total RNA was extracted from yeasts grown to exponential phase (OD 0.6–0.8) using the hot phenol method (Schmitt et al, 1990) and subjected to DNaseI digestion (QIAGEN) at least twice before further procedures. For Northern blot analysis, RNA was electrophoresed in 1.2% formaldehyde agarose gels, transferred to positively charged nylon membranes (GE Osmonics). Membranes were incubated in CHURCH buffer (250 mM sodium phosphate buffer pH 7.2, 1 mM EDTA, 1% BSA, 7% SDS) and hybridized to double-stranded DNA probes 32P radiolabeled by random priming with Klenow Fragment (New England Biolabs) or DNA oligonucleotides 32P radiolabeled using T4 Polynucleotide Kinase (New England Biolabs). The single- and double-stranded telomeric probe detecting TERRA and ARIA and the ones detecting ARRET and αARRET were previously described (Bah et al, 2012). Tf2 and Act1 probes were generated by random priming labeling of a genomic PCR product. After stringency washes in 2–0.2× SSC, 0.2% SDS, radioactive signals were visualized using a Typhoon FLA 9000 Biomolecular Imager (GE Healthcare). For RT–PCR experiments, total RNA was reverse-transcribed with random hexamer oligonucleotides and SuperScript II (Invitrogen) according to manufacturer instructions. cDNA was PCR-amplified using Taq DNA Polymerase (New England Biolabs) for 28–31 cycles in order to allow semi-quantitative measurements. For TERRA qRT–PCR experiments, RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen), and TERRA and act1+ oligonucleotides. cDNA was PCR-amplified using the LightCycler SYBR Green I Master mix (Roche) on a Rotor-Gene Q instrument (QIAGEN). Oligonucleotide sequences and cycling conditions are listed in Supplementary Table S4.
Deletion mutant library screening
The Schizosaccharomyces pombe Haploid Deletion Mutant Set version 2.0 comprising 3,004 strains provided in 96-well format (M-2030H) was purchased from Bioneer Corporation. Deletion mutant strains were grown at 30°C in YES medium in 96-deep well plates. In each plate, CAF1 (wtA), CAF2 (wtB), and CAF39 (rap1Δ) control strains were included (Supplementary Table S3). Once control strains reached mid-exponential phase, total RNA was isolated using the Norgen's total RNA purification 96-well Kit (Norgen Biotek). Purified RNA was dot-blotted onto nylon membranes (GE Osmonics) and hybridized as described for Northern blot analysis using radioactive probes. Radioactive signals were quantified using ImageJ.
Whole transcriptome tiling array analysis
Labeled cDNA libraries were constructed according to the GeneChip Eukaryotic Double Strand Whole Transcript Protocol (Affymetrix). In short, total RNA from three biological replicates was reverse-transcribed with random primers and SuperScript II (Invitrogen) before performing second-strand cDNA synthesis with Klenow fragment (New England Biolabs). Reverse transcription and second-strand synthesis were performed in the presence of dUTP to allow subsequent fragmentation with uracil–DNA glycosylase and human apurinic/apyrimidinic endonuclease 1 (Affymetrix). Fragmented double-stranded cDNA was labeled with DNA labeling reagent and terminal deoxynucleotidyl transferase and hybridized to GeneChip S. pombe Tiling 1.0FR Array containing in total 1,160,624 probes at an average resolution of 20 bp (Affymetrix). Based on poor quality array hybridization, one cay1Δ replicate was excluded from further analysis. Tiling array data have been deposited in the GEO repository (accession number: GSE61792). To accommodate recent changes to the genome, we mapped all probes to the genome build from May 2011 (ftp://ftp.ebi.ac.uk/pub/databases/pombase/pombe/Archived_directories/GFF/). The impact of probe GC content and overall nucleotide composition on expression values was eliminated using the normalization approaches implemented in the rMAT R package (Droit et al, 2010). Gene expression estimates were defined as the median expression value of all probes assigned to the gene. When illustrating the relative fold enrichment pattern along a given genomic region, we applied a smoothing spline to the fold enrichments of individual probes within that region.
DNA isolation and analysis
Genomic DNA was prepared as described previously (Sabatinos & Forsburg, 2010). After digestion with ApaI or HindIII (New England Biolabs), DNA was separated on 0.7–1.2% agarose gels. For telomere overhang analysis, gels were dried and hybridized in non-denaturing conditions to an oligonucleotide probe corresponding to the telomeric C-strand. After signal detection, gels were denatured and hybridized to the same probe. For Southern blotting, DNA was denatured in gel and transferred to a positively charged nylon membrane (GE Osmonics). Hybridizations were performed as described for Northern blot analysis with a random priming labeled double-stranded telomeric probe. Pulsed-field gel electrophoresis was carried out as described previously with some modifications (Nakamura et al, 1998; Ferreira & Cooper, 2001). Cells were washed in TSE (10 mM Tris–HCl (pH 7.5), 0.9 M D-Sorbitol, 45 mM EDTA) and resuspended at 5 × 107 cells with 10 mg/ml zymolyase-20T (Seikagaku Biobusiness) and 50 mg/ml lysing enzyme (Sigma). One volume of 2% low melt agarose (Bio-Rad) in TSE was added and the suspension dispensed into plug molds. Plugs were incubated in TSE containing 5 mg/ml zymolyase-20T and 25 mg/ml lysing enzyme at 37°C for 1 h. Plugs were incubated for 90 min at 50°C in 0.25 M EDTA, 50 mM Tris–HCl (pH 7.5), 1% SDS and then for 3 days at 50°C in 0.5 M EDTA, 10 mM Tris–HCl (pH 9.5), 1% lauroyl sarcosine, 1 mg/ml proteinase K. Plugs were washed in 10 mM Tris–HCl (pH 7.5), 10 mM EDTA and incubated with 0.04 mg/ml PMSF for 1 h at 50°C. Plugs were washed again in 10 mM Tris–HCl (pH 7.5), 10 mM EDTA and then digested with 100 U of NotI (New England Biolabs) for 24 h at 37°C. Plugs were equilibrated in 0.5× TAE, loaded on a 1% agarose gel, and ran in a CHEF-DR III system (Bio-Rad) for 16 h at 14°C using the following program: 60-120 s switch time, 120° angle, 6 V/cm electric field. After running, gels were processed as for Southern blot and hybridized with a mix of L, I, M, and C probes recognizing unique sequences from the most terminal NotI fragments of the left arm of chromosome I, right arm of chromosome I, left arm of chromosome II, and right arm of chromosome II, respectively (Supplementary Fig S7A; Nakamura et al, 1998).
Chromatin immunoprecipitation
ChIPs were carried out as described previously with some modifications (Bah et al, 2012). 150 ml of yeast culture (OD 0.6) was cross-linked in 1% formaldehyde for 30 min and successively quenched in 125 mM glycine for 5 min. Cross-linked material was resuspended in 400 μl lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitor cocktail (Roche)) and subjected to mechanical lysis with glass beads using the FastPrep FP120 apparatus (Bio101 Thermo Savant, Qbiogene) three times 6 m/s for 30 s. Lysates were centrifuged for 30 min at 16,000 g, and pellets were re-suspended in 500 μl lysis buffer and sonicated in a Bioruptor UCD-200 (Diagenode) three times at high power with 30-s intervals for 15 min. Sonicated material was centrifuged for 15 min at 10,000 g, and supernatant containing fragmented chromatin was recovered. Approximately 1 mg of proteins was aliquoted in 400 μl per IP. Immunoprecipitations were performed on a rotating wheel at 4°C in the presence of 25 μl protein A/G sepharose beads (GE Healthcare) and 3 μl of either anti-Myc, anti-H3K9ac, anti-H3K9me3 or 2.5 μl of anti-H3 antibodies. Beads were washed three times in lysis buffer, once in lysis buffer containing 500 mM NaCl, once in wash buffer (10 mM Tris–HCl pH 7.5, 0.25 M LiCl, 0.5% Nonidet P40, 0.5% sodium deoxycholate) and once in lysis buffer. Immunoprecipitated chromatin was eluted in elution buffer (1% SDS, 100 mM sodium bicarbonate, 40 mg/ml RNase A) and incubated at 37°C for 1 h. Cross-links were reversed at 65°C for 16 h and DNA purified with the Wizard SV Gel and PCR Clean-Up System (Promega). Immunoprecipitated DNA was quantified by real-time PCR using the LightCycler SYBR Green I Master mix (Roche) on a Rotor-Gene Q instrument (QIAGEN) or by dot blot hybridization with a radiolabeled telomeric probe. Oligonucleotide sequences and cycling conditions are listed in Supplementary Table S4.
Western blotting and antibodies
Cells were collected in exponential phase, and total protein extracts were prepared by mechanical lysis in the presence of 20% cold TCA. Western blot analysis was performed according to standard protocols. Antibodies were as follows: a rabbit polyclonal anti-Rap1 and a rabbit polyclonal anti-Taz1 antibody (kind gifts from J. Kanoh and J. P. Cooper, respectively); a mouse monoclonal anti-Myc tag antibody (Cell Signaling, #2276); a mouse monoclonal anti-GFP antibody (Roche, #11814460001); rabbit polyclonal anti-H3K9ac and anti-H3K9me3 antibodies (Millipore, #07-352 and #07-442); a rabbit polyclonal anti-H3 and a mouse monoclonal anti-actin antibody (Abcam, ab1791 and ab8224); and HRP-conjugated donkey anti-mouse and anti-rabbit secondary antibodies (Bethyl Laboratories).
Microscopy
Cells were prepared following standard protocols. Images were acquired with a Leica DM6000B microscope or an Olympus Cell-R microscope equipped with Orca ER cameras (Hamamatsu). For visualization and quantification of telomeric foci, cells were imaged with a DeltaVision system (Applied Precision) on an Olympus IX71 microscope equipped with a CoolSnap HQ camera (Roper Scientific).
Acknowledgments
We thank Martin Moravec and Giacomo Robbiani for help during the development of the project; Julie P. Cooper, Miguel G. Ferreira, Robin Allshire, Junko Kanoh, and Vikram Panse for yeast strains and reagents; and members of the Azzalin laboratory for discussions. We also thank the Functional Genomics Center Zürich (FGCZ) for tiling array services and the Scientific Center for Optical and Electron Microscopy (SCOPEM) of Zürich for microscopy services. This work was supported by grants to C.M.A. from the European Research Council (BFTERRA), the Swiss National Science Foundation (PP00P3-123356/144917), ETH Zurich (ETH-01 12-2) and Fondazione Cariplo (2008-2507).
Author contributions
LEL, AB, and CMA planned the experiments; LEL, AB, HW, VS, MS, and CMA performed the experiments and analyzed the data; CS analyzed the tiling array data; LEL and CMA wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Information
Review Process File
References
- Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat Struct Mol Biol. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- Atzei P, Gargan S, Curran N, Moynagh PN. Cactin targets the MHC class III protein IkappaB-like (IkappaBL) and inhibits NF-kappaB and interferon-regulatory factor signaling pathways. J Biol Chem. 2010a;285:36804–36817. doi: 10.1074/jbc.M110.139113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzei P, Yang F, Collery R, Kennedy BN, Moynagh PN. Characterisation of expression patterns and functional role of Cactin in early zebrafish development. Gene Expr Patterns. 2010b;10:199–206. doi: 10.1016/j.gep.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- Bah A, Wischnewski H, Shchepachev V, Azzalin CM. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:2995–3005. doi: 10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Baldwin KL, Dinh EM, Hart BM, Masson PH. CACTIN is an essential nuclear protein in Arabidopsis and may be associated with the eukaryotic spliceosome. FEBS Lett. 2013;587:873–879. doi: 10.1016/j.febslet.2013.02.041. [DOI] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, Hiraoka Y, Shore D, Hu HY, Chang S, Lei M. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol. 2011;18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Cusanelli E, Romero CA, Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell. 2013;51:780–791. doi: 10.1016/j.molcel.2013.08.029. [DOI] [PubMed] [Google Scholar]
- Droit A, Cheung C, Gottardo R. rMAT–an R/Bioconductor package for analyzing ChIP-chip experiments. Bioinformatics. 2010;26:678–679. doi: 10.1093/bioinformatics/btq023. [DOI] [PubMed] [Google Scholar]
- Durand-Dubief M, Sinha I, Fagerström-Billai F, Bonilla C, Wright A, Grunstein M, Ekwall K. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J. 2007;26:2477–2488. doi: 10.1038/sj.emboj.7601690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung BO, Brun CM, Arora R, Lorenzi LE, Azzalin CM. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS One. 2012;7:e35714. doi: 10.1371/journal.pone.0035714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Fujita I, Tanaka M, Kanoh J. Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe. PLoS One. 2012;7:e49151. doi: 10.1371/journal.pone.0049151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EB, Espinosa JM, Forsburg SL. Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing. Mol Cell Biol. 2005;25:8887–8903. doi: 10.1128/MCB.25.20.8887-8903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J, Cooper JP. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012;40:2956–2963. doi: 10.1093/nar/gkr1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- Lin P-H, Huang LH, Steward R. Cactin, a conserved protein that interacts with the Drosophila IκB protein Cactus and modulates its function. Mech Dev. 2000;94:57–65. doi: 10.1016/s0925-4773(00)00314-2. [DOI] [PubMed] [Google Scholar]
- Lu L, Hu S, Wei R, Qiu X, Lu K, Fu Y, Li H, Xing G, Li D, Peng R, He F, Zhang L. The HECT type ubiquitin ligase NEDL2 is degraded by anaphase-promoting complex/cyclosome (APC/C)-Cdh1, and its tight regulation maintains the metaphase to anaphase transition. J Biol Chem. 2013;288:35637–35650. doi: 10.1074/jbc.M113.472076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol. 2003;13:1748–1757. doi: 10.1016/j.cub.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Miller KM, Cooper JP. The telomere protein Taz1 Is required to prevent and repair genomic DNA breaks. Mol Cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin J, Li R, Frendewey D. Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J. 1989;8:551–559. doi: 10.1002/j.1460-2075.1989.tb03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyes cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep. 2012;2:855–865. doi: 10.1016/j.celrep.2012.08.031. [DOI] [PubMed] [Google Scholar]
- Szatanek T, Anderson-White BR, Faugno-Fusci DM, White M, Saeij JP, Gubbels MJ. Cactin is essential for G1 progression in Toxoplasma gondii. Mol Microbiol. 2012;84:566–577. doi: 10.1111/j.1365-2958.2012.08044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannoury H, Rodriguez V, Kovacevic I, Ibourk M, Lee M, Cram EJ. CACN-1/Cactin interacts genetically with MIG-2 GTPase signaling to control distal tip cell migration in C. elegans. Dev Biol. 2010;341:176–185. doi: 10.1016/j.ydbio.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, Tran NL, Speiser LJ, Wong M, Saez E, Schultz P, Chanda SK, Verma IM, Tergaonkar V. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol. 2010;12:758–767. doi: 10.1038/ncb2080. [DOI] [PubMed] [Google Scholar]
- Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J. Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast. 2005;22:1061–1068. doi: 10.1002/yea.1293. [DOI] [PubMed] [Google Scholar]
- Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TY, Kao YW, Lin JJ. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci USA. 2014;111:3377–3382. doi: 10.1073/pnas.1307415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zhou ZX, Zhang MJ, Peng X, Takayama Y, Xu XY, Huang LZ, Du LL. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Res. 2013;23:705–715. doi: 10.1101/gr.146357.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Information
Review Process File