Kulic et al. show that RBPJ, a transcriptional repressor of Notch, is frequently deleted in human cancers and can function as a tumor suppressor. Loss of RBPJ acts to derepress target gene promoters, allowing Notch-independent activation by alternate transcription factors that promote tumor growth.
Abstract
Aberrant Notch activity is oncogenic in several malignancies, but it is unclear how expression or function of downstream elements in the Notch pathway affects tumor growth. Transcriptional regulation by Notch is dependent on interaction with the DNA-binding transcriptional repressor, RBPJ, and consequent derepression or activation of associated gene promoters. We show here that RBPJ is frequently depleted in human tumors. Depletion of RBPJ in human cancer cell lines xenografted into immunodeficient mice resulted in activation of canonical Notch target genes, and accelerated tumor growth secondary to reduced cell death. Global analysis of activated regions of the genome, as defined by differential acetylation of histone H4 (H4ac), revealed that the cell death pathway was significantly dysregulated in RBPJ-depleted tumors. Analysis of transcription factor binding data identified several transcriptional activators that bind promoters with differential H4ac in RBPJ-depleted cells. Functional studies demonstrated that NF-κB and MYC were essential for survival of RBPJ-depleted cells. Thus, loss of RBPJ derepresses target gene promoters, allowing Notch-independent activation by alternate transcription factors that promote tumorigenesis.
Notch signaling is directed through RBPJ, the DNA-binding component of the pathway (Kovall and Hendrickson, 2004; Borggrefe and Oswald, 2009). RBPJ is ubiquitously expressed and acts as a transcriptional repressor in the absence of active Notch (Hamaguchi et al., 1992; Bray, 2006). Binding of active Notch to RBPJ results in expulsion of a histone deacetylase-containing corepressor complex and recruitment of histone acetyltransferases to the Notch–RBPJ ternary complex to facilitate chromatin remodeling and transcriptional activation (Borggrefe and Oswald, 2009). Increased expression of Notch1 or its ligand Jagged1 is associated with poor survival in breast and other cancers (Reedijk et al., 2005; Koch and Radtke, 2007). To evaluate the relevance of RBPJ in tumor promotion, we examined RBPJ mRNA and protein levels in primary human cancers and modeled RBPJ depletion in tumor xenograft studies.
RESULTS
RBPJ is frequently lost in human cancers
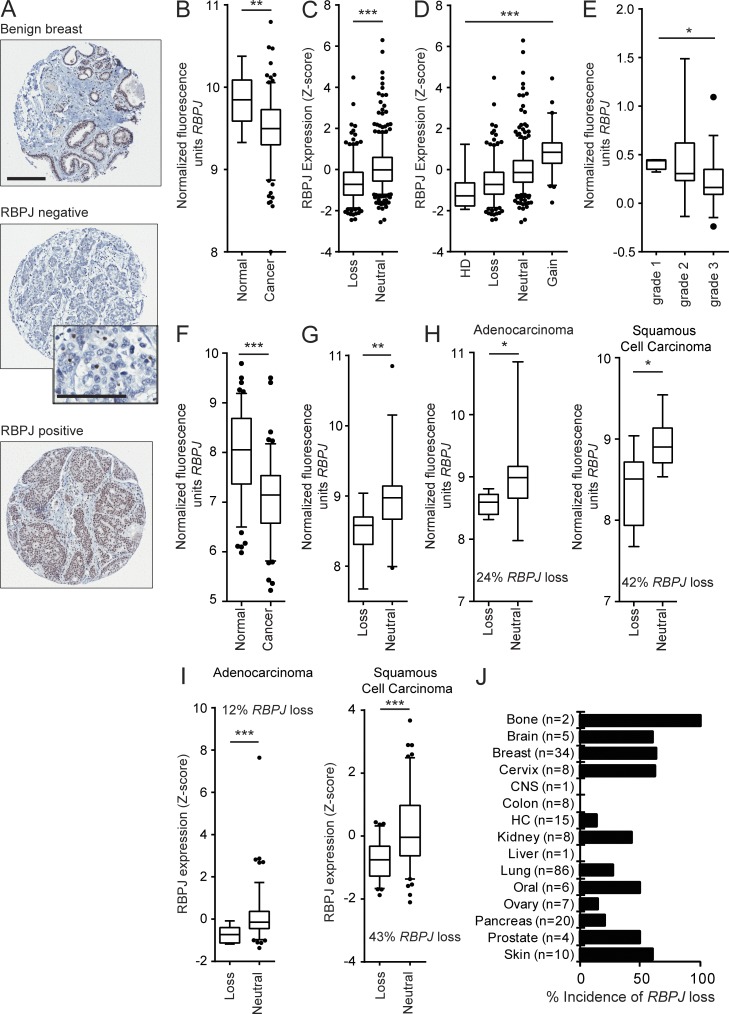
To determine whether altered RBPJ expression is associated with oncogenesis, we performed immunohistochemical staining of 264 human breast carcinoma cases. Immunostaining revealed lack of RBPJ protein in 15% (40/264) of cases, whereas nonmalignant breast tissue showed high levels of epithelial expression (Fig. 1 A). RBPJ loss did not correlate with hormone receptor or human epidermal growth factor receptor 2 status (unpublished data). Examination of microarray data from independent studies confirmed significantly reduced RBPJ mRNA expression in breast cancers (Fig. 1 B; Yu et al., 2008). Using TCGA data (Network, 2012), we evaluated RBPJ copy loss and mRNA expression in invasive breast cancers. Genomic loss of RBPJ occurred in 33% (277/828) of cases, and this coincided with significantly reduced transcript levels (Fig. 1 C). Cases either with RBPJ homozygous deletion (HD; n = 7) and loss (n = 270) showed the lowest RBPJ expression (Fig. 1 D). Analysis of microarray data from a study in which tumors were classified by grade showed that RBPJ expression was preferentially reduced in higher-grade breast cancers (Fig. 1 E; Ginestier et al., 2006), suggesting that reduced RBPJ expression may be associated with more aggressive tumors. Of interest, a significant negative correlation between expression of RBPJ and its canonical target gene, HEY2, was observed in the grade 3 subset (n = 39, R2 = 0.2, Pearson P = 0.003). A separate invasive lobular breast carcinoma dataset also showed a negative correlation between RBPJ and HEY2 mRNA expression (n = 18, R2 = 0.4, Pearson P = 0.005; Rhodes et al., 2004; Zhao et al., 2004).
Figure 1.
RBPJ is frequently lost in human cancers. (A) Examples of RBPJ immunohistochemical staining in benign breast tissue (n = 8) and breast cancer tissue microarray cores (RBPJ negative, n = 40; RBPJ positive, n = 224; bar, 200 µm). High power inset (bar = 100 µm) of the RBPJ-negative tumor core shows positive staining in internal control cells in the tumor microenvironment. (B) RBPJ mRNA expression in breast tumors (n = 183) and adjacent normal breast tissue (n = 13; Yu et al., 2008). (C) Analysis of RBPJ expression and RBPJ genomic copy loss (n = 277) versus no loss (neutral, n = 551) in invasive breast cancers (TCGA data). (D) Data from C plotted by RBPJ copy number status; HD (n = 7), loss (n = 270), neutral (n = 489), and amplification (gain, n = 62). P < 0.0001 by Kruskal–Wallis followed by Dunn’s multiple comparisons post-test showed significant differences in all comparisons except between the HD versus loss group. (E) RBPJ mRNA expression in human breast cancers stratified by tumor grade; grade 1 (n = 4), grade 2 (n = 12), and grade 3 (n = 39; Ginestier et al., 2006). (F) RBPJ mRNA expression in normal bronchial epithelium collected from healthy individuals (n = 67) versus non–small cell lung carcinoma (n = 111; Bild et al., 2006; Lockwood et al., 2010). (G) Analysis of lung cancers of mixed type with RBPJ genomic copy loss (n = 14) versus no loss (n = 30) with paired mRNA expression and aCGH data (Lockwood et al., 2008, 2010). (H) Analysis of lung cancers from G broken down into tumor subtypes; adenocarcinoma RBPJ genomic loss (n = 6) versus no loss (n = 19); squamous cell carcinoma RBPJ genomic loss (n = 8) versus no loss (n = 11; Lockwood et al., 2008, 2010). (I) Analysis of RBPJ mRNA expression and genomic copy number in TCGA lung cancer adenocarcinomas (RBPJ genomic loss [n = 15] versus no loss [n = 114]) and squamous cell carcinomas (RBPJ genomic loss [n = 77] versus no loss [n = 101]; Cerami et al., 2012; Lockwood et al., 2012; Gao et al., 2013). (J) RBPJ copy number alteration evaluated using aCGH across a panel of 215 cancer cell lines (CNS: central nervous system, HC: hematopoietic cell lines). At least one allele of RBPJ is lost at an overall frequency of 35% (also see Table S1). P-values were obtained using a Mann-Whitney test for B, C, and F–I and a Kruskal-Wallis one-way analysis of variance test for D and E. Data are presented as box-and-whisker plots. The bottom and top bars of the whisker indicate the 5th and 95th percentiles respectively with points outside the whiskers shown as individual dots. *, P < 0.05, **, P < 0.005, ***, P < 0.0001.
We also evaluated RBPJ in non–small cell lung cancers (n = 111) compared with normal bronchial epithelium from healthy individuals (n = 67) and observed reduced expression in tumors (Fig. 1 F; Bild et al., 2006; Lockwood et al., 2010). Genomic loss of at least one copy of RBPJ was associated with significantly reduced transcript levels in primary lung tumors (Fig. 1 G; Lockwood et al., 2008; Lockwood et al., 2010). Classification of tumors in Fig. 1 G into subtypes showed that RBPJ copy number loss occurred more frequently in lung squamous cell carcinomas compared with adenocarcinomas, consistent with our analysis of TCGA lung tumor data (Fig. 1, H and I; Cerami et al., 2012; Lockwood et al., 2012; Gao et al., 2013).
Examination of 215 different tumor cell lines representing 15 tumor types showed that deletion of at least one copy of RBPJ occurred in 35% of all cell lines (Fig. 1 J and Table S1). Oncomine analysis of additional cancer datasets revealed significant RBPJ underexpression in tumor tissue relative to matched normal tissue, including colon, bladder, ovarian, and gastric cancers (Rhodes et al., 2004; unpublished data). In contrast to NOTCH1, our analysis supports that RBPJ expression is reduced in several tumor types.
RBPJ deficiency promotes tumor growth
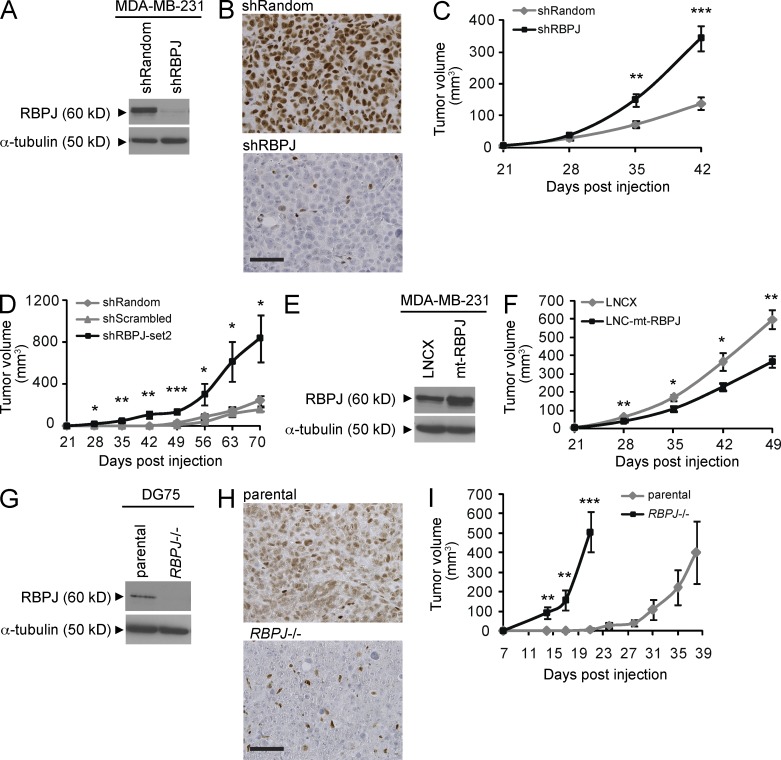
To determine whether RBPJ depletion promotes tumor progression, we knocked down RBPJ in MDA-MB-231 human breast cancer cells and subcutaneously implanted the cells into immunodeficient mice (Fig. 2, A and B). Stable RBPJ knockdown using either of two independent short hairpin (sh) RNAs significantly increased MDA-MB-231 tumor growth compared with two independent control cell lines expressing distinct nonspecific shRNAs (Fig. 2, C and D). In contrast, expression of a mutant (mt)-RBPJ (MacKenzie et al., 2004), which blocks Notch-driven transcription but permits endogenous RBPJ to bind DNA and retain repression, inhibited MDA-MB-231 tumor growth similar to direct blockade of Notch (Fig. 2, E and F; Leong et al., 2007). Before tumor seeding, the mt-RBPJ transcript was expressed 5.5-fold above vector control cells but only 3.5-fold in excised endpoint tumors (unpublished data). Modest overexpression of mt-RBPJ protein in endpoint tumors (Fig. 2 E) is likely due to preferential outgrowth of cells not expressing the construct. We confirmed increased tumor growth consequent to RBPJ loss in a Burkitt lymphoma cell line, DG75, in which RBPJ has been genetically ablated (Fig. 2, G–I; Maier et al., 2005). Thus, two independent methods demonstrate that RBPJ depletion promotes tumor growth in various malignancies.
Figure 2.
RBPJ deficiency promotes tumor growth. (A and B) Analysis of MDA-MB-231 tumors transduced with an RBPJ knockdown construct (shRBPJ) or a nonspecific shRNA (shRandom). (A) Immunoblot of endpoint tumor lysates. (B) RBPJ immunohistochemistry (bar, 50 µm). (C) MDA-MB-231 tumor growth after expression of shRBPJ (n = 16) compared with shRandom control (n = 22). (D) Depletion of RBPJ in MDA-MB-231 tumors using an alternate RBPJ targeting construct (set2) and a different silencing control (shScrambled; n = 4 for each group). (E) Immunoblot analysis of endpoint MDA-MB-231 tumor lysates transduced with LNCX empty vector or an mt RBPJ construct (LNC-mt-RBPJ). (F) MDA-MB-231 tumor growth after expression of LNC-mt-RBPJ compared with the LNCX control (n = 8 for each group). (G and H) Analysis of parental or RBPJ−/− DG75 Burkitt lymphoma tumors. (G) Immunoblot of endpoint tumor lysates. (H) RBPJ immunohistochemistry (bar, 50 µm). (I) DG75 RBPJ−/− tumor growth (n = 10) compared with parental DG75 tumors (n = 9). For C, F, and I, p-values were obtained using a Mann-Whitney test and for D a one-way ANOVA. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.005; ***, P ≤ 0.0005.
RBPJ deficiency results in a Notch-like gene signature
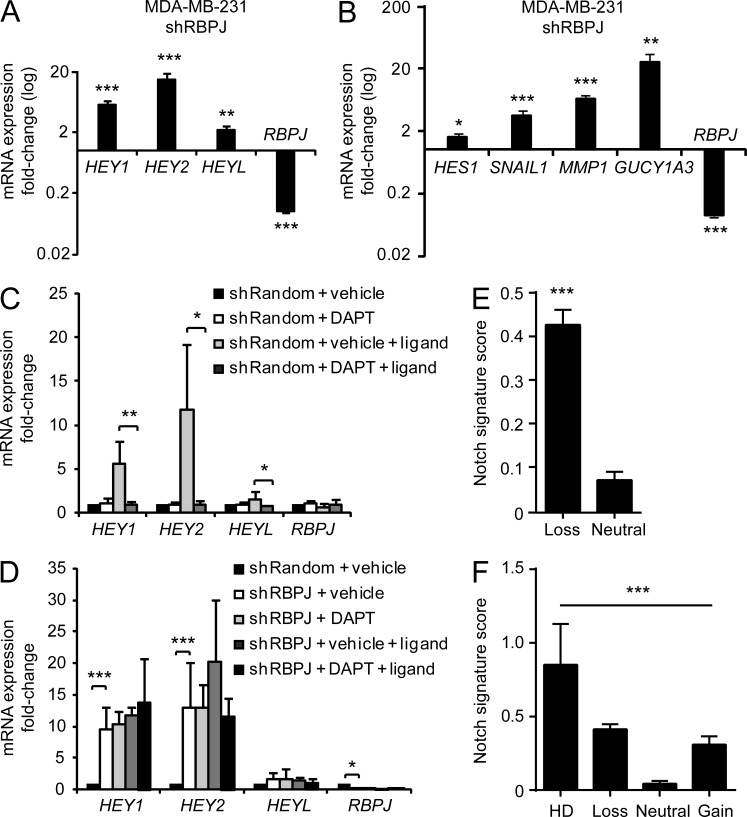
Because RBPJ acts as a transcriptional repressor, depletion can result in induction of target genes (Raafat et al., 2009; Liefke et al., 2010; Meier-Stiegen et al., 2010). We confirmed that RBPJ knockdown in MDA-MB-231 xenografts caused significant up-regulation of well characterized target genes of the HEY gene family (Fig. 3 A). HES1, SNAIL1, and GUCY1A3, additional known Notch target genes (Sahlgren et al., 2008; Borggrefe and Oswald, 2009; Chang et al., 2011), along with MMP1 were also up-regulated in RBPJ-depleted tumors (Fig. 3 B). To determine whether Notch activity contributes to transcriptional activation in the absence of RBPJ, we blocked activation of all Notch receptors in vitro using the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) to prevent Notch receptor cleavage. We co-cultured MDA-MB-231 cells with Jagged1 ligand-expressing mouse cells to activate endogenous Notch. This induced HEY genes in control cells, and the induction was blocked by DAPT (Fig. 3 C). In contrast, up-regulation of HEY genes by RBPJ knockdown in ligand-stimulated cells was not affected by DAPT treatment, indicating up-regulation of these target genes independent of Notch receptor activation (Fig. 3 D). Similarly, HEY expression was significantly increased in DG75 RBPJ−/− tumors compared with RBPJ-expressing controls and was not affected by DAPT-mediated Notch inhibition (unpublished data). These results imply that target gene expression is not solely an indicator of Notch receptor activity, as RBPJ depletion represents an alternate mechanism for promoting tumor growth and HEY expression.
Figure 3.
RBPJ depletion induces Notch target genes in a Notch-independent fashion. (A and B) Gene expression analysis of endpoint MDA-MB-231 tumors with data expressed as fold-change relative to the shRandom control, which corresponds to 1 on the y-axis. (A) Induction of HEY genes in shRBPJ tumors (n = 15, 90% RBPJ knockdown) compared with shRandom tumors (n = 14). (B) Induction of HES1, SNAIL1, MMP1, and GUCY1A3 in shRBPJ tumors (n ≥ 8, >90% RBPJ knockdown) compared with shRandom tumors (n ≥ 8). (C and D) Endogenous Notch signaling was induced in MDA-MB-231 cells by Jagged1 ligand stimulation, followed by gene expression analysis (n = 5). Data are expressed as fold change relative to shRandom + vehicle (DMSO). (C) HEY induction in shRandom cells is abolished by blocking Notch activation using 10 µM DAPT. (D) DAPT treatment on the magnitude of HEY gene induction in shRBPJ cells (≥80% RBPJ knockdown). (E) Mean Notch signature score in a total of 828 invasive TCGA breast cancers comparing the subset with RBPJ loss (n = 277) to those without (neutral, n = 551). The Notch gene signature identified in T-ALL cell lines (Margolin et al., 2009) was used to assign a score. (F) Data from E plotted by RBPJ copy number status; HD (n = 7), loss (n = 270), neutral (n = 489), and amplification (gain, n = 62). P < 0.0001 by Kruskal–Wallis followed by Dunn’s multiple comparisons post-test showed a significant difference only between the HD versus neutral, loss versus neutral and gain versus neutral group. For A, B, and E, p-values were obtained using a Mann-Whitney test and a Kruskal-Wallis one-way analysis of variance test for F and data are presented as mean + SEM. P-values for C and D were obtained using a Student’s t tests and data are presented as mean + SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
To evaluate whether RBPJ loss produces a Notch-like gene signature, we identified genes differentially expressed in 10 human T-ALL cell lines with Notch activating mutations treated with vehicle or a Notch inhibitor (Benjamini-Hochberg corrected P ≤ 0.05; Margolin et al., 2009). 170 genes showing a change in response to the Notch inhibitor across all T-ALL lines were defined as Notch-regulated genes (Table S2). Based on this signature, we assigned a Notch signature score (see Materials and methods) for each of the 828 invasive breast cancers from the TCGA dataset (shown in Fig. 1 C) comparing mean scores between samples with RBPJ loss to those without. Breast cancers with RBPJ loss had a significantly increased Notch signature score compared with tumors without RBPJ loss (Fig. 3 E). The breast cancer cases with RBPJ HD, copy loss, or copy gain had a significantly higher Notch signature score compared with samples with neutral RBPJ copy number (Fig. 3 F). These data imply that RBPJ activates a subset of Notch target genes.
RBPJ depletion causes epigenetic changes that correspond to promoter activity
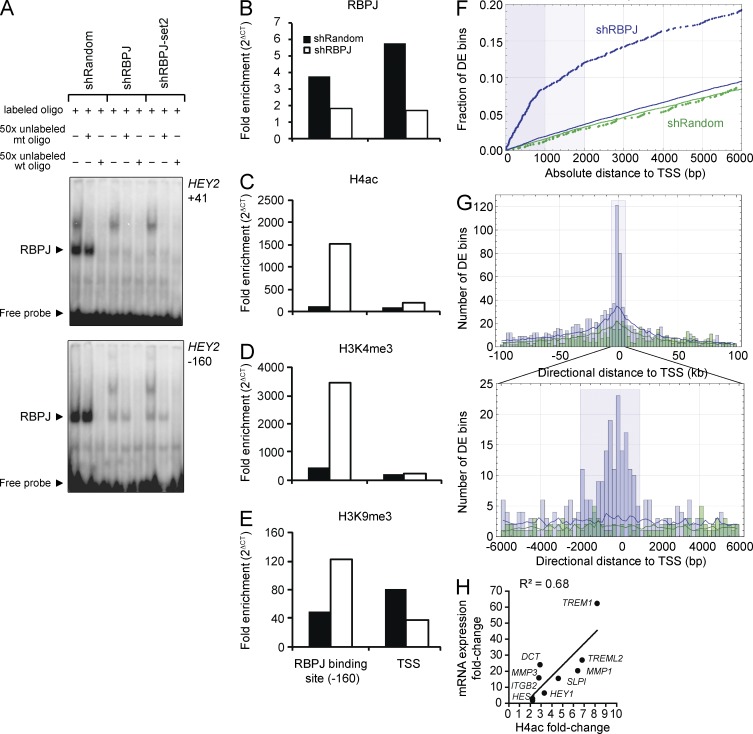
To address how RBPJ loss leads to target gene activation, we first assessed whether another transcriptional activator binds the vacant RBPJ motif in RBPJ-depleted cells. Electrophoretic mobility shift assays (EMSAs) in control and shRBPJ MDA-MB-231 cells did not reveal binding of alternate transcription factors to the RBPJ consensus sequences of the HEY1 (not depicted) and HEY2 promoters of RBPJ-depleted cells (Fig. 4 A). We also performed local chromatin immunoprecipitation (ChIP) in these cells confirming reduced RBPJ occupancy at the HEY2 promoter, corroborating the EMSA data (Fig. 4 B). Transcriptional repression by the RBPJ corepressor complex depends on histone deacetylase activity, which condenses chromatin and silences gene expression (Borggrefe and Oswald, 2009). To confirm HEY promoter activation, we demonstrated that RBPJ depletion resulted in histone acetylation by acetylated histone H4 (H4ac) ChIP followed by real-time quantitative PCR (RT-qPCR; Fig. 4 C). Histone H3 lysine4 tri-methylation (H3K4me3), an alternate mark of active gene promoters, was similarly enriched at the RBPJ binding site of the HEY2 promoter in RBPJ-depleted cells (Fig. 4 D). Histone H3 lysine9 tri-methylation (H3K9me3), a modification associated with transcriptional repression, was reduced across the HEY2 transcription start site (TSS) in RBPJ-depleted cells (Fig. 4 E). The modest increase in H3K9me3 at the RBPJ binding site may result from feedback mechanisms after RBPJ depletion and HEY2 induction. Hence, RBPJ loss effects histone marks at promoters of expressed genes.
Figure 4.
RBPJ depletion in MDA-MB-231 cells results in histone changes corresponding to promoter activity. (A) EMSA was performed using MDA-MB-231 nuclear lysates (n = 1) and labeled probes representing two RBPJ binding sites in the HEY2 promoter (HEY2: −160 to −167 and +41 to +48 bp from the TSS) in the absence or presence of excess unlabeled mutated probe (+mt) or excess wild-type probe (wt). Comparable results were obtained using binding sites in the HEY1 promoter (not depicted). (B–E) ChIP analysis of the HEY2 promoter (n = 1) with fold enrichment calculated as 2(Ct IgG − Ct target). Bound fragments were quantified by RT-qPCR with primers spanning the −160 bp RBPJ binding site in the promoter and the TSS. (B) RBPJ ChIP. (C) H4ac ChIP. (D) H3K4me3 ChIP. (E) H3K9me3 ChIP. (F and H) Locations of genome-wide 500 bp H4ac-differentially enriched regions identified by ChIP-seq (corrected P ≤ 0.005) relative to 38,405 nonredundant UCSC gene TSSs. Solid lines show random expectations (n = 20) for shRBPJ (blue) and shRandom (green). (F) Empirical distribution functions for the distance from the nearest TSS to the center of a differentially enriched (DE) region in the shRBPJ (blue) and the shRandom (green) control. Shaded blue regions that extend to 1 and 2 kb mark distances within which, for shRBPJ data, differentially enriched regions are spatially concentrated relative to TSSs. (G) Overview (±100 kb) and detailed (±6 kb) histograms of directional distance between a differentially enriched region center and its nearest TSS. The shaded blue rectangle in the detailed view marks the −2/+1 kb TSS regions used in analyses. (H) Fold change in the H4ac signal correlates with fold change in mRNA expression (RT-qPCR) evaluated for a subset of the 114 TSSs that were significantly differentially enriched in the knockdown relative to shRandom cells, which correspond to 1 (Pearson correlation P = 0.006).
To define derepressed/activated promoter regions in RBPJ-depleted cells genome-wide, we profiled H4ac enrichment by ChIP-sequencing (seq), and then used edgeR (Robinson et al., 2010) to assess differential H4ac enrichment in 500 bp genome-wide regions in shRBPJ (knockdown) versus shRandom (control) MDA-MB-231 cells and retained regions that had a Benjamini-Hochberg corrected P < 0.005 (Benjamini and Hochberg, 1995). H4ac was differentially enriched in shRBPJ cells relative to shRandom in regions that extended from approximately −2 kb to +1 kb relative to TSSs, whereas for regions that were differentially enriched in the shRandom versus shRBPJ cells the spatial distribution was comparable to random expectation (Fig. 4, F and G). Consistent with this, in human T lymphoblastic leukemia cells the majority of RBPJ and Notch1 binding sites were found within 2 kb of TSSs (Wang et al., 2011). Assessing differential H4ac enrichment in −2/+1 kb regions for all 38,405 nonredundant UCSC gene TSSs, and retaining only regions that had an H4ac fold change of at least 2 and a Benjamini-Hochberg corrected P < 0.005, we identified 114 TSS regions that were differentially enriched in RBPJ knockdown cells (Table S3). For nine of the differentially expressed genes, we used RT-qPCR to confirm that H4ac enrichment was associated with increased mRNA expression (Fig. 4 H). Within regions −2/+1 kb from the TSS, eight of the nine genes had an RBPJ binding affinity score that was above the median for the global set of 38,405 regions, and for all nine TSS regions we identified a putative RBPJ binding site as a high-scoring match (>0.96) to a TGGGAA de novo sequence motif (Aerts et al., 2005; Roider et al., 2007; unpublished data). To further evaluate whether promoters differentially marked with H4ac are associated with genes previously identified as bound by RBPJ, we obtained 2,112 RBPJ binding sites in the CUTLL1 T-lymphoblastic leukemia/lymphoma cell line (Wang et al., 2011). Evaluating these RBPJ ChIP-seq peaks against our genome-wide H4ac differentially enriched regions at an absolute distance of 2 kb from the TSS showed concordance in RBPJ knockdown regions (P = 2.5 × 10−4) compared with control H4ac regions or random expectations (P = 0.11). Eight (HEY1, HES1, UCKL1, PMEPA1, DGAT1, CR612603, MCF2L2, and ZDHHC14) of the 114 genes that we identified as differentially enriched in RBPJ knockdown cells at −2/+1 kb from the TSS have an RBPJ peak within 2 kb of the TSS. These results indicate that RBPJ depletion enriches for histone H4 acetylation near TSSs.
RBPJ deficiency increases tumor cell survival
To clarify how genes whose promoters were associated with H4ac changes contribute to increased tumorigenicity, we used Ingenuity Pathway Analysis software to identify statistically significant biological functions for the genes corresponding to the 114 differentially enriched promoter regions in RBPJ knockdown cells. The 92 genes mapped by Ingenuity Pathway Analysis returned five significant cellular functions (Benjamini-Hochberg corrected P ≤ 0.025): lipid metabolism, molecular transport, small molecule biochemistry, cell death, and cellular compromise.
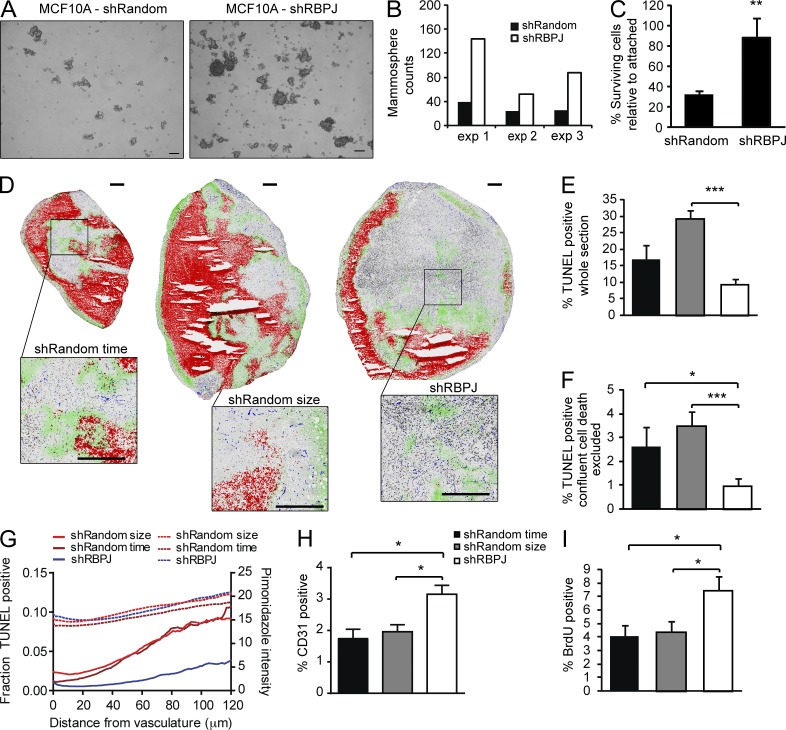
Because all of the induced genes that were marked with H4ac, except TREML2, appeared in the cell death function, we evaluated whether the apoptotic response was altered in RBPJ-depleted cells. We first performed mammosphere formation assays, which enrich for breast cancer stem cells with aberrant self-renewal, using a nontumorigenic breast epithelial cell line MCF10A transduced with shRandom and shRBPJ constructs. RBPJ knockdown increased mammosphere formation up to 3.7-fold (Fig. 5, A and B). Next, we incubated MDA-MB-231 cells in suspension for 48 h to induce anoikis. Compared with control cells, RBPJ knockdown markedly protected cells from death resulting from loss of cell matrix attachment (Fig. 5 C). To confirm that RBPJ deficiency also conferred a survival advantage in vivo, we used terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) to mark all dead cells in tumor cryosections. Both MDA-MB-231 size-matched shRandom control tumors, which were collected when they reached a comparable size to the RBPJ-knockdown tumors, and shRandom time-matched control tumors, which were harvested at the same time as the RBPJ-knockdown tumors, showed increased TUNEL staining compared with RBPJ-depleted tumors (Fig. 5, D–F).
Figure 5.
RBPJ deficiency increases mammosphere formation and confers resistance to cell death in breast cancer cells. (A and B) Mammosphere formation evaluated in MCF10A, a breast epithelial cell line. (A) Representative images of mammospheres formed on day 7 per 10,000 initiating cells from shRandom control cells and shRBPJ cells (bars, 100 µm). (B) Quantification of mammospheres from three independent experiments after 7 d of culture. (C) MDA-MB-231 cell survival in vitro after suspension for 48 h (n = 4). (D) Staining and quantitative mapping of multiple markers in MDA-MB-231 whole tumor cryosections. shRandom control tumors were harvested on the same day (time-matched), or when they reached comparable size (size-matched) to shRBPJ tumors. Representative images with TUNEL staining of dead cells (red), hypoxic areas with pimonidazole (green), S-phase proliferating cells with BrdU (black), vasculature with CD31 (blue) and gray tumor tissue boundaries with hematoxylin (bars, 500 µm). (E and F) Quantification of TUNEL staining shown in D (n = 7 for each group) in the whole section (E) or with confluent cell death excluded (F). (G) TUNEL labeling of cell death and pimonidazole staining of hypoxic cells was evaluated as a function of the distance from nearest CD31 stained vasculature. Lines represent averaged data for each group. Solid lines are TUNEL staining and dashed line are pimonidazole data (n = 7 for each group). (H and I) Quantification of CD31- (H) and BrdU (I)-positive staining in MDA-MB-231 viable tumor tissue (n = 7 for each group). P-values for C were obtained from a Student’s t test and data are presented as mean + SD. P-values for E, F, H, and I were obtained using a Mann-Whitney test and bars are presented as mean + SEM. *, P ≤ 0.05; **, P < 0.005; ***, P ≤ 0.001.
We also stained for and quantified vasculature, hypoxic areas, and proliferating cells in tumor sections (Fig. 5, D and G–I). The difference in cell death between control and RBPJ-depleted tumors was especially pronounced in regions at further distances from blood vessels where hypoxia is highest (Fig. 5 G). Microvessel density (quantified by CD31 staining) and proliferation (quantified by BrdU staining) was also significantly increased in shRBPJ tumors compared with both time-matched and size-matched control tumors (Fig. 5, H and I).
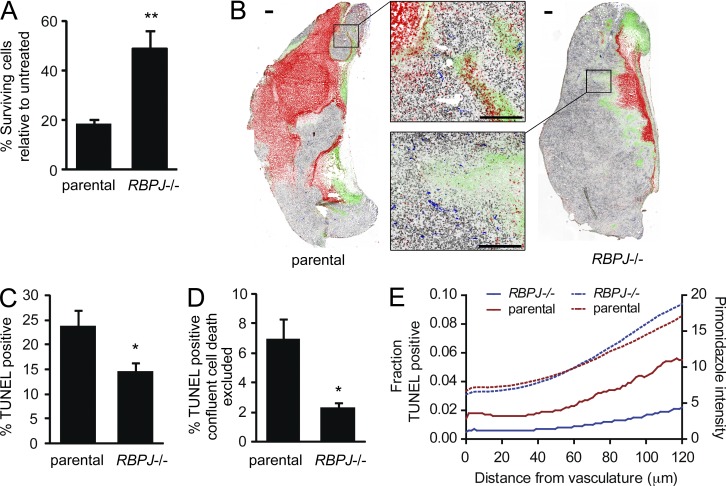
We confirmed our findings in the DG75 lymphoma line, where RBPJ deficiency significantly reduced apoptosis in vitro and in vivo compared with parental cell xenografts (Fig. 6, A–E). This cell death was more pronounced at increasing distances from the vasculature, consistent with the findings in MDA-MB-231 xenografts (Fig. 6 E). However, no difference was observed in CD31 or BrdU staining in DG75 tumors (unpublished data). These data suggest that a common phenotype with RBPJ loss is an increase in survival of tumor cells in vivo.
Figure 6.
Complete RBPJ loss confers resistance to cell death in DG75 lymphoma cells. (A) DG75 cell survival after induction of cell death with 100 µM tert-Butyl hydroperoxide (TBHP, n = 3). (B) Staining and quantitative mapping of multiple markers in DG75 whole tumor cryosections. Parental tumors were harvested when they reached a comparable size to RBPJ−/− tumors. Representative images with TUNEL staining of dead cells (red), hypoxic areas with pimonidazole (green), S-phase proliferating cells with BrdU (black), vasculature with CD31 (blue), and gray tumor tissue boundaries with hematoxylin (bars, 300 µm). (C and D) Quantification of TUNEL staining shown in B (n = 7 for each group) in the whole section (C) or with confluent cell death excluded (D). (E) TUNEL and pimonidazole staining was evaluated as a function of the distance from nearest vasculature. For TUNEL data, n = 7 for each group. For pimonidazole data, n = 5 for DG75 RBPJ−/− and n = 4 for DG75 parental. P-values for A were obtained from a Student’s t test and data are presented as mean + SD. P-values for C and D were obtained using a Mann-Whitney test and bars are presented as mean + SEM. *, P < 0.05; **, P < 0.005.
MYC and NF-κB contribute to increased survival in RBPJ-deficient cancer cells
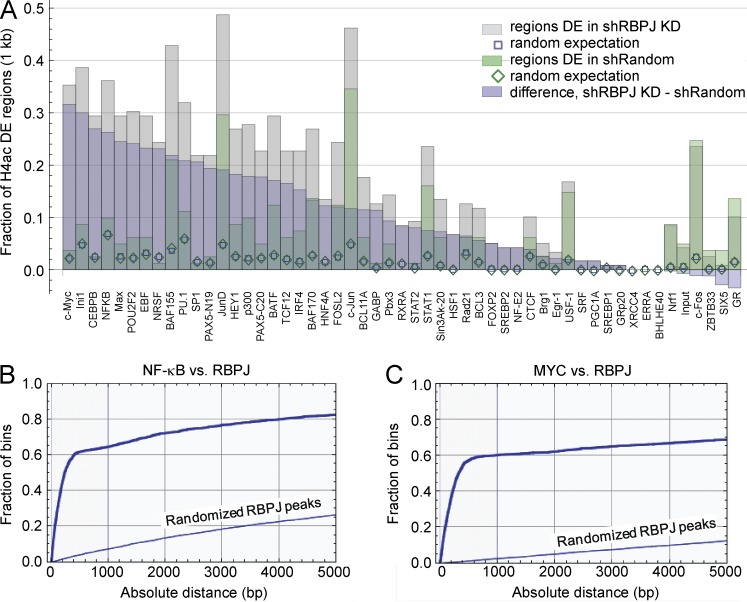
To identify transcription factors that may contribute to gene activation in the context of RBPJ depletion, we compared the spatial concordance between the genome-wide 500 bp H4ac differentially enriched regions in MDA-MB-231 cells and the in vivo binding regions reported for 52 transcription factors in 24 cell lines (Fig. 7 A; Birney et al., 2007). Binding regions for MYC and NF-κB were concentrated within ∼1,000 bp of the H4ac regions that were differentially enriched in the RBPJ knockdown cells (Materials and methods; Fig. 7 A). Evaluating 2,112 RBPJ binding sites from CUTLL T lymphoblastic leukemia/lymphoma cells (Wang et al., 2011), we also found enrichment of NF-κB and MYC binding regions (within ∼500 bp of the binding site; Fig. 7, B and C). For the 114 genes that were differentially enriched in RBPJ knockdown cells, binding motifs for these two factors were among the most highly enriched within regions up to 1 kb upstream of the TSS (Zambelli et al., 2009; unpublished data). Both transcription factors have previously been shown to cooperatively regulate transcription of a common set of target genes with Notch (Lee et al., 2000; Margolin et al., 2009; Barbarulo et al., 2011).
Figure 7.
MYC and NF-κB binding regions are associated with H4ac changes and are near RBPJ binding regions. (A) Spatial association between genome-wide 500 bp H4ac regions in MDA-MB-231 cells and ENCODE ChIP-seq data for 52 transcription factors and input DNA in 24 cell lines (Birney et al., 2007). The bar chart shows the fractions of H4ac differentially enriched (DE) regions with centers within 1 kb of the center of an enriched transcription factor region. (B and C) Concordance between RBPJ ChIP-Seq peaks and NF-κB and MYC binding regions. Spatial relationship between 2112 ChIP-seq enriched RBPJ peaks in CUTLL cells (Wang et al., 2011) and ChIP-seq enriched regions reported for NF-κB p65 (B) and MYC (C) in ENCODE data (Birney et al., 2007). Empirical distribution functions for distances between the center of a thresholded ENCODE peak and the center of the nearest RBPJ peak region, contrasted to the distribution function when RBPJ peak locations on each chromosome were randomized.
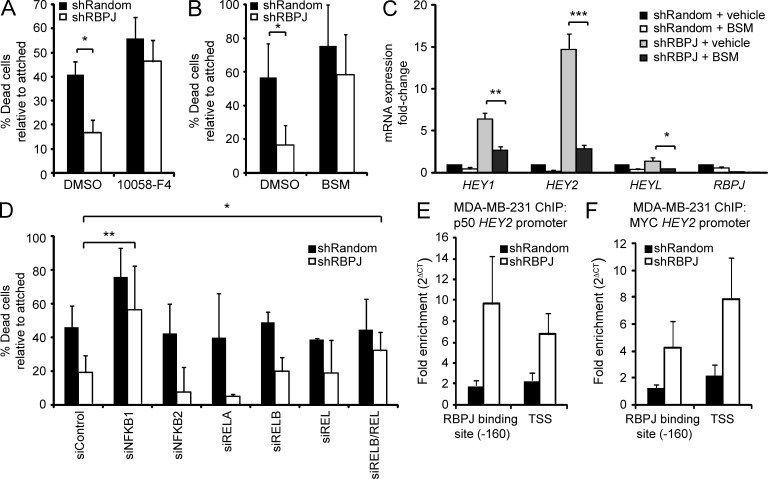
To determine whether MYC and NF-κB–mediated activation are required to promote survival in RBPJ-depleted cells, we performed anoikis assays in the presence of inhibitors against MYC (10058-F4) or NF-κB (BSM-345541, an I-Kappa-B kinase α inhibitor; Burke et al., 2003). Inhibition of either pathway abolished the survival advantage observed with RBPJ loss, resulting in cell death comparable to shRandom MDA-MB-231 cells (Fig. 8, A and B). BSM-345541 also significantly reduced HEY gene expression up-regulated by RBPJ knockdown (Fig. 8 C). To further define which NF-κB family members are required for survival activity in MDA-MB-231 cells, we used siRNA to deplete NF-κB1 (processed to p50), NF-κB2 (processed to p52), RELA/p65, RELB, and REL (c-Rel). Knockdown of either NFKB1 or the combined knockdown of RELB and REL abolished the survival advantage conferred by RBPJ depletion, suggesting that a combination of RELB/p50 and REL/p50 heterodimers may be responsible for the survival activity in shRBPJ MDA-MB-231 cells (Fig. 8 D). Finally, we observed a trend for enriched NF-κB (p50) and MYC binding at the HEY2 promoter in RBPJ-depleted MDA-MB-231 cells (Fig. 8, E and F). Hence, loss of RBPJ-mediated repression may enable nuclear signaling by NF-κB and MYC activating prosurvival signaling pathways.
Figure 8.
MYC and NF-κB contribute to survival in RBPJ-deficient MDA-MB-231 cells. (A and B) Cell death after forced suspension and treatment with indicated agents for 48 h. (A) Vehicle control (DMSO) or 80 µM 10058-F4 (n = 3). (B) Vehicle control (DMSO) or 25 µM BSM-345541 (BSM, n = 4). (C) Gene expression analysis in MDA-MB-231 cells transduced with shRBPJ or shRandom after treatment with vehicle (DMSO) or 25 µM BSM (n = 3). Data are expressed as fold-change relative to shRandom + vehicle. (D) Transfection with control small inhibitory (si)-RNA (siControl), siNFKB1, siNFKB2, siRELA, siRELB, and/or siREL (n ≥ 3). P-values are reported for treatments that significantly increased cell death in shRBPJ-expressing cells relative to the siControl. (E and F) ChIP analysis of the HEY2 promoter with fold enrichment calculated as 2(Ct IgG − Ct target). Bound fragments were quantified by RT-qPCR with primers spanning the −160 bp RBPJ binding site in the HEY2 promoter and the HEY2 TSS (E) NF-κB (p50) ChIP (n = 5). (F) MYC ChIP (n = 3). P-values for A–D were obtained from a Student’s t test and data are presented as mean + SD. For E and F, data are presented as SEM. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
DISCUSSION
Our results indicate that depletion of RBPJ leads to gene induction of RBPJ-bound promoters, increased cell survival, and enhanced tumorigenicity. In contrast to NOTCH1, RBPJ expression is frequently reduced in several primary human tumor types including up to 33% of invasive breast cancer cases. We further show that RBPJ deficiency promotes tumor growth in two cancer cell lines derived from distinct tumor types in which RBPJ is targeted using two methods, shRNA-mediated knockdown or genetic ablation. Although we present data on breast cancer and lymphoma as model systems, we provide additional clinical data supporting RBPJ loss in other human malignancies, such as lung cancers, through either or both RBPJ copy number loss or reduced mRNA expression. Supporting our observations, depletion of RBPJ was also reported to enhance eye tumor growth and metastases in Drosophila (Liefke et al., 2010).
RBPJ depletion causes a similar outcome to Notch activation in promoting tumor growth. Our current and previous work supports that retaining RBPJ transcriptional repression through blockade of Notch (by preventing Notch cleavage or transcriptional activation) inhibits MDA-MB-231 tumor growth (Leong et al., 2007). Conversely, RBPJ deficiency causes up-regulation of recognized Notch target genes, such as those of the HEY gene family, even in the presence of DAPT which inhibits Notch receptor processing and activation. We used a gamma secretase inhibitor to block signaling from all Notch receptors by preventing receptor activation at the membrane. Our aim was to perturb the Notch axis at a site upstream of the NotchIC–MAML–RBPJ ternary complex and show that the signal triggered by RBPJ loss is independent of Notch receptor activation. Derepression upon RBPJ removal also extends to other Notch receptor target genes. In our analysis of 828 invasive human breast cancers, cases with RBPJ loss were associated with an increased Notch signature score compared with tumors without RBPJ loss. Of these cases, breast cancers with RBPJ HD and copy loss had a significantly higher Notch signature score compared with samples with neutral RBPJ copy number. However, cases with RBPJ copy gain also showed an increased Notch signature score compared with samples with neutral RBPJ copy number, although RBPJ HD samples tended to have a higher Notch score than either of those with RBPJ copy loss or gain. In samples with gene amplification, increased RBPJ presumably facilitates transcriptional activation in response to Notch receptor activity. Together, our data support that target gene expression is not solely an indicator of Notch receptor activity, as RBPJ depletion represents an alternate mechanism for promoting tumor growth and HEY expression.
RBPJ is ubiquitously expressed in normal tissue having a promoter sequence characteristic of a housekeeping gene (Hamaguchi et al., 1992; Kawaichi et al., 1992). In the absence of Notch activation, RBPJ is a default transcriptional repressor and enters the nucleus precommitted to this function, which depends on histone deacetylase activity to condenses chromatin and silences gene expression (Zhou and Hayward, 2001; Borggrefe and Oswald, 2009). Conversely, our findings show that depletion of the RBPJ protein at RBPJ binding sites caused enrichment of both H3K4me3 and H4ac, histone marks associated with active promoters. H3K9me3, a modification associated with transcriptional repression, was more modestly increased at the HEY2 RBPJ binding site but was reduced across the HEY2 TSS in RBPJ knockdown cells. Our data support that RBPJ loss results in epigenetic changes that allow promoter activity. As preexisting RBPJ occupancy is required for gene repression in the absence of Notch activation at these promoters, these genes would be expected to be particularly susceptible to derepression upon RBPJ removal.
In Drosophila, active Notch can recruit RBPJ to promoters where it was previously not bound (Krejcí and Bray, 2007). RBPJ is likely not required for the repression of these genes but only for transcriptional activation via Notch. Thus loss of RBPJ may both derepress a subset of Notch target genes and potentially block Notch signaling at other targets. Further, Notch-independent RBPJ signaling has been reported. During early pancreatic and neural development, the Ptf1a–RBPJ complex regulates gene transcription independently of Notch (Beres et al., 2006; Hori et al., 2008). In a human T lymphoblastic leukemia/lymphoma cell line, RBPJ sites were identified that do not bind Notch1. Only 36% of Notch1 binding sites were found to overlap with RBPJ binding sites and 66% of RBPJ sites overlapped with Notch1 sites as evaluated by ChIP-seq (Wang et al., 2011). Viral proteins can also co-opt RBPJ to allow for their own propagation via induction of viral and cellular promoters (Hayward, 2004; Persson and Wilson, 2010). Interestingly, fungal species lack Notch but have RBPJ genes (Prevorovský et al., 2009) providing additional support for a Notch-independent role for RBPJ. These Notch-independent genes may also be susceptible to derepression upon RBPJ depletion.
Several lines of evidence support that Notch is associated with conferring protection against cell death and is an important mediator of tumor cell survival (Capaccione and Pine, 2013). We have previously shown that expression of constitutively active Notch increases the resistance of MCF10A, a breast epithelial cell line, to anoikis and others have demonstrated induction of hyperproliferative acinar structures and colonies in soft agar (Leong et al., 2007; Mazzone et al., 2010). Compatible with a role in inhibiting apoptosis, mammary-specific deletion of Rbpj did not block Notch-induced tumor formation (Raafat et al., 2009). Rather, Notch-induced mammary tumors on a mammary-specific Rbpj−/− background showed reduced apoptosis compared with tumors containing functional Rbpj. Although the interpretation in this study was that Notch has Rbpj-independent oncogenic activity, our findings suggest that RBPJ loss prevents tumor cell death independent of Notch receptor activation, thereby promoting tumorigenesis. We observed reduced cell death as a common phenotype resulting from RBPJ deficiency in two distinct cancer models in vivo, which was especially pronounced in hypoxic areas of tumor tissue. These harsher tumor microenvironments have limited oxygen and nutrient supply and have been proposed to be the origin of more aggressive cells (Vaupel, 2004). RBPJ depletion may also contribute to increased proliferation and angiogenesis, as shRBPJ MDA-MB-231 tumors showed significantly increased BrdU and CD31 staining.
Loss of RBPJ-mediated repression may enable nuclear signaling by MYC and NF-κB, which allows increased survival. Both these signaling pathways are thought to be activated in MDA-MB-231 cells and MYC and NF-κB have previously been shown to cooperatively regulate transcription of a common set of target genes with Notch (Nakshatri et al., 1997; Lee et al., 2000; Cappellen et al., 2007; Margolin et al., 2009; Barbarulo et al., 2011). In RBPJ-deficient cells, NF-κB and MYC binding was enriched at the HEY2 promoter and each of these pathways was required for protection against anoikis. Further, treatment with an inhibitor of NF-κB signaling reduced HEY gene expression up-regulated by RBPJ knockdown. Recently MYC has been shown to be a general amplifier of actively transcribed genes in tumor cells rather than binding and regulating a new set of genes (Lin et al., 2012). Hence MYC may amplify the output of the existing derepressed genes resulting from RBPJ knockdown. Our data suggest that in the absence of RBPJ, enhanced tumor cell survival is associated with MYC- and NF-κB–driven prosurvival mechanisms.
Here, we describe a novel mechanism of cancer progression in mammalian cells that involves loss of RBPJ, the primary transcriptional mediator of canonical Notch signaling. The signal associated with loss of RBPJ overlaps with but is not identical to Notch activation. Indeed, Notch receptor activation is not required to drive the signal generated with RBPJ loss, but rather the signals are mediated in part by NF-κB and MYC. Although Notch activation and RBPJ depletion may not be equivalent, loss of RBPJ represents an alternate means of deregulating a component of the Notch signaling cascade, and so has therapeutic implications, as specific inhibition of Notch processing or function would be predicted to not have beneficial effects on tumors with RBPJ loss. Further, in cancers lacking RBPJ, targeting other oncogenic signals that contribute to survival, such as MYC or NF-κB, may be a therapeutic option.
MATERIALS AND METHODS
Cell culture.
The human DG75 Burkitt’s lymphoma parental and RBPJ−/− cell lines (Maier et al., 2005) were grown in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 2 mM glutamine (Invitrogen), and 100 U of each penicillin and streptomycin. MDA-MB-231 human breast carcinoma cells, viral producer line 293T, and AmphoPhoenix were cultured in DMEM (Sigma-Aldrich) and supplemented as above. For co-culture experiments, 105 MDA-MB-231 shRandom or shRBPJ cells were co-cultured with 105 mouse SVEC4-10 endothelial cells transduced with MSCV-IRES-yellow fluorescent protein (MIY) control vector or MIY-Jagged1. Cells were plated in 12-well tissue culture dishes in media containing either 10 µM DAPT (EMD Millipore) or DMSO vehicle control (Invitrogen). Cells were harvested after 48 h of co-culture and RT-qPCR was performed with human-specific primers to avoid amplification of mouse transcripts. The normal breast epithelial cell line MCF10A was culture in a 1:1 mixture of DMEM/F12 supplemented with 5% horse serum, 2 mM glutamine, 20 ng/ml epidermal growth factor (all from Sigma-Aldrich), 100 ng/ml cholera toxin (Cederlane), 10 µg/ml insulin (Sigma-Aldrich), 500 ng/ml hydrocortisone (Sigma-Aldrich), and 100 U/ml each of penicillin and streptomycin (Invitrogen). All cells were maintained at 37°C in a 5% CO2 atmosphere.
Mammosphere culture.
MCF10A human breast epithelial cells were suspended in MammoCult complete medium containing growth supplements heparin sulfate and hydrocortisone according to the manufacturer’s instructions (Stem Cell Technologies). Mammospheres were cultured according to the MammoCult product sheet. In brief, the cell suspension was passed through a 45-µm cell strainer to obtain single cells. 10,000 MCF10A cells were seeded into each well of a 6-well ultra-low adherence dish (Stem Cell Technologies) containing 2 ml MammoCult complete medium. After 7 d in culture at 5% CO2 at 37°C, spheres >60 µm in diameter were counted.
Cell death assay.
Anoikis experiments were performed with MDA-MB-231 cells by plating 2.5 × 105 cells/well in Costar low-attachment 6-well plates (Stem Cell Technologies). Cells were then incubated in suspension for 48 h. BSM-345541 (EMD Biosciences) or 10058-F4 (Sigma-Aldrich) was added upon cell plating. In experiments where siRNA was used, cells were first transfected using Lipofectamine RNAiMax (Invitrogen) and then collected 24 h later for plating for the anoikis assay. AllStars negative control, NF-κB1 (Hs_NFkB1_7), NF-κB2 (Hs_NFkB2_9), RELA (Hs_RELA_7), RELB (Hs_RELB_5), and cREL (Hs_REL_5) siRNA were obtained from QIAGEN. For treatment with 100 µM tert-butyl hydroperoxide (TBHP; Molecular Probes; Invitrogen), DG75 cells were plated at 7.5 × 105 cells/well in a 6-well plate and treated for 2 h to induce death. To quantify the surviving fraction, cells were harvested (for MDA-MB-231 cells the supernatant, PBS wash, and adherent cells were pooled). Cells were then pelleted, washed once with cold PBS, and resuspended in 100 µl Annexin V binding buffer (10 mM Hepes, 140 mM NaCl, and 2.5 mM CaCl2, pH 7.4; all from Sigma-Aldrich). After addition of 5 µl Annexin V–488 to DG75 cells or Annexin V–APC to MDA-MB-231 cells (both fluorescent conjugates from Invitrogen) and 5 µl propidium iodide (1 mg/ml; Sigma-Aldrich) per tube, the reaction was incubated for 15 min. 500 µl of ice-cold Annexin V binding buffer was added to each tube and flow cytometry analysis was performed immediately using the EPIC Elite flow cytometer (Beckman Coulter) and FCS Express (De Novo Software). Data are expressed as the proportion of surviving cells (defined as Annexin V/propidium iodide negative) or dying cells (Annexin V and/or propidium iodide positive) relative to untreated attached (MDA-MB-231) or untreated (DG75) cells within each group.
RNA interference and gene transfer.
shRNA-encoding DNA oligonucleotides (Integrated DNA Technologies) were cloned into the HpaI and XhoI site of the pLentilox3.7 green fluorescent protein (GFP) lentiviral vector (gift from L. Van Parijs, Massachusetts Institute of Technology, Cambridge, MA; Rubinson et al., 2003). The targeting sequences are as follows: shRandom, human shRBPJ, and shRBPJ-set2 have been previously described (Niessen et al., 2008; NM_015874); shScrambled, 5′-GATTAGAACCCTCACGGTACG-3′. For overexpression, the following retroviral vectors, all of which encoded human genes, were used: pLNCX (gift from A.D. Miller, Fred Hutchinson Cancer Research Centre, Seattle, WA), pLNC-FlagRBP2N, pLNC-FlagRBP2NR179H (the mutated RBPJ; MacKenzie et al., 2004), MIY, and pMIY-Jagged1. Target cells were transduced and purified based on GFP or YFP expression by flow sorting using a FACS-440 flow-sorter (BD). Cell lines expressing LNCX constructs were selected with 300 µg/ml G418 (Invitrogen). Knockdown and overexpression were assessed by RT-qPCR and immunoblotting.
RNA isolation and RT-qPCR.
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturers’ recommendations. First-strand cDNA was synthesized using Superscript II Reverse transcription (Invitrogen) and RT-qPCR was performed using the ABI Prism 7900HT detection system (Applied Biosystems) and the SYBR Green PCR Master Mix kit (Applied Biosystems), with gene-specific primer pairs (Table S4). Data obtained from the RT-qPCR reaction was analyzed using the comparative threshold cycle (Ct) method (User Bulletin No. 2; PerkinElmer Life Sciences) with GAPDH used as the reference gene for normalization.
Immunoblotting.
Cultured cells and tumor tissues were lysed, resolved by SDS-PAGE, and probed overnight with primary antibodies using the following dilutions: 1:1,000 rat anti-RBPJ clone number 76719 (Institute of Immunology), and 1:10,000 mouse anti α-tubulin (Sigma-Aldrich).
EMSA.
Nuclear lysates were collected from shRandom, shRBPJ, and shRBPJ-set2 overexpressing MDA-MB-231 cells for the RBPJ EMSA assays. In brief, the cells were washed two times with PBS, resuspended in four pellet volumes of buffer A (10 mM Hepes-KOH, pH 7.8, 10 mM KCl, 1.5 mM MgCl2, 5 mM with addition of a protease inhibitor cocktail; all from Sigma-Aldrich), pelleted for 15 s at 12,000 g and resuspended in 375 µl buffer A containing 0.5% NP-40 (Sigma-Aldrich). After mixing, cells were incubated at 4°C for 10 min to lyse the cells, centrifuged for 10 min at 12,000 g at 4°C, and the cytosolic extract was removed. The nuclei were washed once with buffer A and resuspended in three pellet volumes of buffer B (50 mM Hepes-KOH, pH 7.8, 10% glycerol, 50 mM KCl, 300 mM NaCl2, and 0.1 mM EDTA, with addition of a protease inhibitor cocktail; all from Sigma-Aldrich) and incubated at 4°C for 20 min. The nuclear lysates were collected after centrifugation for 10 min at 12,000 g at 4°C and stored at −80°C. The oligonucleotides containing RBPJ wild-type or mt motifs in the HEY2 promoter (Table S4) were end-labeled with 32P dCTP using the Klenow fragment of DNA Polymerase I. The binding reaction (10 mM TrisHCl, 50 mM NaCl, 1 mM EDTA, pH 8, 4% glycerol, and 2 µg PolydI-dC binding buffer; all from Sigma-Aldrich), and 10 µg of nuclear protein was performed by preincubating with either 50-fold excess wild-type or mt (Noseda et al., 2006) nonradioactive duplex oligos for 15 min on ice, and then adding a 150,000-cpm 32P-labeled double-stranded probe and incubating for 30 min at room temperature. DNA–protein complexes were electrophoresed on 5% Tris-Borate EDTA gels, dried, and exposed to a phosphorimager plate for 16 h.
Tumor xenografts.
All protocols involving mice were evaluated and approved by the University of British Columbia Animal Care and Ethics Committee. Female non-obese diabetic/severe combined immunodeficient mice (NOD/SCID, Animal Resource Centre of the British Columbia Cancer Research Centre) were injected subcutaneously into the dorsa with human tumor cell lines. Tumor growth was monitored at the indicated times by external measurements with calipers, and tumor volume calculated using the formula (π/6 × length × width × height). For MDA-MB-231 tumor studies, 5 × 106 stably transduced cells were injected in 100 µl unsupplemented DMEM. For DG75 tumor studies, 107 cells were injected in 200 µl PBS. Individual tumors were split for either fixation in 4% paraformaldehyde or freezing, and then used for histology, immunostaining, or RNA and protein analysis.
Whole section tumor staining.
Tumor staining study protocols were similar to those previously reported (Kyle et al., 2003; Huxham et al., 2004). Mice were administered 1,500 mg/kg BrdU (Sigma-Aldrich) in 0.9% NaCl and 60 mg/kg pimonidazole (Hypoxyprobe) by intraperitoneal injection 2 h before tumor excision. Harvested tumors were frozen at −20°C on an aluminum block, covered in embedding medium (O.C.T.) and stored at −80°C until sectioning. Cryosections 10 µm thick were obtained using a Cryosar HM560 (Microm International GmbH; Thermo Fisher Scientific), dried overnight, and fixed in 50:50 acetone-methanol for 10 min at room temperature. Incorporated BrdU was detected using a monoclonal mouse anti-BrdU antibody (clone BU33; Sigma-Aldrich), followed by an anti–mouse peroxidase-conjugated antibody (Sigma-Aldrich) and a metal-enhanced 3,3′-diaminobenzidine substrate (Pierce; Thermo Fisher Scientific). Hypoxic cells labeled with pimonidazole were detected using a polyclonal rabbit anti-pimonidazole antibody (Hypoxiprobe) and visualized with a fluorescent Alexa Fluor 488 tagged anti–rabbit secondary antibody. Vasculature was stained using a rat anti–mouse monoclonal antibody to CD31 (clone MEC 13.3; BD) and fluorescent Alexa Fluor 546 anti–rat secondary antibody (Invitrogen). TUNEL staining was performed using the in situ cell death detection kit (Roche) with a TMR red-tagged dUTP. After imaging of fluorescence, slides were placed in distilled water for 10 min and treated with 2 mol/liter HCl at room temperature for 1 h, followed by neutralization for 5 min in 0.1 mol/liter sodium borate and rinsing in PBS. Slides were counterstained with hematoxylin, dehydrated, and mounted using Permount (Thermo Fisher Scientific) before being imaged. Images were acquired using a fluorescence microscope (Imager Z1; Carl Zeiss), a cooled, monochrome CCD camera (Retiga 4000R; QImaging), and motorized x-y stage (Ludl Electronic Products). This system allows tiling of adjacent fields of view to compile images of entire tumor cryosections at a resolution of 0.75 µm/pixel; sections were repeat-imaged at various points during staining. Using Image (National Institutes of Health) and user-supplied algorithms, images of BrdU, CD31, pimonidazole, TUNEL, and hematoxylin staining, all from the same tumor section, were overlaid and staining artifacts removed. Grayscale images of CD31 and TUNEL were inverted, thresholded, colorized (blue and red, respectively), and overlaid onto images of BrdU and hematoxylin (black and gray, respectively). Grayscale images of pimonidazole (green channel) were overlaid using Photoshop CS (version 8.0; Adobe) and combined using the multiply mask. Tumor tissues were analyzed either as whole tumor sections or as viable tissue only after crop and removal of confluent necrosis. TUNEL-positive staining was identified by selecting pixels a minimum of five standard deviations above tissue background levels and is reported as the fraction of TUNEL positive staining for whole sections or as the fraction of TUNEL-positive staining in viable tissue. BrdU-positive staining is reported as the fraction of positive pixels in viable tissue. Distribution analyses were performed in viable tissue only; each pixel in an image cropped to remove necrosis was sorted based on its distance relative to CD31-positive vasculature. For pimonidazole staining the mean pixel intensity is reported as a function of distance from vasculature, and for TUNEL analyses the fraction of pixels meeting or exceeding the stain threshold is reported as a function of distance from nearest vasculature.
Immunohistochemistry.
The breast cancer tissue microarray sections were cut from formalin-fixed, paraffin-embedded small-core biopsies placed in the same block. Xenograft tumors excised from mice were fixed overnight in 10% phosphate-buffered formalin, progressively dehydrated through gradients of alcohol, and embedded in paraffin. Further analysis was performed by PMI Labs. In brief, 4-µm sections were cut, deparaffinized in xylene, rehydrated, and then stained with hematoxylin and eosin. For immunohistochemical analysis, antigen retrieval was performed with a Pascal decloaker (Dako) using citrate buffer, pH 6.0 (Dako). The rat anti-RBPJ7A11 antibody (Maier et al., 2005) and IgG2Bkappa control (BD) were used as primary antibodies at a 1:10 dilution. The slides were stained using the avidin-biotin-peroxidase method with the EnVision Flex kit (Dako) as per manufacturer’s instructions, a 1:50 dilution of Biotin-SP-AffiniPure donkey anti–rat secondary (Medicorp Inc.), and 1:500 peroxidase-conjugated streptavidin (Medicorp Inc.) followed by the chromagen 3,3′-diaminobenzidine (Dako) to develop the immunostain. All sections were counterstained with hematoxylin. 1.5% sodium bicarbonate was used as a bluing agent. Sections were imaged using a light microscope (BX41; Olympus). Tissue microarray slides were scanned with the imaging system (Aperio Technologies) and images were imported into ImageScope software (Aperio Technologies) for viewing and acquisition of micrographs. Immunophenotypic evaluation was performed independently by two evaluators who scored the tissue microarrays for presence or absence of RBPJ.
Microarray data.
Published microarray data were obtained from the following sources: Gene Expression Omnibus (GEO) accession nos. GSE5364 (human breast cancer and adjacent nonmalignant tissue; Yu et al., 2008), GSE4824 (lung cancer cell lines; Lockwood et al., 2008), GSE3141 (111 NSCLC tumors; Bild et al., 2006); and the Oncomine database (normalized breast cancer microarray data; Ginestier et al., 2006). In addition, data for 67 normal human bronchial epithelial cells analyzed with the U133 plus array platform (Affymetrix), and 49 lung cancer samples analyzed with a custom chip (Affymetrix; Lockwood et al., 2010), were obtained from the System for Integrative Genomic Microarray Analysis (SIGMA) website (Chari et al., 2006). Microarray data were log2 transformed.
TCGA data.
RNA-seq RPKM (z-score) and copy number (GISTIC alteration status) data for all genes of interest were downloaded for 828 invasive breast cancers, 178 lung SqCC, and 129 lung AC from the MSKCC cBIO Cancer Genomics Portal (Cerami et al., 2012; Gao et al., 2013). For each tumor type, the expression of RBPJ was compared between samples with RBPJ copy number loss versus those without using the Mann-Whitney U-Test (two-tailed).
Array comparative genomic hybridization (aCGH).
DNA copy number profiles were generated for 49 lung cancer samples using whole genome tiling path array comparative genomic hybridization. Details of the genomic array, DNA extraction, labeling and hybridization, image analysis, and normalization have been described previously (Ishkanian et al., 2004). Data are available through the SIGMA website.
Notch signature score.
We analyzed published microarray data (GEO accession no. GSE12868; Margolin et al., 2009) comparing 10 human T-ALL cell lines with Notch activating mutations treated with either DMSO or compound E. A total of 182 probes were identified as significantly down-regulated by compound E treatment (Benjamini-Hochberg corrected P ≤ 0.05 paired t test, one-tailed; Table S2). Probes were mapped to 170 annotated genes, 162 of which had an RNA-seq RPKM z-score in TCGA. Expression data for each gene was compiled for 828 invasive breast cancers using a method modified from Loboda et al. (2010) to determine the Notch signature score. In brief, the mean expression for compound E down-regulated genes (i.e., Notch-induced genes) was determined for each sample. The mean expression of compound E up-regulated genes was then subtracted to give a composite Notch pathway score for each sample. The scores were then compared between breast cancer samples with RBPJ loss versus those without using the Mann-Whitney U-Test.
Histone, NF-κB, and MYC ChIP.
shRandom or shRBPJ ChIP samples were prepared from stable MDA-MB-231 cell lines grown to 70% confluence. In brief, cells were cross-linked with 1% formaldehyde for 10 min at room temperature, the reaction was quenched by addition of 0.125 M glycine, and the cells were then washed in PBS and harvested in the presence of protease inhibitors. ChIP was performed as previously described (Robertson et al., 2008). Chromatin DNA was fragmented by sonication for 10 min using the Sonic Dismembrator 550 (cup horn; Thermo Fisher Scientific) to produce chromatin fragments ranging from 100 to 800 bp. The chromatin was precleared with 40 µl of blocked Protein A/G Sepharose beads (GE Healthcare) at 4°C for 2 h. After centrifugation, supernatant was removed from the beads and transferred to fresh tubes. Each immunoprecipitation was performed with 100 µg (histone IPs) or 25 µg (transcription factor IPs) of precleared chromatin and 2.5 µg of anti-H4ac antibody (H4K5/K8/K12/K16, product number 06–866; EMD Millipore), 4 µl rabbit anti-histone H3 trimethyl lysine 4 clone MC315 (Millipore), rabbit anti-histone H3 trimethyl lysine 9 (ab8898; Abcam), 2 µg anti–NF-κB p50 antibody (H-119; Santa Cruz Biotechnology, Inc.), 2 µg anti-c-MYC antibody (N-262; Santa Cruz Biotechnology, Inc.), or 4 µg rabbit IgG from (EMD Millipore), and was incubated at 4°C for 1 h. To each immunoprecipitation reaction, 20 µl of protein A/G Sepharose beads were added and incubated by rotating at 4°C overnight. Beads were recovered by centrifugation and washed twice with ChIP wash buffer (20 mM Tris-HCl, pH 8.0, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 150 mM NaCl) and once with ChIP final wash buffer (20 mM Tris-HCl, pH 8.0, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 500 mM NaCl). DNA–antibody complexes were eluted using 100 µl Elution Buffer (100 mM NaHCO3 and 1% SDS), and 5 µg DNase-free RNase (Roche) was added and incubated at 68°C for 2 h with shaking on a thermomixer to reverse the cross-links. The beads were pelleted by centrifugation and the supernatant was collected. Elution was repeated with the addition of 100 µl of Elution Buffer and incubation at 68°C for 5 min with shaking on a thermomixer. After pooling the two elutions, DNA was recovered from the eluate using the QIAquick PCR Purification kit (QIAGEN). The immunoprecipitated DNA was validated by qPCR using 0.25 µM final concentration GAPDH primers (Forward Part# 101221; Reverse Part # 101222 from Active Motif), HEY2-160, HEY2 TSS (Table S4), and ZNF3 as a negative control. qPCR was set up on a 7900HT Fast Real-Time PCR System (Applied Biosystems) using 1 µl of immunoprecipitated eluate and the SYBR Green PCR Master Mix (Applied Biosystems). Cycling parameters were 95°C for 10 min, 40× (95°C for 10 s, 59°C for 30 s, and 72°C for 30 s), 72°C for 5 min, 95°C for 15 s, 59°C for 15 s, and 95°C for 15 s. Fold enrichment was calculated as 2(Ct IgG − Ct target).
RBPJ ChIP.
shRandom or shRBPJ MDA-MB-231 cell lines were grown and processed for ChIP as described above with the following changes: chromatin preparation and immunoprecipitation steps were performed as described in the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology) except using Protein A/G agarose beads (Promega). Each immunoprecipitation was performed with 20 µg of precleared chromatin and 10 µl rabbit anti-RBPSUH (Cell Signaling Technology), rabbit anti-histone H3 antibody, and rabbit anti-IgG (provided by kit). The immunoprecipitated DNA was validated by qPCR using primers for HEY2-160, HEY2 TSS (Table S4), and RPL30 as a negative control (provided by the kit). Fold enrichment was calculated as 2(Ct IgG − Ct target).
acH4 ChIP library construction and sequencing analysis.
Sequencing libraries for the acH4 ChIP were prepared as previously described (Robertson et al., 2007). In brief, ChIP DNA (roughly 50 ng) was run in 8% PAGE and the 100–300 bp fraction was excised, eluted overnight at 4°C in 300 µl of elution buffer (5:1, LoTE buffer [3 mM Tris-HCl, pH 7.5, 0.2 mM EDTA]-7.5 M ammonium acetate), and purified by ethanol precipitation using a Spin-X Filter Tube (Thermo Fisher Scientific). Libraries were prepared using the Illumina single-end (SE) library construction protocol. This involved DNA end-repair, and phosphorylation by T4 DNA polymerase, Klenow DNA Polymerase, and T4 polynucleotide kinase, respectively, in a single reaction and subsequent 3′ A overhang generation by Klenow fragment (3′ to 5′ exo minus) and ligation to Illumina SE adapters (with 5′ overhangs). Adapter-ligated products were purified on Qiaquick spin columns (QIAGEN) and PCR-amplified using Phusion DNA polymerase for 10 cycles and the SE primer set (Illumina). The PCR product was purified using 8% PAGE gels, and DNA quality was assessed and quantified using an Agilent DNA 1000 series II assay (Agilent Technologies) and a 7500 spectrophotometer (Nanodrop), and subsequently diluted to 10 nM. The final concentration was confirmed using a Quant-iT dsDNA HS assay kit and Qubit fluorometer (Invitrogen). Clusters were generated on Illumina cluster stations and were sequenced to 51 bp for shRBPJ and 76 bp for shRandom using GAIIx (Illumina), according to the manufacturer’s instructions. The sequence read data were submitted to the Sequence Read Archive with accession no. SRP012092.
Identifying regions differentially enriched in H4ac.
Single-end Illumina reads for shRBPJ (50 bp, 14.50 M reads after MAPQ>10 filtering and duplicate removal) and shRandom (75 bp, 19.70 M reads after MAPQ>10 filtering and duplicate removal) were aligned to the NCBI36/hg18 reference genome with BWA version 0.5.7 (Li et al., 2009). Differentially enriched 500-bp genome-wide regions were identified with edgeR (Robinson et al., 2010) v2.0.3 and R v2.11.1, using edgeR’s TMM library size normalization to adjust for the different sequencing depths in the two libraries, and calculating B-H–corrected p-values for differential enrichment. Considering both a concentration versus fold change plot that we annotated with adjusted p-values and results from regions that we defined around transcriptional start sites (below), we set an adjusted p-value threshold of 0.005, a log2 fold change threshold of 1.0, and a log2 concentration threshold of −21.5, and generated a set of filtered differentially enriched regions. The concentration threshold rejected regions with relatively low read coverages that would be most affected by variability in the ChIP-seq data.
Spatial relationship between differentially enriched regions and TSSs.
For the above 500 bp differentially enriched regions, we determined distances between a region center and the nearest UCSC gene (Hsu et al., 2006) TSS. Spatial distributions were initially assessed as an empirical distribution function for the absolute value of distance from a differentially enriched region center to the nearest TSS (Fig. 4 F). The region in which H4ac was enriched was refined to −2/+1 kb by considering directional distances to a TSS, i.e., by considering transcript strand (Fig. 4 G). The calculations were done separately for regions that were differentially enriched in the shRBPJ versus the shRandom data, then for regions that were differentially enriched in the shRandom versus the shRBPJ data. Random expectations for both spatial distributions were generated as the mean of 20 distance calculations in which TSS locations on each chromosome were randomly relocated.
Identifying genes with TSS regions differentially enriched in H4ac.
We used edgeR (Robinson et al., 2010) to identify which −2/+1 kb TSS regions were differentially enriched in the H4ac knockdown data. For this calculation, we addressed redundant TSS coordinates due to a gene having multiple transcripts by assigning sets of identical TSS coordinates to a single TSS coordinate, and merging any TSSs that were separated by <500 bp into a single coordinate (R script available on request). Adjusted p-values for published RPBJ target genes were assessed in the context of a scatter plot of regional read concentration versus fold change. From this, we set an adjusted p-value threshold of 0.005, a log2 fold change threshold of 1.0, and a log2 concentration threshold of −16, and generated a set of 114 filtered, differentially expressed TSS regions. More stringent concentration filtering was chosen for TSS regions than for genome-wide bins because genome-wide bins were used to identify the extent of TSS regions, whereas thresholded TSS regions were used to define genes that were likely regulated by RBPJ binding.
Spatial relationship between differentially expressed regions and CHIP-seq data.
55 ENCODE (Birney et al., 2007) ChIP-seq threshold region datasets were downloaded from the UCSC hg18 genome browser. Excluding Pol3 and TAF1 because they are not transcription factors (TFs), and using custom Mathematica scripts (Wolfram Research), we filtered each of the remaining 52 TF datasets and 1 input DNA dataset by removing records for any region that had a score in the lowest 10% for that TF or a length in the upper 10% for that TF. For each filtered dataset, we calculated distances between the center of a filtered, genome-wide 500 bp H4ac differentially enriched region and the center of the nearest TF region. To identify spatial relationships that were shared or differed between TFs, we assessed empirical distribution functions. To compare results for H4ac regions that were differentially enriched in the knockdown or the control, and random expectations for each, we summarized the spatial relationships as a bar chart showing the Y-coordinate of each empirical distribution function at a center-to-center distance of 1 kb (Fig. 7 A).
We used block bootstrap v0.8.1 (Encode Statistics) to estimate p-values for the concordance between locations of 2,112 genome-wide strong enriched regions from Wang et al. (2011) and locations of genome-wide 500-bp regions that edgeR called as differentially enriched in H4ac in the RBPJ knockdown (BH-corrected P < 0.05) and in the control (BH-corrected P < 0.01). For each pairwise comparison, we set R = 0.05 and reported the mean p-value from thirty 10,000-iteration runs.
Transcription factor position frequency matrix (PFM) analysis.
We generated a set of genome sequences for the 38,405 nonredundant −2/+1 kb TSS regions for NCBI36/hg18 UCSC genes that were used for the TSS-region H4ac differential enrichment calculation (above). We used a command-line version of TRAP (Roider et al., 2007) to assign binding affinity scores to each TSS region, for each transcription factor binding matrix (PFM; D’Haeseleer, 2006) in a set consisting of: all matrices in TRANSFAC Professional v9.3 (Wingender, 2008); a TGGGAA RBPJ motif and an NF-κB–like RBPJ motif, both returned by de novo discovery with MEME v3.5.7 (Bailey et al., 2009) from a set of 68 published, experimentally determined RBPJ binding site regions (Table S5); four NF-κB–related PFMs from the nonredundant subset of the JASPAR 2010 database (MA0107.1 RELA, MA0061.1 NF-κB, MA0105.1 NFKB1, and MA0101.1 REL; Portales-Casamar et al., 2010); and a published RelB-p52 PFM (Britanova et al., 2008). To identify high-scoring PFM matches, we scanned TSS region sequences with PFMs using MotifLocator v3.2, with default settings, and a third order background model that we generated from the 38405 nonredundant TSS regions using CreateBackgroundModel v3.2. We assessed enrichment of PFMs available in both JASPAR and the public TRANSFAC databases for the 114 knockdown differentially enriched genes, using the Pscan webserver (Zambelli et al., 2009) and −1/0 kb TSS regions. Pscan ranks PFMs for enrichment of in a target set of TSS regions via the P values of z-scores calculated for that target set versus a background whole-genome region set.
Ingenuity pathway analysis.
IPA (Ingenuity Systems) was used to interpret acH4 data by performing Core Analysis using default parameters in the context of biofunctions (molecular and cellular functions). The 114 TSS regions that were identified as knockdown differentially enriched by the H4ac analysis were defined as input parameters for the analysis. Significance was tested by with the B-H correction for multiple testing using P ≤ 0.025.
Statistical analyses.
Specific tests are noted in the text and figure legends. For pairwise comparisons, in vitro data were analyzed using a Student’s t test; otherwise, we used the Mann-Whitney U test. The parametric one-way ANOVA and nonparametric Kruskal-Wallis one-way analysis of variance tests were used to compare multiple groups. Prism 6 (GraphPad Software) was used for statistical analysis and generating box-and-whisker plots. P-values <0.05 were considered significant unless otherwise stated.
Online supplemental material.
Table S1 shows RBPJ copy number alteration in 216 cancer cell lines. Table S2 shows differentially expressed probes in duplicate cultures of 10 human T-ALL cell lines with Notch-activating mutations. Table S3 shows 114 transcriptional start sites differentially enriched in H4ac in RBPJ knockdown MDA-MB-231 cells. Table S4 shows a list of primer sequences used. Table S5 shows RBPJ binding regions used for PFM analysis Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121192/DC1.
Supplementary Material
Acknowledgments
We thank Fred Wong and Denise McDougal for assistance with flow cytometry and cell sorting and Patrick Plettner for assistance with data submission.
This research was supported by grants to A. Karsan from the Canadian Institutes of Health Research (MOP 64354) and the Cancer Research Society. I. Kulic, L. Chang, and J.H.E. Baker were supported by Studentships from the Michael Smith Foundation for Health Research. I. Kulic was also supported by a Doctoral Research Award from the Canadian Institutes of Health Research and a Postgraduate Master’s Scholarship from the Natural Sciences and Engineering Research Council of Canada. B. Kempkes was supported by a grant from the Deutsche Jose Carreras Leukämie-Stiftung (R02/11) and the Deutsche Krebshilfe (106899).
The authors declare no competing financial interests.
Contributions: I. Kulic designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. G. Robertson wrote the manuscript and conducted bioinformatic analysis of ChIP-seq data with assistance from I. Kulic, L. Chang, and M.D. Robinson using the tool developed by M.D. Robinson. H-J. Chun and S. Jones also contributed to bioinformatic analysis. W.W. Lockwood conducted analysis on the Notch signature, TCGA, and aCGH data. L. Chang, J.H.E. Baker, W. Mok, M. Fuller, M. Fournier, and V. Chou performed experiments. A.I. Minchinton enabled whole section tumor staining. Cell lines were provided by B. Kempkes and W.L. Lam and tissue microarrays by B. Gilks and T.A. Thomson. M. Hirst and M. Marra enabled generation of ChIP-seq data. A. Karsan conceived and supervised the project, interpreted data, and wrote the manuscript.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DAPT
- N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- EMSA
- electrophoretic mobility shift assay
- H3K4me3
- histone H3 lysine4 tri-methylation
- H3K9me3
- histone H3 lysine9 tri-methylation
- H4ac
- acetylated histone H4
- HD
- homozygous deletion
- mt
- mutant
- sh
- short hairpin
- TSS
- transcription start site
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
References
- Aerts S., Van Loo P., Thijs G., Mayer H., de Martin R., Moreau Y., and De Moor B.. 2005. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 33:W393–W396 10.1093/nar/gki354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., and Noble W.S.. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarulo A., Grazioli P., Campese A.F., Bellavia D., Di Mario G., Pelullo M., Ciuffetta A., Colantoni S., Vacca A., Frati L., et al. . 2011. Notch3 and canonical NF-κB signaling pathways cooperatively regulate Foxp3 transcription. J. Immunol. 186:6199–6206 10.4049/jimmunol.1002136 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y.. 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society, Series B. 57:289–300. [Google Scholar]
- Beres T.M., Masui T., Swift G.H., Shi L., Henke R.M., and MacDonald R.J.. 2006. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 26:117–130 10.1128/MCB.26.1.117-130.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild A.H., Yao G., Chang J.T., Wang Q., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., et al. . 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 439:353–357 10.1038/nature04296 [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. . Children’s Hospital Oakland Research Institute. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 447:799–816 10.1038/nature05874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T., and Oswald F.. 2009. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 66:1631–1646 10.1007/s00018-009-8668-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S.J.2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Britanova L.V., Makeev V.J., and Kuprash D.V.. 2008. In vitro selection of optimal RelB/p52 DNA-binding motifs. Biochem. Biophys. Res. Commun. 365:583–588 10.1016/j.bbrc.2007.10.200 [DOI] [PubMed] [Google Scholar]
- Burke J.R., Pattoli M.A., Gregor K.R., Brassil P.J., MacMaster J.F., McIntyre K.W., Yang X., Iotzova V.S., Clarke W., Strnad J., et al. . 2003. BMS-345541 is a highly selective inhibitor of IκB kinase that binds at an allosteric site of the enzyme and blocks NF-κB-dependent transcription in mice. J. Biol. Chem. 278:1450–1456 10.1074/jbc.M209677200 [DOI] [PubMed] [Google Scholar]
- Capaccione K.M., and Pine S.R.. 2013. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 34:1420–1430 10.1093/carcin/bgt127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellen D., Schlange T., Bauer M., Maurer F., and Hynes N.E.. 2007. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 8:70–76 10.1038/sj.embor.7400849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. . 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.C., Fu Y., Garside V.C., Niessen K., Chang L., Fuller M., Setiadi A., Smrz J., Kyle A., Minchinton A., et al. . 2011. Notch initiates the endothelial-to-mesenchymal transition in the atrioventricular canal through autocrine activation of soluble guanylyl cyclase. Dev. Cell. 21:288–300 10.1016/j.devcel.2011.06.022 [DOI] [PubMed] [Google Scholar]
- Chari R., Lockwood W.W., Coe B.P., Chu A., Macey D., Thomson A., Davies J.J., MacAulay C., and Lam W.L.. 2006. SIGMA: a system for integrative genomic microarray analysis of cancer genomes. BMC Genomics. 7:324 10.1186/1471-2164-7-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer P.2006. What are DNA sequence motifs? Nat. Biotechnol. 24:423–425 10.1038/nbt0406-423 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. . 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C., Cervera N., Finetti P., Esteyries S., Esterni B., Adélaïde J., Xerri L., Viens P., Jacquemier J., Charafe-Jauffret E., et al. . 2006. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin. Cancer Res. 12:4533–4544 10.1158/1078-0432.CCR-05-2339 [DOI] [PubMed] [Google Scholar]
- Hamaguchi Y., Yamamoto Y., Iwanari H., Maruyama S., Furukawa T., Matsunami N., and Honjo T.. 1992. Biochemical and immunological characterization of the DNA binding protein (RBP-Jκ) to mouse J kappa recombination signal sequence. J. Biochem. 112:314–320. [DOI] [PubMed] [Google Scholar]
- Hayward S.D.2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14:387–396 10.1016/j.semcancer.2004.04.018 [DOI] [PubMed] [Google Scholar]
- Hori K., Cholewa-Waclaw J., Nakada Y., Glasgow S.M., Masui T., Henke R.M., Wildner H., Martarelli B., Beres T.M., Epstein J.A., et al. . 2008. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 22:166–178 10.1101/gad.1628008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., and Haussler D.. 2006. The UCSC Known Genes. Bioinformatics. 22:1036–1046 10.1093/bioinformatics/btl048 [DOI] [PubMed] [Google Scholar]
- Huxham L.A., Kyle A.H., Baker J.H., Nykilchuk L.K., and Minchinton A.I.. 2004. Microregional effects of gemcitabine in HCT-116 xenografts. Cancer Res. 64:6537–6541 10.1158/0008-5472.CAN-04-0986 [DOI] [PubMed] [Google Scholar]
- Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., et al. . 2004. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 36:299–303 10.1038/ng1307 [DOI] [PubMed] [Google Scholar]
- Kawaichi M., Oka C., Shibayama S., Koromilas A.E., Matsunami N., Hamaguchi Y., and Honjo T.. 1992. Genomic organization of mouse Jκ recombination signal binding protein (RBP-Jκ) gene. J. Biol. Chem. 267:4016–4022. [PubMed] [Google Scholar]
- Koch U., and Radtke F.. 2007. Notch and cancer: a double-edged sword. Cell. Mol. Life Sci. 64:2746–2762 10.1007/s00018-007-7164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R.A., and Hendrickson W.A.. 2004. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23:3441–3451 10.1038/sj.emboj.7600349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcí A., and Bray S.. 2007. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21:1322–1327 10.1101/gad.424607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle A.H., Huxham L.A., Baker J.H., Burston H.E., and Minchinton A.I.. 2003. Tumor distribution of bromodeoxyuridine-labeled cells is strongly dose dependent. Cancer Res. 63:5707–5711. [PubMed] [Google Scholar]
- Lee S.H., Wang X., and DeJong J.. 2000. Functional interactions between an atypical NF-κB site from the rat CYP2B1 promoter and the transcriptional repressor RBP-Jκ/CBF1. Nucleic Acids Res. 28:2091–2098 10.1093/nar/28.10.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K.G., Niessen K., Kulic I., Raouf A., Eaves C., Pollet I., and Karsan A.. 2007. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 204:2935–2948 10.1084/jem.20071082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Gao F., Li N., Li S., Yin G., Tian G., Jia S., Wang K., Zhang X., Yang H., et al. . 2009. An improved method for genome wide DNA methylation profiling correlated to transcription and genomic instability in two breast cancer cell lines. BMC Genomics. 10:223 10.1186/1471-2164-10-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., Rodriguez P., Dominguez M., and Borggrefe T.. 2010. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 24:590–601 10.1101/gad.563210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., and Young R.A.. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 151:56–67 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Nebozhyn M., Klinghoffer R., Frazier J., Chastain M., Arthur W., Roberts B., Zhang T., Chenard M., Haines B., et al. . 2010. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med. Genomics. 3:26 10.1186/1755-8794-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood W.W., Chari R., Coe B.P., Girard L., Macaulay C., Lam S., Gazdar A.F., Minna J.D., and Lam W.L.. 2008. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 27:4615–4624 10.1038/onc.2008.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood W.W., Chari R., Coe B.P., Thu K.L., Garnis C., Malloff C.A., Campbell J., Williams A.C., Hwang D., Zhu C.Q., et al. . 2010. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med. 7:e1000315 10.1371/journal.pmed.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood W.W., Wilson I.M., Coe B.P., Chari R., Pikor L.A., Thu K.L., Solis L.M., Nunez M.I., Behrens C., Yee J., et al. . 2012. Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development. PLoS ONE. 7:e37775 10.1371/journal.pone.0037775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie F., Duriez P., Larrivée B., Chang L., Pollet I., Wong F., Yip C., and Karsan A.. 2004. Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats and involves signaling through RBP-Jκ. Blood. 104:1760–1768 10.1182/blood-2003-12-4244 [DOI] [PubMed] [Google Scholar]
- Maier S., Santak M., Mantik A., Grabusic K., Kremmer E., Hammerschmidt W., and Kempkes B.. 2005. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J. Virol. 79:8784–8792 10.1128/JVI.79.14.8784-8792.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin A.A., Palomero T., Sumazin P., Califano A., Ferrando A.A., and Stolovitzky G.. 2009. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc. Natl. Acad. Sci. USA. 106:244–249 10.1073/pnas.0806445106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M., Selfors L.M., Albeck J., Overholtzer M., Sale S., Carroll D.L., Pandya D., Lu Y., Mills G.B., Aster J.C., et al. . 2010. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc. Natl. Acad. Sci. USA. 107:5012–5017 10.1073/pnas.1000896107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Stiegen F., Schwanbeck R., Bernoth K., Martini S., Hieronymus T., Ruau D., Zenke M., and Just U.. 2010. Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PLoS ONE. 5:e11481 10.1371/journal.pone.0011481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H., Bhat-Nakshatri P., Martin D.A., Goulet R.J. Jr, and Sledge G.W. Jr. 1997. Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell. Biol. 17:3629–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network T.C.G.A.Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen K., Fu Y., Chang L., Hoodless P.A., McFadden D., and Karsan A.. 2008. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 182:315–325 10.1083/jcb.200710067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda M., Fu Y., Niessen K., Wong F., Chang L., McLean G., and Karsan A.. 2006. Smooth Muscle α-actin is a direct target of Notch/CSL. Circ. Res. 98:1468–1470 10.1161/01.RES.0000229683.81357.26 [DOI] [PubMed] [Google Scholar]
- Persson L.M., and Wilson A.C.. 2010. Wide-scale use of Notch signaling factor CSL/RBP-Jκ in RTA-mediated activation of Kaposi’s sarcoma-associated herpesvirus lytic genes. J. Virol. 84:1334–1347 10.1128/JVI.01301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E., Thongjuea S., Kwon A.T., Arenillas D., Zhao X., Valen E., Yusuf D., Lenhard B., Wasserman W.W., and Sandelin A.. 2010. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 38:D105–D110 10.1093/nar/gkp950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevorovský M., Grousl T., Stanurová J., Rynes J., Nellen W., Půta F., and Folk P.. 2009. Cbf11 and Cbf12, the fission yeast CSL proteins, play opposing roles in cell adhesion and coordination of cell and nuclear division. Exp. Cell Res. 315:1533–1547 10.1016/j.yexcr.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Raafat A., Lawson S., Bargo S., Klauzinska M., Strizzi L., Goldhar A.S., Buono K., Salomon D., Vonderhaar B.K., and Callahan R.. 2009. Rbpj conditional knockout reveals distinct functions of Notch4/Int3 in mammary gland development and tumorigenesis. Oncogene. 28:219–230 10.1038/onc.2008.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedijk M., Odorcic S., Chang L., Zhang H., Miller N., McCready D.R., Lockwood G., and Egan S.E.. 2005. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 65:8530–8537 10.1158/0008-5472.CAN-05-1069 [DOI] [PubMed] [Google Scholar]
- Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., and Chinnaiyan A.M.. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 6:1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., et al. . 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 4:651–657 10.1038/nmeth1068 [DOI] [PubMed] [Google Scholar]
- Robertson A.G., Bilenky M., Tam A., Zhao Y., Zeng T., Thiessen N., Cezard T., Fejes A.P., Wederell E.D., Cullum R., et al. . 2008. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 18:1906–1917 10.1101/gr.078519.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roider H.G., Kanhere A., Manke T., and Vingron M.. 2007. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics. 23:134–141 10.1093/bioinformatics/btl565 [DOI] [PubMed] [Google Scholar]
- Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J., Rooney D.L., Zhang M., Ihrig M.M., McManus M.T., et al. . 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401–406 10.1038/ng1117 [DOI] [PubMed] [Google Scholar]
- Sahlgren C., Gustafsson M.V., Jin S., Poellinger L., and Lendahl U.. 2008. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA. 105:6392–6397 10.1073/pnas.0802047105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P.2004. The role of hypoxia-induced factors in tumor progression. Oncologist. 9:10–17 10.1634/theoncologist.9-90005-10 [DOI] [PubMed] [Google Scholar]
- Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W.S., Schug J., Blacklow S.C., Arnett K.L., et al. . 2011. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. USA. 108:14908–14913 10.1073/pnas.1109023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E.2008. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 9:326–332 10.1093/bib/bbn016 [DOI] [PubMed] [Google Scholar]
- Yu K., Ganesan K., Tan L.K., Laban M., Wu J., Zhao X.D., Li H., Leung C.H., Zhu Y., Wei C.L., et al. . 2008. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet. 4:e1000129 10.1371/journal.pgen.1000129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F., Pesole G., and Pavesi G.. 2009. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37:W247–W252 10.1093/nar/gkp464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Langerød A., Ji Y., Nowels K.W., Nesland J.M., Tibshirani R., Bukholm I.K., Kåresen R., Botstein D., Børresen-Dale A.L., and Jeffrey S.S.. 2004. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell. 15:2523–2536 10.1091/mbc.E03-11-0786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., and Hayward S.D.. 2001. Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol. 21:6222–6232 10.1128/MCB.21.18.6222-6232.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.