Yang et al. show that a disulfide isoform of HMGB1, with a role in TLR4 signaling, physically interacts with and binds MD-2. MD-2 deficiency in macrophage cell lines or in primary mouse macrophages stimulated with HMGB1 implicates MD-2 in TLR4 signaling. They also identify an HGMB1 peptide inhibitor, P5779, which when administered in vivo can protect mice from acetaminophen-induced hepatoxicity, ischemia/reperfusion injury, and sepsis.
Abstract
Innate immune receptors for pathogen- and damage-associated molecular patterns (PAMPs and DAMPs) orchestrate inflammatory responses to infection and injury. Secreted by activated immune cells or passively released by damaged cells, HMGB1 is subjected to redox modification that distinctly influences its extracellular functions. Previously, it was unknown how the TLR4 signalosome distinguished between HMGB1 isoforms. Here we demonstrate that the extracellular TLR4 adaptor, myeloid differentiation factor 2 (MD-2), binds specifically to the cytokine-inducing disulfide isoform of HMGB1, to the exclusion of other isoforms. Using MD-2–deficient mice, as well as MD-2 silencing in macrophages, we show a requirement for HMGB1-dependent TLR4 signaling. By screening HMGB1 peptide libraries, we identified a tetramer (FSSE, designated P5779) as a specific MD-2 antagonist preventing MD-2–HMGB1 interaction and TLR4 signaling. P5779 does not interfere with lipopolysaccharide-induced cytokine/chemokine production, thus preserving PAMP-mediated TLR4–MD-2 responses. Furthermore, P5779 can protect mice against hepatic ischemia/reperfusion injury, chemical toxicity, and sepsis. These findings reveal a novel mechanism by which innate systems selectively recognize specific HMGB1 isoforms. The results may direct toward strategies aimed at attenuating DAMP-mediated inflammation while preserving antimicrobial immune responsiveness.
After infection or injury, the immediate host inflammatory response is mediated by receptors on innate immune cells that can efficiently recognize pathogen- or damage-associated molecular patterns (PAMPs or DAMPs). For instance, the mammalian response to bacterial endotoxin (LPS) is mediated by the LPS-binding protein (LBP), CD14, MD-2, and TLR4. Upon capturing LPS, LBP transfers it to CD14 and MD-2, which then delivers LPS to the signaling, high-affinity transmembrane receptor TLR4 (Nagai et al., 2002). The engagement of LPS with TLR4 triggers the sequential release of “early” (e.g., TNF, IL-1, and IFN-β) and “late” proinflammatory mediators (e.g., HMGB1; Wang et al., 1999).
As a ubiquitous nuclear protein, HMGB1 can be passively released from damaged cells (Scaffidi et al., 2002) after sterile tissue injury as a result of ischemia/reperfusion (I/R; Tsung et al., 2005) or chemical toxicity (Antoine et al., 2013). HMGB1 can signal through a family of receptors, including RAGE (Huttunen et al., 1999), TLR4 (Yang et al., 2010), and cluster of differentiation 24 (CD24)/Siglec-10 (Chen et al., 2009), thereby functioning as a DAMP that alerts, recruits, and activates innate immune cells to produce a wide range of cytokines and chemokines. Thus, seemingly unrelated conditions such as infection and sterile injury can converge on a common process: inflammation, which is orchestrated by HMGB1 actively secreted from innate immune cells or passively released from damaged tissues (Zhang et al., 2010; Andersson and Tracey, 2011). Extracellular HMGB1 has been established as a pathogenic mediator of both infection- and injury-elicited inflammatory diseases (Yang et al., 2013).
HMGB1 is a redox-sensitive protein as it contains three conserved cysteine residues at position 23, 45, and 106. The redox status of the cysteines dictates its extracellular chemokine- or cytokine-inducing properties. Specifically, HMGB1 with all cysteine residues reduced (fully reduced HMGB1) binds to CXCL12 and stimulates immune cell infiltration via the CXCR4 receptor in a synergistic fashion. Partially oxidized HMGB1, with a Cys23-Cys45 disulfide bond and a reduced Cys106 (disulfide HMGB1), activates immune cells to produce cytokines/chemokines via the TLR4 receptor. Once all cysteines are terminally oxidized (sulfonyl HMGB1), HMGB1 is devoid of chemotactic and cytokine activities (Tang et al., 2012; Venereau et al., 2012). Previously we showed that HMGB1 induces inflammatory responses via the TLR4–MD-2 signaling pathway and that the interaction with TLR4–MD-2 requires a specific HMGB1 redox form with a distinct atomic structure of thiol-cysteine 106 (Yang et al., 2012). Ample evidence suggests that HMGB1, when actively secreted by activated immune cells or passively released from dying cells, is a mixture of several isoforms with distinct posttranslational modifications (Yang et al., 2013). Paradoxically, it is unknown how the immune system uses the TLR4–MD-2 receptor system to distinguish between different isoforms of HMGB1, specifically recognizing the disulfide HMGB1 molecule to the exclusion of other isoforms.
MD-2 carries a hydrophobic pocket folded by two antiparallel β-sheets for binding LPS (Park et al., 2009) and confers molecular specificity for LPS interaction and TLR4 signaling (Nagai et al., 2002; Meng et al., 2010). Accordingly, here we reasoned that MD-2 may similarly discriminate different HMGB1 isoforms to facilitate TLR4-dependent signaling. Our current findings reveal that only the disulfide HMGB1 binds to MD-2, and this interaction is critically important for HMGB1-mediated cytokine/chemokine production and the development of subsequent tissue injury. Screening of HMGB1 peptide libraries identified a tetramer (FSSE, P5779) as a specific MD-2–targeting antagonist that prevents HMGB1–MD-2 interaction and cytokine induction, thereby protecting animals against liver I/R injury, chemical toxemia, and sepsis.
RESULTS AND DISCUSSION
Cytokine-inducing (disulfide) HMGB1 effectively binds to MD-2
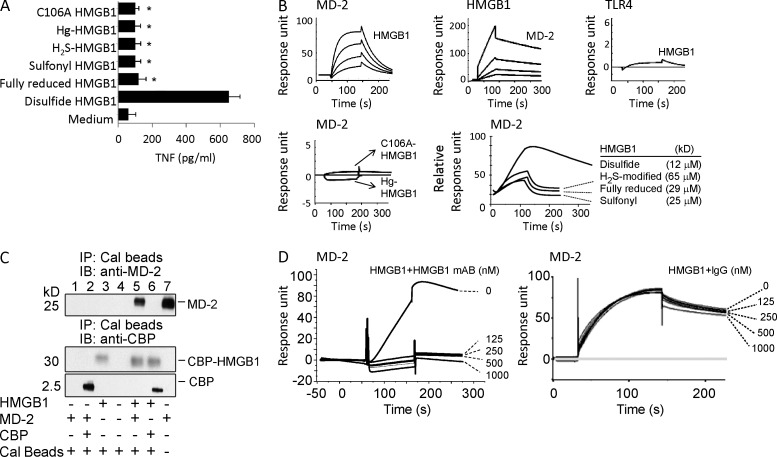
HMGB1 contains three redox-sensitive cysteine residues that are modified by redox reactions to produce multiple HMGB1 isoforms that extracellularly express or lack chemokine or cytokine activities. To elucidate the underlying molecular mechanisms, we examined whether MD-2, an extracellular adaptor receptor of the TLR4 signalosome, can discriminate various HMGB1 isoforms with their distinct inflammatory properties. Different forms of HMGB1, produced by point mutations or chemical modifications by exposure to mercury thiolates or the reducing agent dithiothreitol, were tested for their MD-2–binding properties. Consistent with previous studies (Venereau et al., 2012; Yang et al., 2012), only the disulfide HMGB1 isoform induced TNF secretion (Fig. 1 A). Biosensor-based surface plasmon resonance (SPR) analysis (Biacore) confirmed that only the disulfide HMGB1 binds to MD-2 with high affinity (apparent Kd = 12 nM) regardless of whether MD-2 or HMGB1 was immobilized on the sensor chip (Fig. 1 B). In contrast, HMGB1 was incapable of directly binding to TLR4 (Fig. 1 B) in the absence of MD-2, although TLR4 was functionally active in MD-2 binding in Biacore analysis (not depicted), implicating MD-2 as an essential participant in the HMGB1–TLR4 signaling pathway. Unlike the disulfide isoform of HMGB1, H2S modified, fully reduced, or sulfonyl HMGB1 failed to induce TNF release from macrophage cultures (Fig. 1 A), with >1,000-fold reduction in MD-2 binding as compared with disulfide HMGB1 (Fig. 1 B). Notably, chemical modification of the cysteine 106 of the disulfide HMGB1 also abolished the TNF-stimulating and MD-2–binding properties, indicating a critical role of the thiol-cysteine 106 in the regulation of HMGB1 cytokine activity (Fig. 1, A and B).
Figure 1.
Disulfide HMGB1 binds to MD-2. (A) TNF release was measured from RAW 264.7 cells stimulated with various isoforms of HMGB1 as indicated (1 µg/ml, 16 h). Data are presented as means ± SEM. *, P < 0.05 versus disulfide HMGB1. n = 3–5 experiments. (B) SPR (Biacore) analysis was performed to assess HMGB1 binding to MD-2 or TLR4 (coated on the chip). (top) HMGB1 binding to human MD-2 was tested at different HMGB1 concentrations (12.5, 25, 50, and 100 nM) with an apparent Kd of 12 nM (left). Human MD-2 (12.5, 25, 50, and 100 nM) binding to HMGB1 (coated on the chip; middle) and disulfide HMGB1 (100 nM) binding to TLR4 (coated on the chip; right) were tested. (bottom) Noncytokine-inducing HMGB1 (C106A, Hg-HMGB1, 1 µM) was tested for binding to MD-2 (coated on the chip; left). HMGB1 isoforms were tested for binding to MD-2 (coated on the chip; right). Data are presented as response units or relative response units over time (seconds) and are representative of three experiments. (C) Mixture of CBP-tagged HMGB1 or CBP alone with supernatant of yeast Sf9 cells expressing MD-2 was immunoprecipitated with calmodulin beads (immunoprecipitation [IP]), and immunoblotted (IB) with anti–human MD-2 or CBP antibodies. Recombinant MD-2 protein was included as positive control (right lane). Data shown are representative of three repeats. (D) SPR analysis of HMGB1 binding to human MD-2 (coated on the chip) was performed in the presence of anti-HMGB1 mAb (left) or irrelevant mouse IgG (right) as shown. Data are representative of three repeats.
To further study HMGB1–MD-2 interactions, immunoprecipitation assays were used to pull down MD-2 from HMGB1-expressing cell lysates. Co-incubation with calmodulin-binding protein (CBP)–tagged disulfide HMGB1, but not CBP tag alone, pulled down MD-2 protein from yeast cells transfected with an MD-2–expressing construct (Fig. 1 C), confirming that MD-2 binds disulfide HMGB1. Furthermore, this interaction was blocked by anti-HMGB1 mAbs, but not by irrelevant IgG, demonstrating that the HMGB1–MD-2 interaction is specific and targetable by antagonists (Fig. 1 D).
MD-2 is required for HMGB1-mediated inflammatory responses
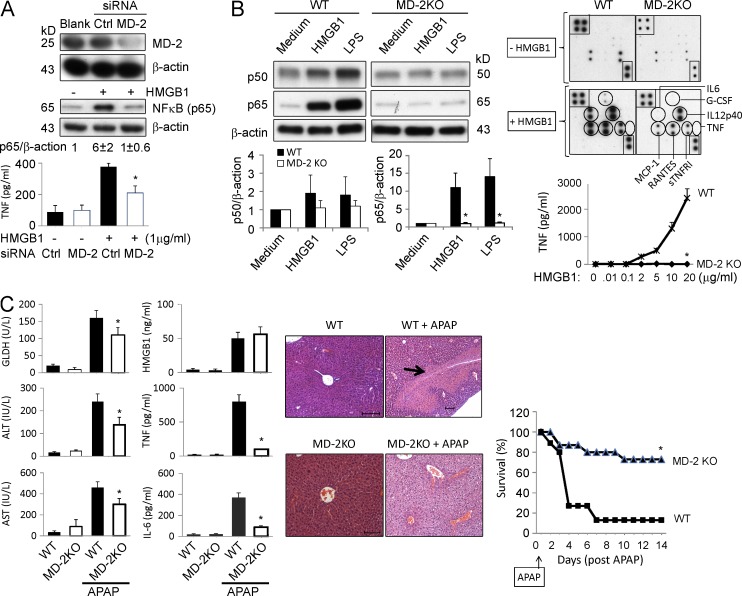
To further assess the importance of MD-2 in HMGB1-mediated cytokine induction, we used siRNA to knock down MD-2 expression in murine macrophage-like RAW 264.7 cells or human (THP-1) monocytes. The silencing of MD-2 expression (by 80–90%) was accompanied by a significant reduction of HMGB1-stimulated NF-κB activation and TNF release in both murine macrophages and human monocytes (Fig. 2 A and not depicted). To confirm the requirement for MD-2 in HMGB1-induced innate immune activation, thioglycollate-elicited peritoneal macrophages were isolated from WT and MD-2 KO mice and stimulated by disulfide HMGB1. Disruption of MD-2 expression resulted in complete impairment of both LPS- and HMGB1-induced activation of NF-κB and secretion of cytokines (TNF and IL-6) and chemokines (e.g., RANTES and MCP-1; Fig. 2 B). The release of IL-12/p40 stimulated with HMGB1 is via an MD-2–independent mechanism, likely attributable to signaling via other receptors.
Figure 2.
MD-2 is indispensable for HMGB1-dependent TLR4 signaling. (A, top) Knockdown of MD-2 (siRNA) was performed on RAW 264.7 cells. MD-2 and NF-κB levels (p65) were assessed by Western blotting. The level of NF-κB (p65) protein was normalized relative to the level of β-actin (ratio) by densitometry and expressed as the fold change over unstimulated cells. (bottom) HMGB1-induced TNF release from RAW 264.7 cells with MD-2 knockdown (open bars) or control siRNA (closed bars). *, P < 0.05 versus control siRNA group. n = 4–5 experiments. (B, left) HMGB1 (2 µg/ml) or ultrapure LPS (200 ng/ml) was used to stimulate primary peritoneal macrophages from WT or MD-2 KO mice for 16 h, and NF-κB (p50 and p65) protein levels in nuclear extracts were assessed by Western blotting (top left). NF-κB activation is expressed as of p50 or p65 relative to β-actin and calculated as the fold change over unstimulated cells (bottom left). (right) Mouse macrophages were stimulated with HMGB1 and cytokine release was measured using mouse cytokine antibody array (G-CSF, IL-12p40, IL-6, TNF, RANTES, MCP-1, and sTNFR1; top right) or ELISA (for TNF; bottom right). *, P < 0.05 versus WT group. n = 5 separate experiments. (C, left) WT or MD-2 KO mice were challenged with APAP in a liver injury model and were euthanized 24 h later to measure serum levels of liver enzymes (GLDH, ALT, and AST; left column of graphs) and cytokines (HMGB1, TNF, and IL-6; right column of graphs). *, P < 0.05 versus WT APAP group. n = 5–13 mice per group. (middle) Representative H&E staining of liver tissues from these mice are shown. n = 5–8 mice per group (the arrow indicates necrosis region). Bars, 100 µm. (right) Animal survival after receiving a lethal dose of APAP in WT and MD-2 KO mice was assessed (percent survival). *, P < 0.05 versus WT. n = 15 mice per group. (A–C) Data are presented as means ± SEM.
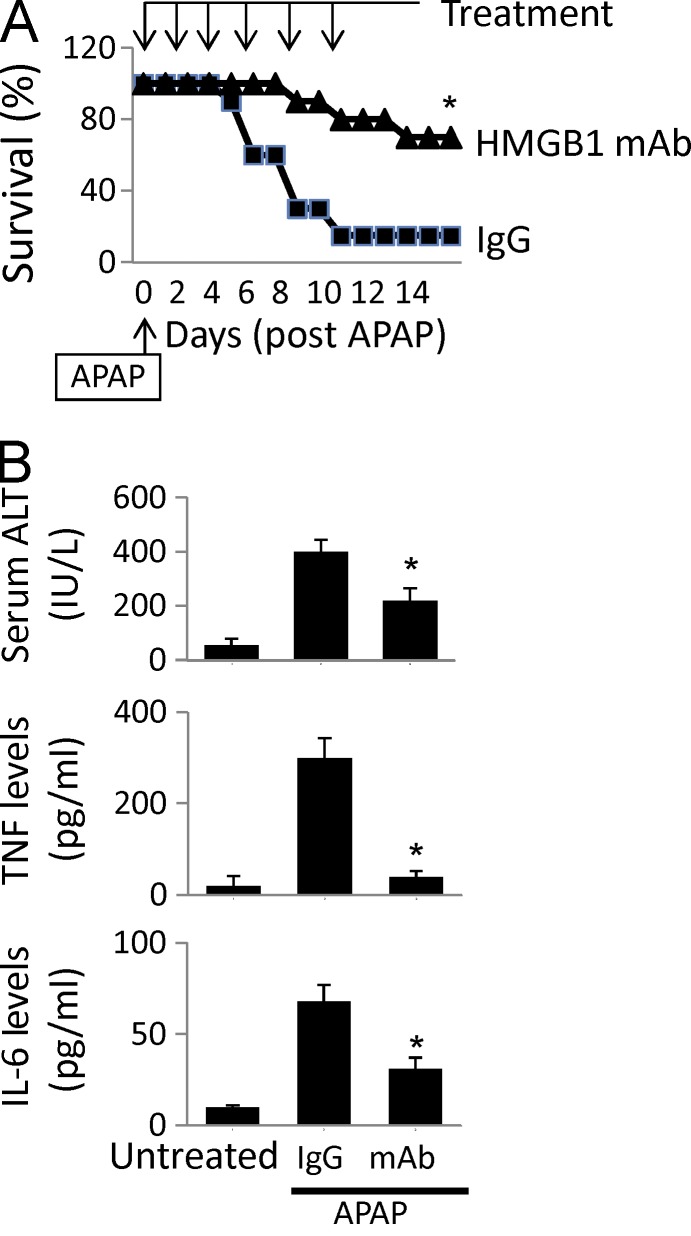
HMGB1 is an important mediator of acetaminophen (APAP)-induced hepatotoxicity (Antoine et al., 2012). To evaluate the in vivo importance of MD-2 in HMGB1-induced inflammatory responses, we studied the impact of MD-2 deficiency on sterile inflammation using the APAP intoxication model. The disruption of MD-2 expression resulted in a significant reduction in acute hepatic injury, as assessed by liver enzyme release (glutamate dehydrogenase [GLDH], aspartate aminotransferase [AST], and alanine aminotransferase [ALT]) and histological analysis of liver necrotic lesions compared with WT mice subjected to APAP injection (Fig. 2 C, arrow). Furthermore, the lessened hepatic damage in MD-2 KO mice was accompanied by significant reduction in cytokine (TNF and IL-6) release and APAP-induced animal lethality, confirming an essential role for MD-2 in sterile inflammation and injury (Fig. 2 C). Notably, serum HMGB1 levels were comparably elevated in WT and MD-2 KO mice at 24 h after APAP administration (Fig. 2 C). The central role of HMGB1 in APAP-induced liver toxicity was further confirmed by using an HMGB1-neutralizing mAb, which significantly inhibited APAP-induced release of hepatic enzymes (ALT) and proinflammatory cytokines (TNF and IL-6) and improved survival (Fig. 3). Collectively, these in vivo experimental data reveal an essential role for MD-2 and HMGB1 in the pathogenesis of sterile injury.
Figure 3.
Anti-HMGB1 mAb administration ameliorates APAP-induced liver injury in mice. (A) Mice received an APAP injection (i.p.) followed by treatment with an anti-HMGB1 antibody or control IgG injection (i.p.; see Materials and methods). Animal survival (percent survival) was assessed. n = 20 mice/group. *, P < 0.05 versus IgG group. (B) Serum levels of liver enzyme (ALT) and cytokines (TNF and IL-6) at 24 h after APAP were measured in mice receiving treatment of anti-HMGB1 antibody or control IgG (see Materials and methods). *, P < 0.05 versus IgG group. n = 10 mice/group. (A and B) Data are presented as means ± SEM.
Development of a novel MD-2–binding peptide as an HMGB1-specific inhibitor
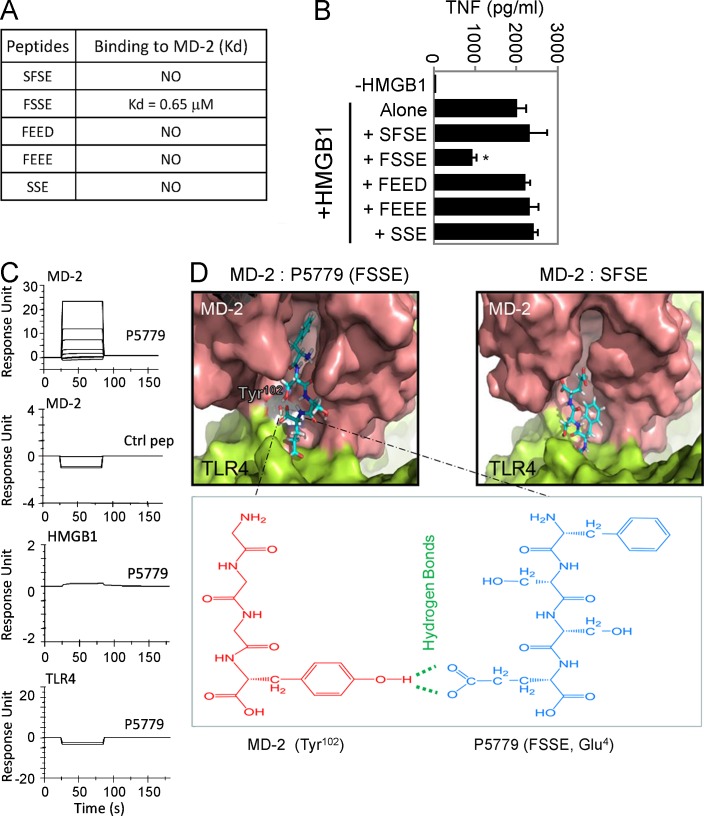
Having identified a critical involvement of the cysteine 106 region of HMGB1 in HMGB1–MD-2 interaction and HMGB1–TLR4 signaling, we used a rational strategy to screen for mimetic peptide inhibitors. A series of trimer and tetramer peptides spanning the cysteine 106 region and incorporating cysteine homologues were screened for MD-2–binding properties using the Biacore technology and molecular docking technique (Fig. 4, A, C, and D). Although most peptides lacked MD-2–binding capacity, we identified one epitope within the HMGB1 B box domain that acted as a potent HMGB1-specific inhibitor. Molecular docking simulation revealed that the FSSE (P5779) tetramer fully extended into the hydrophobic pocket of MD-2, thereby forming maximal van der Waals interaction with surrounding hydrophobic residues along with an additional hydrogen bond with the Tyr102 (Fig. 4 D). Consequently, it bound to MD-2 with a Kd value of 0.65 µM and significantly inhibited HMGB1-induced TNF release from human macrophages (Fig. 4, A and B). This interaction was specific, as P5779 failed to bind to other proteins such as HMGB1 and TLR4 in the absence of MD-2 (Fig. 4 C). Similarly, scrambling the amino acid sequence of P5779 (control peptide) abolished the MD-2–binding capacity in Biacore experiments (Fig. 4 C) and in molecular docking analysis (Fig. 4 D).
Figure 4.
Screening for HMGB1 inhibitors. (A) SPR analysis was performed to test the interaction of MD-2 (coated on the chip) with P5779 (FSSE) and other peptides (100 nM). Kd values are shown. Data are representative of three experiments. (B) Primary human macrophages were stimulated in vitro with HMGB1 (1 µg/ml) plus different peptides (50 µg/ml) for 16 h, and TNF release was measured by ELISA. Data are presented as means ± SEM. *, P < 0.05 versus HMGB1 alone. n = 4–5 experiments. (C) SPR analysis was performed to measure binding of P5779 (12.5, 25, 50, and 100 nM) or scrambled control (ctrl) peptide (100 nM) to human MD-2 (Kd = 0.65 µM for P5779), HMGB1, or TLR4 (coated on the chip). Data are representative of three experiments. (D) Schematic illustration showing molecular docking of MD-2 with tetramer peptides FSSE (left) and SFSE (right). The pink area represents the surface of the peptide-binding pocket of MD-2, and the green area denotes the TLR4 protein surface. The bottom panel shows hydrogen bonds and van der Waals interactions. P5779, with a stronger van der Waals interaction than control, is fully extended into the hydrophobic pocket of MD-2 and forms an additional hydrogen bond with Tyr102 of MD-2.
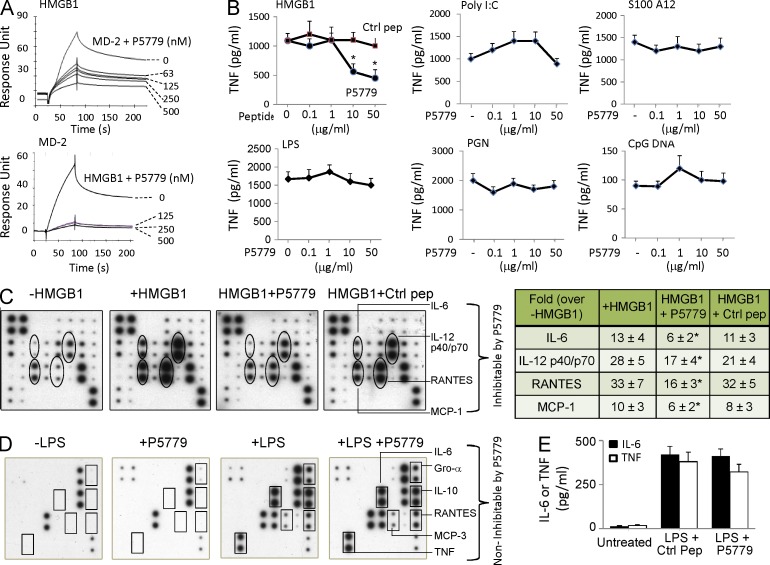
To evaluate the therapeutic potential of the MD-2–binding peptide, we next studied whether P5779 was capable of disrupting MD-2–HMGB1 interactions, thereby inhibiting HMGB1-induced cytokine production. P5779 inhibited the MD-2–HMGB1 interaction in a concentration-dependent manner when either MD-2 or HMGB1 was coated onto the Biacore sensor chip (Fig. 5 A). Furthermore, P5779 inhibited HMGB1-induced TNF release in primary human macrophages in a concentration-dependent fashion (Fig. 5 B). The effective concentration of P5779 that suppressed 50% TNF release (IC50) was ∼5 µg/ml in the presence of HMGB1 at 1 µg/ml. Scrambling the amino acid sequence of P5779 abolished the capacity to inhibit HMGB1-induced TNF release (Fig. 5 B). Exposure of macrophages to P5779 failed to inhibit TNF release mediated by polyinosinic-polycytidylic acid (Poly I:C), S100A12, LPS, peptidoglycan (PGN), and CpG DNA (Fig. 5 B). P5779 also significantly reduced HMGB1-induced release of other cytokines including IL-6 and IL-12p40/p70 and chemokines such as RANTES and MCP-1 (Fig. 5 C). P5779 did not inhibit LPS-stimulated cytokine/chemokine release in vitro in macrophages (Fig. 5 D) and failed to attenuate LPS-induced systemic cytokine levels in vivo, even when administered at high doses (8 mg/kg) in mice (Fig. 5 E). Thus, P5779 selectively attenuates HMGB1–MD-2–TLR4 signaling without inhibiting macrophage activation in response to PAMPs.
Figure 5.
Development of a tetramer peptide (P5779) as an MD-2–binding HMGB1-specific inhibitor. (A) On SPR analysis, HMGB1 was coated on the chip and MD-2 (1 µM) was flowed over as analyte, plus different amounts of P5779 as shown. Inhibition of HMGB1 binding to MD-2 by P5779 (IC50 = 29 nM) was assessed (top). In the reverse experiment, human MD-2 was coated on the chip, and HMGB1 (1 µM) plus different amounts of P5779 were added as analytes. HMGB1 binding to MD-2 was inhibited by P5779 (IC50 = 2 nM; bottom). Data are representative of three separate experiments. (B) Human primary macrophages, isolated from human blood, were stimulated with HMGB1 (1 µg/ml) or other stimuli (Poly I:C, S100A12, LPS, PGN, and CpG DNA) in vitro, plus increasing amounts of P5779 (or scrambled control peptide) for 16 h. TNF release was measured by ELISA. *, P < 0.05 versus HMGB1 plus control peptide (ctrl). n = 4–5 experiments. (C) Thioglycollate-elicited peritoneal mouse macrophages were stimulated in vitro with HMGB1 (1 µg/ml) plus P5779 or control peptide (50 µg/ml) for 16 h, and extracellular levels of various cytokines were analyzed by mouse cytokine antibody array (left). Data are representative of three to four experiments, each performed in duplicate and expressed as fold increase over unstimulated cells using densitometry (-HMGB1; right table). *, P < 0.05 versus +HMGB1 group. (D) Primary human macrophages, isolated from blood, were stimulated in vitro with LPS (2 ng/ml) for 16 h in the absence or presence of P5779 (50 µg/ml) or control peptide, and extracellular levels of various cytokines were analyzed by human cytokine antibody array. Data are representative of three repeats. (E) Male C57BL/6 mice received an LPS injection (8 mg/kg, i.p.) plus P5779 or control peptide (500 µg/mouse, i.p.). Animals were euthanized 90 min later. Serum TNF and IL-6 levels were measured by ELISAs. n = 5 mice per group. (B and E) Data are presented as means ± SEM.
Therapeutic efficacy of MD-2–targeted P5779 in APAP toxicity, ischemia, and sepsis
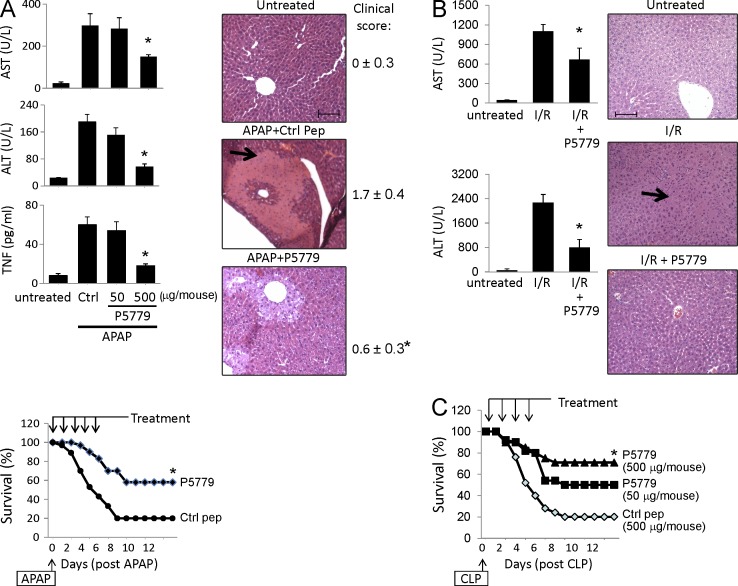
In the APAP-induced liver toxicity model, P5779 treatment dose-dependently reduced APAP-induced elevation of hepatic serum enzymes (AST and ALT), proinflammatory cytokines (TNF), liver necrosis, and improved survival (Fig. 6 A, arrow). In sterile injury mediated by hepatic I/R, P5779 also significantly blunted hepatic serum enzyme release (AST and ALT) and neutrophil infiltration (Fig. 6 B, arrow). In addition, treatment with P5779 in a sepsis model induced by cecal ligation and puncture (CLP) significantly and dose-dependently improved survival rates as compared with scrambled peptide-treated controls (Fig. 6 C). Importantly, P5779 was effective even when administered 24 h after onset of the peritonitis, consistent with the known delayed pathogenic role of HMGB1 in sepsis sequelae. Collectively, these results indicate that P5779 disrupts binding of disulfide HMGB1 to MD-2, thereby attenuating HMGB1-mediated organ failure and mortality in vivo.
Figure 6.
Treatment with the HMGB1 inhibitor P5779 ameliorates APAP-mediated toxicity, I/R injury, and sepsis mortality in vivo. (A, top left) C57BL/6 mice received an APAP injection (i.p.; see Materials and methods) and were administered with P5779 (at doses indicated) or control peptide (ctrl; 500 µg/mouse, i.p.). Mice were euthanized at 24 h after APAP, and serum enzyme (ASL and ALT) and cytokine (TNF) levels were measured by ELISAs. n = 6–10 mice per group. (bottom left) In survival experiments, mice received an APAP injection (i.p.) and were administered with P5779 or control peptide (i.p.; see Materials and methods). Survival was monitored for 2 wk (percent survival). n = 30 mice/group. (right) Representative H&E images of liver tissue sections are shown for normal (untreated) or APAP-injected mice receiving P5779 or control peptides. Clinical scores were assessed and are shown on the right. Liver necrosis is demonstrated by an arrow. n = 6–10 mice/group. *, P < 0.05 versus control peptide group. (B, left) P5779 or control peptide was administered (500 µg/mouse, i.p.) at the time of I/R surgery, and mice were euthanized 6 h later to measure serum levels of ALT and AST and to evaluate histological liver injury. *, P < 0.05 versus I/R group. n = 5–7 mice/group. (right) Representative H&E liver tissue sections are shown (neutrophil infiltration: arrow). n = 3–5 mice per group. (A and B) Data are presented as means ± SEM. Bars, 100 µm. (C) Mice received CLP surgery, and P5779 or control peptide was administered i.p. at the doses indicated. Animal survival was monitored for 2 wk (percent survival). *, P < 0.05 versus control peptide group. n = 20 mice/group.
These results reveal a novel mechanism of selective innate immune cell recognition of HMGB1 by MD-2 that discriminates the HMGB1 isoforms. By screening HMGB1 peptide libraries, we identified a novel tetramer peptide (FSSE, P5779) that specifically prevents the MD-2–HMGB1 interaction without impairing the MD-2/LPS/TLR4 signaling in innate immune cells. This peptide conferred protection not only in animal models of sterile injury-elicited inflammatory diseases but also after a lethal infection challenge, opening the possibility of developing novel therapeutic strategies to attenuate DAMP-mediated injurious inflammatory responses without inhibiting PAMP-elicited innate immunity.
MD-2 carries a β-cup fold structure composed of two antiparallel β-sheets that form a large hydrophobic pocket for binding to LPS (Park et al., 2009). The estimated affinity of MD-2 binding to HMGB1 (12 nM) is comparable with MD-2 binding to LPS (65 nM; Visintin et al., 2006). Further structural analysis is required to reveal the disulfide HMGB1-binding site on MD-2.
HMGB1-neutralizing antibodies are protective against sterile injury (Tsung et al., 2005), and agents capable of inhibiting HMGB1 release or its extracellular activities (Wang et al., 1999; Yang et al., 2004) also confer protection against sepsis. During early stages of sepsis, PAMP-mediated inflammatory responses are essential to host defense. At later stages, the release of DAMPs amplifies the cytokine storm and organ dysfunction (Wang et al., 2014). This notion is supported by recent observations that HMGB1 levels are persistently elevated during later stages of sepsis, despite termination of the initial infection, and contribute to long-term pathological consequences of sepsis (Valdés-Ferrer et al., 2013). Microbial-induced sepsis can be clinically indistinguishable from the sterile injury-elicited systemic inflammatory response syndrome (SIRS; Sursal et al., 2013). Based on the findings that TLR4–MD-2 acts as a mutually exclusive signaling receptor complex for disulfide HMGB1, it is possible to develop strategies that selectively attenuate DAMP-mediated inflammatory responses while preserving PAMP-mediated signaling.
Substantial evidence supports the necessity to preserve early PAMP-mediated innate immune responses to counteract microbial infections. For instance, defective TLR4 signaling in C3H/HeJ mice is associated with aggravated disease severity and increased mortality in animal models of infection (Khanolkar et al., 2009). LPS enhances macrophage phagocytic activity through TLR4, and selective deletion of TLR4 on myeloid cells impairs bacterial clearance in the CLP model (Deng et al., 2013). These findings emphasize the importance of generating therapeutic approaches to selectively target damage-mediated inflammation while preserving physiological protective immune responses. The discovery of P5779 as an MD-2–targeting selective inhibitor for the DAMP- but not the PAMP-elicited inflammatory responses provides such a novel therapeutic tool.
MATERIALS AND METHODS
Reagents.
Human TLR4–MD-2 complex, human MD-2, TLR2, and soluble RAGE were obtained from R&D Systems. LPS (Escherichia coli; 0111:B4), APAP, Triton X-114, PGN from Bacillus subtilis, blasticidin S, NaSH, mouse IgG, and human macrophage-CSF (M-CSF) were purchased from Sigma-Aldrich. Protein A/G agarose and isopropyl-d-thiogalactopyranoside (IPTG) were from Thermo Fisher Scientific. NHS-activated Sepharose 4 fast flow beads were obtained from GE Healthcare. Thioglycollate medium was purchased from BD. Ultrapure LPS, Poly I:C, and type B CpG oligonucleotide were obtained from InvivoGen. Human S100 A12 was from Circulex Co. Anti–human and –mouse MD-2 antibodies were obtained from Imgenex. Anti-CBP tag antibody was from GenScript. Anti-p50 antibody (E381) and anti-p65 antibody were obtained from Epitomics and Santa Cruz Biotechnology, Inc., respectively. Serum ALT and AST levels were determined by color endpoint assay kits from BIOO Scientific.
Preparations of HMGB1 proteins, antibodies, and peptides.
Recombinant HMGB1 was expressed in E. coli and purified to homogeneity as described previously (Wang et al., 1999; Li et al., 2004). This cytokine-stimulating HMGB1 contains a disulfide bond between cysteines 23 and 45 and reduced thiol on cysteine 106, characterized by liquid chromatography tandem mass spectrometric analysis (LC-MS/MS; Yang et al., 2012). HMGB1 with redox modifications was created chemically by a synthetic formation of mercury thiolate on cysteine at position 106 (Hg-HMGB1), by S-sulfhydration (H2S) to convert cysteine thiol (-SH) group to -SSH or by mutation of cysteine 106 to alanine (C106A HMGB1) as described previously (Yang et al., 2010, 2012). HMGB1 with cysteine modified by H2S was generated by incubating HMGB1 with 5 mM NaSH for 3 h at room temperature. Oxidized or DTT-reduced HMGB1 was prepared as previously described (Yang et al., 2012). The LPS content in HMGB1 was measured by the Chromogenic Limulus Amebocyte Lysate Assay (Lonza). HMGB1 was extracted with Triton X-114 to remove any contaminating LPS as described previously (Li et al., 2004). The purity and integrity of all recombinant proteins were verified by Coomassie blue staining after SDS-PAGE, with a purity predominantly >85%. The LPS content in all HMGB1 protein preparations is undetectable or <10 pg/mg protein as measured by Limulus assay. Anti-HMGB1 mAb (2g7) was generated as reported previously (Qin et al., 2006). Trimer or tetramer peptides (FSSE, FSSEY, FEEE, FEED, SSE, and SFSE) and CBP (MKRRWKKNFIAVSAANRFKKISSSGAL) were all custom-made from GeneMed Inc. The peptides were purified to 90% purity as determined by HPLC. Endotoxin was not detectable in the synthetic peptide preparations as measured by Limulus assay. The peptides were first dissolved in DMSO and further diluted in PBS as instructed by the manufacturer and prepared freshly before use. Pre-casted mini-protean Tris-Tricine gels were from Bio-Rad Laboratories.
Cell isolation and culture.
Thioglycollate-elicited peritoneal macrophages were obtained from mice (C57BL/6 or gene knockout, male, 10–12 wk old) injected with 2 ml of sterile 4% thioglycollate broth i.p. as previously described (Yang et al., 2010). Murine macrophage-like RAW 264.7 and human leukemia monocytes THP-1 were obtained from ATCC. Human primary monocytes were purified by density gradient centrifugation through Ficoll from blood donated by normal individuals as reported previously (Yang et al., 2010). Human primary macrophages in 96-well plates were stimulated with HMGB1 at 1 µg/ml, TLR4 agonist LPS at 4 ng/ml, TLR3 agonist Poly I:C at 50 µg/ml, TLR2 agonist PGN at 5 µg/ml, RAGE agonist S100A12 at 50 µg/ml, and TLR9 agonist CpG DNA at 1 µM, plus increasing amounts of P5779 (or scrambled control peptide) as indicated for 16 h. TNF release was measured by ELISA.
Immunoprecipitation assay.
Recombinant rat HMGB1 with a CBP tag or 10 µg CBP peptide alone was incubated overnight with 50 µl human MD-2 supernatant (precleared with calmodulin beads) at 4°C with gentle shaking. Human MD-2 supernatant was obtained from sf9 insect cells transfected with human MD-2 (Teghanemt et al., 2008). Both HMGB1 and MD-2 supernatant contained nondetectable amounts of LPS as measured by Limulus amebocyte lysate assay. The mixture of CBP-HMGB1 or CBP and MD-2 was then incubated with calmodulin beads (30 µl drained beads) for 1 h at 4°C. After extensive washing with PBS containing 0.1% Triton X-100, proteins bound to the beads were analyzed by Western blot probed with anti–human MD-2 or anti-CBP antibodies.
Cytokine and NF-κB measurements.
Levels of TNF and IL-6 released in the cell culture or from mice serum were measured by ELISA kits (R&D Systems). Serum HMGB1 levels were measured by ELISA kit (IBL International). Cytokine expression profile from thioglycollate-elicited peritoneal macrophages of mice or primary human macrophages was determined by mouse or human cytokine array C1 (Raybiotech) according to the manufacturer’s instructions. 22 cytokines or chemokines were determined simultaneously. NF-κB activation was analyzed by detecting p50 and p65 expression in the nuclear fraction by Western blot. β-Actin expression was also measured as control for equal loading of samples. Western blots were scanned with a silver image scanner (Silver-scanner II; Lacie Limited), and the relative band intensity was quantified using ImageJ software (v1.59; National Institutes of Health) and expressed as a ratio to the amount of β-actin.
SPR analysis.
The Biacore T200 instrument was used for real-time binding interaction experiments. For HMGB1–MD-2 binding analyses, human MD-2 was immobilized onto a CM5 series chip (GE Healthcare). One flow cell was used as a reference and thus immediately blocked upon activation by 1 M ethanolamine, pH 8.5. The sample flow cell was injected with disulfide HMGB1 (or isoforms; in 10 mM acetate buffer, pH 5.2) at a flow rate of 10 µl/min for 7 min at 25°C. Increasing concentrations of disulfide HMGB1 or isoforms of HMGB1 (C106A, sulfonyl, fully reduced, Hg, or H2S-modified HMGB1 at 1 µM) were flowed over immobilized MD-2. In reverse fashion, HMGB1 was coated on the chip and various amounts of MD-2 were added as analyte. Findings were confirmed by using two additional human MD-2 proteins from D.T. Golenbock and T.R. Billiar. For TLR4-HMGB1 binding experiments, human TLR4 was coated on the chip, and 100 nM disulfide HMGB1 was added as analyte. For peptide screening experiments, human MD-2 was coated on the sensor chip, trimer, or 100 nM tetramer peptides (FSSE, FSSEY, FEEE, FEED, SSE, and SFSE) were added as analytes. The dissociation time was set for 2 min, followed by a 1-min regeneration using a 10 mM NaOH solution. The Kd was evaluated using the Biacore evaluation software. For experiments using HMGB1 antibody to block MD-2–HMGB1 interaction, human MD-2 was coated on the chip, HMGB1 was added as analyte (100 nM) plus increasing amounts of HMGB1 mAb or control IgG, and response units were recorded.
Molecular docking of MD-2 with peptides.
The crystal structure of the MD-2–TLR4 was obtained from the Protein Data Bank (PDB code: 3VQ2), and molecular docking was performed by using the MOE software as previously described (Zan et al., 2012). A molecular visualization system, the PyMOL 0.99, was used to construct the three-dimensional figures.
Knockdown MD-2 in RAW 264.7 and THP-1 cells using siRNA.
For MD-2 knockdown in RAW 264.7 cells, cells were transfected with mouse MD-2 or 50 nM control siRNA (on-target plus smart pool; GE Healthcare) using DharmaFect1 transfection reagent. To knock down MD-2 in THP-1 cells, transfection with MD-2–specific siRNA was performed by using an Amaxa Nucleofector kit (Lonza). The efficiency of knockdown was confirmed by Western blot probed with anti–MD-2 antibody at 48 h after transfection. At 48 h after transfection, cells were stimulated with 1 µg/ml HMGB1 for 16 h. Cell lysate and supernatant were collected and analyzed by Western blot or ELISA. NF-κB measurements on RAW 264.7, THP-1, or primary mouse macrophages from MD-2 KO mice were performed using NE-PER Protein Extraction kit (Thermo Fisher Scientific).
Animals.
Male C57BL/6 mice were obtained from the Jackson Laboratory. MD-2 KO (on C57BL/6 background) mice were purchased from the RIKEN Bio-Resource Center. All animals were maintained at The Feinstein Institute for Medical Research or University of Pittsburgh under standard temperature and light cycles, and all animal procedures were approved by the institutional animal care and use committee.
For genotyping of MD-2 KO mice from tail snips, PCR primers were designed by the RIKEN Bio-Resource Center and were obtained from Invitrogen. The same primers were used to identify WT (PCR product = 2,000 bp) and MD-2 KO (PCR product = 800 bp) in genotyping.
For murine hepatic warm I/R, a 70% warm liver I/R model was performed as previously described (Tsung et al., 2005). Mice received i.p. injections of P5779 (500 µg/mouse) or vehicle at the time of surgery and were euthanized at 6 h afterward. Whole blood was collected by cardiac puncture, and liver was harvested and fixed in 10% formalin for analysis.
For CLP, C57BL/6 mice (male, 8–12 wk of age) were subjected to CLP procedure as described previously (Yang et al., 2004). P5779 or scrambled control peptide was administered i.p. at 50 or 500 µg/mouse, and treatment was given once a day for 4 d starting at 24 h after CLP surgery. Survival was monitored for 2 wk.
For the APAP hepatic toxicity model, three sets of experiments were conducted. In all experiments, mice were routinely fasted overnight and received i.p. injection of APAP (350 mg/kg for survival experiments and 400 mg/kg for serum measurements when mice were euthanized at 24 h after APAP) as previously described (Antoine et al., 2012; Yang et al., 2012). The first set of experiments was performed using male MD-2 KO or C57BL/6 mice (8–12 wk old). Mice had APAP injection and were euthanized 24 h later (for serum measurements) or monitored for 2 wk (for survival experiments). The second set of experiments was performed using anti-HMGB1 antibody in the APAP model in WT male (C57BL/6) mice. In survival experiments, mice had APAP injection and received anti-HMGB1 antibodies (5 µg/mouse, i.p. once daily for 4 d followed by two additional doses once every other day beginning at 2 h after APAP). Irrelevant nonimmune IgG was used as a control. For serum measurements, mice were subjected to APAP injection and received injection of anti-HMGB1 mAb (5 µg/mouse, injected i.p. at 2 and 7 h after APAP) and euthanized at 24 h after APAP. The third set of experiments was used to assess the efficacy of P5779 in the APAP model in WT mice. Male C57BL/6 mice received APAP injection plus P5779 (at 50 or 500 µg/mouse) or scrambled control peptide (500 µg/mouse, i.p. injected at 2 and 7 h after APAP) and euthanized at 24 h after APAP. In survival experiments, mice had injection of APAP and received treatment of P5779 or control peptide (500 µg/mouse, i.p. once a day for 5 d starting at 2 h after APAP), and survival was monitored for 2 wk. Hepatotoxicity was determined by serum levels of GLDH, ALT, and AST as described previously (Antoine et al., 2013).
Histological evaluation.
Harvested livers were fixed in 10% formalin and embedded in paraffin. 5-µm sections were stained with hematoxylin and eosin (H&E). H&E staining of livers was performed by AML Laboratory. The liver histology was evaluated in a blinded fashion, and clinical scores were calculated based on the amount of necrosis and inflammation (cell swelling, loss of tissue structure, and congestion) using a previously reported method with modifications (Desmet, 2003). Score 0 = no evidence of necrosis or inflammation as assessed from three to four representative sections from each animal; 1 = mild necrosis or inflammation (<25% of the total area examined); 2 = notable necrosis and inflammation (25–50% of the total area); 3 = severe necrosis and inflammation (>50% area).
Statistical analysis.
Data are presented as means ± SEM unless otherwise stated. Differences between treatment groups were determined by Student’s t test and one-way ANOVA followed by the least significant difference test. Differences between groups in animal survival experiments were determined using two-tailed Fisher’s exact test. Cytokine array experiments were analyzed using the software UN-Scan-it from Silk Scientific Inc. P-values <0.05 were considered statistically significant.
Acknowledgments
We would like to thank Patricia Loughran (University of Pittsburgh, Pittsburgh, PA) for liver histology and acknowledge The Radiation Medicine Institute of Chinese Academy of Medical Sciences and Peking Union Medical College (Tianjin, China) for allowing us to use the molecular docking simulation software.
This work was supported by grants from The Feinstein Institute for Medical Research, General Clinical Research Center (M01RR018535 to K.J. Tracey) and from the National Institutes of Health (RO1GM62508 to K.J. Tracey, RO1GM098446 to H. Yang, 5P50GM053789 to T.R. Billiar, RO1GM107876 to C. Szabo, and R01AT005076 to H. Wang).
The authors declare no competing financial interests.
Author contributions: H. Yang, Z. Ju, P. Lundbäck, J.P. Pribis, S.I. Valdes-Ferrer, and D.J. Antoine performed the experiments. A.A. Ragab, M. He, and Y. Al-Abed conducted SPR analysis. J. Li, J. Meng, and D.T. Golenbock provided HMGB1 or MD-2 reagents. W. Long performed the molecular docking experiments. H. Yang, Y. Al-Abed, B. Lu, S.S. Chavan, D. Gero, C. Szabo, S.I. Valdes-Ferrer, H.E. Harris, T.R. Billiar, U. Andersson, K.J. Tracey, and H. Wang analyzed data. H. Yang, T.R. Billiar, Y. Al-Abed, J. Roth, H. Wang, U. Andersson, and K.J. Tracey wrote the paper.
Footnotes
Abbreviations used:
- ALT
- alanine aminotransferase
- APAP
- acetaminophen
- AST
- aspartate aminotransferase
- CBP
- calmodulin-binding protein
- CLP
- cecal ligation and puncture
- DAMP
- damage-associated molecular pattern
- GLDH
- glutamate dehydrogenase
- I/R
- ischemia/reperfusion
- PAMP
- pathogen-associated molecular pattern
- PGN
- peptidoglycan
- Poly I:C
- polyinosinic-polycytidylic acid
- SPR
- surface plasmon resonance
References
- Andersson U., and Tracey K.J.. 2011. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu. Rev. Immunol. 29:139–162 10.1146/annurev-immunol-030409-101323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine D.J., Jenkins R.E., Dear J.W., Williams D.P., McGill M.R., Sharpe M.R., Craig D.G., Simpson K.J., Jaeschke H., and Park B.K.. 2012. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J. Hepatol. 56:1070–1079 10.1016/j.jhep.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Antoine D.J., Dear J.W., Lewis P.S., Platt V., Coyle J., Masson M., Thanacoody R.H., Gray A.J., Webb D.J., Moggs J.G., et al. 2013. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 58:777–787 10.1002/hep.26294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.Y., Tang J., Zheng P., and Liu Y.. 2009. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 323:1722–1725 10.1126/science.1168988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Scott M.J., Loughran P., Gibson G., Sodhi C., Watkins S., Hackam D., and Billiar T.R.. 2013. Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis. J. Immunol. 190:5152–5160 10.4049/jimmunol.1300496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet V.J.2003. Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, Kiernan TW, Wollman J. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis [Hepatology 1981;1:431-435]. J. Hepatol. 38:382–386 10.1016/S0168-8278(03)00005-9 [DOI] [PubMed] [Google Scholar]
- Huttunen H.J., Fages C., and Rauvala H.. 1999. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-κB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 274:19919–19924 10.1074/jbc.274.28.19919 [DOI] [PubMed] [Google Scholar]
- Khanolkar A., Hartwig S.M., Haag B.A., Meyerholz D.K., Harty J.T., and Varga S.M.. 2009. Toll-like receptor 4 deficiency increases disease and mortality after mouse hepatitis virus type 1 infection of susceptible C3H mice. J. Virol. 83:8946–8956 10.1128/JVI.01857-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang H., Mason J.M., Levine J., Yu M., Ulloa L., Czura C.J., Tracey K.J., and Yang H.. 2004. Recombinant HMGB1 with cytokine-stimulating activity. J. Immunol. Methods. 289:211–223 10.1016/j.jim.2004.04.019 [DOI] [PubMed] [Google Scholar]
- Meng J., Lien E., and Golenbock D.T.. 2010. MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J. Biol. Chem. 285:8695–8702 10.1074/jbc.M109.075127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Akashi S., Nagafuku M., Ogata M., Iwakura Y., Akira S., Kitamura T., Kosugi A., Kimoto M., and Miyake K.. 2002. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 3:667–672. [DOI] [PubMed] [Google Scholar]
- Park B.S., Song D.H., Kim H.M., Choi B.S., Lee H., and Lee J.O.. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 458:1191–1195 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- Qin S., Wang H., Yuan R., Li H., Ochani M., Ochani K., Rosas-Ballina M., Czura C.J., Huston J.M., Miller E., et al. 2006. Role of HMGB1 in apoptosis-mediated sepsis lethality. J. Exp. Med. 203:1637–1642 10.1084/jem.20052203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T., and Bianchi M.E.. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 418:191–195 10.1038/nature00858 [DOI] [PubMed] [Google Scholar]
- Sursal T., Stearns-Kurosawa D.J., Itagaki K., Oh S.Y., Sun S., Kurosawa S., and Hauser C.J.. 2013. Plasma bacterial and mitochondrial DNA distinguish bacterial sepsis from sterile systemic inflammatory response syndrome and quantify inflammatory tissue injury in nonhuman primates. Shock. 39:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Billiar T.R., and Lotze M.T.. 2012. A Janus tale of two active high mobility group box 1 (HMGB1) redox states. Mol. Med. 18:1360–1362 10.2119/molmed.2012.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teghanemt A., Widstrom R.L., Gioannini T.L., and Weiss J.P.. 2008. Isolation of monomeric and dimeric secreted MD-2. Endotoxin·sCD14 and Toll-like receptor 4 ectodomain selectively react with the monomeric form of secreted MD-2. J. Biol. Chem. 283:21881–21889 10.1074/jbc.M800672200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsung A., Sahai R., Tanaka H., Nakao A., Fink M.P., Lotze M.T., Yang H., Li J., Tracey K.J., Geller D.A., and Billiar T.R.. 2005. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J. Exp. Med. 201:1135–1143 10.1084/jem.20042614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Ferrer S.I., Rosas-Ballina M., Olofsson P.S., Lu B., Dancho M.E., Li J., Yang H., Pavlov V.A., Chavan S.S., and Tracey K.J.. 2013. High-mobility group box 1 mediates persistent splenocyte priming in sepsis survivors: evidence from a murine model. Shock. 40:492–495 10.1097/SHK.0000000000000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venereau E., Casalgrandi M., Schiraldi M., Antoine D.J., Cattaneo A., De Marchis F., Liu J., Antonelli A., Preti A., Raeli L., et al. 2012. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 209:1519–1528 10.1084/jem.20120189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin A., Iliev D.B., Monks B.G., Halmen K.A., and Golenbock D.T.. 2006. MD-2. Immunobiology. 211:437–447 10.1016/j.imbio.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Wang H., Bloom O., Zhang M., Vishnubhakat J.M., Ombrellino M., Che J., Frazier A., Yang H., Ivanova S., Borovikova L., et al. 1999. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 285:248–251 10.1126/science.285.5425.248 [DOI] [PubMed] [Google Scholar]
- Wang H., Ward M.F., and Sama A.E.. 2014. Targeting HMGB1 in the treatment of sepsis. Expert Opin. Ther. Targets. 18:257–268 10.1517/14728222.2014.863876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ochani M., Li J., Qiang X., Tanovic M., Harris H.E., Susarla S.M., Ulloa L., Wang H., DiRaimo R., et al. 2004. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc. Natl. Acad. Sci. USA. 101:296–301 10.1073/pnas.2434651100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Hreggvidsdottir H.S., Palmblad K., Wang H., Ochani M., Li J., Lu B., Chavan S., Rosas-Ballina M., Al-Abed Y., et al. 2010. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc. Natl. Acad. Sci. USA. 107:11942–11947 10.1073/pnas.1003893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lundbäck P., Ottosson L., Erlandsson-Harris H., Venereau E., Bianchi M.E., Al-Abed Y., Andersson U., Tracey K.J., and Antoine D.J.. 2012. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 18:250–259 10.2119/molmed.2011.00389 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang H., Antoine D.J., Andersson U., and Tracey K.J.. 2013. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Biol. 93:865–873 10.1189/jlb.1212662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J., He X., Long W., and Liu P.. 2012. Insights into binding modes of tumstatin peptide T7 with the active site of αvβ3 integrin. Mol. Simul. 38:498–508 10.1080/08927022.2011.649428 [DOI] [Google Scholar]
- Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., and Hauser C.J.. 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 464:104–107 10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]