Abstract
An alternative to qPCR was developed for nucleic acid assays, involving signal rather than target amplification. The new technology, Restriction Cascade Exponential Amplification (RCEA), relies on specific cleavage of probe-target hybrids by restriction endonucleases (REase). Two mutant REases for amplification (Ramp), S17C BamHI and K249C EcoRI, were conjugated to oligonucleotides, and immobilized on a solid surface. The signal generation was based on: (i) hybridization of a target DNA to a Ramp-oligonucleotide probe conjugate, followed by (ii) specific cleavage of the probe-target hybrid using a non-immobilized recognition REase. The amount of Ramp released into solution upon cleavage was proportionate to the DNA target amount. Signal amplification was achieved through catalysis, by the free Ramp, of a restriction cascade containing additional oligonucleotide-conjugated Ramp and horseradish peroxidase (HRP). Colorimetric quantification of free HRP indicated that the RCEA achieved a detection limit of 10 aM (10−17 M) target concentration, or approximately 200 molecules, comparable to the sensitivity of qPCR-based assays. The RCEA assay had high specificity, it was insensitive to non-specific binding, and detected target sequences in the presence of foreign DNA. RCEA is an inexpensive isothermal assay that allows coupling of the restriction cascade signal amplification with any DNA target of interest.
DNA detection techniques typically involve specific hybridization between a DNA target of interest from a test sample with a complementary single-stranded (ssDNA) oligonucleotide probe. In most cases, the test sample represents a mixture of DNA sequences that can produce high false positive detection rates due to non-specific binding and cross-hybridization. In addition, DNA hybridization assays generally have low sensitivity, and typically only work well for relatively high, pico- to nanomolar target concentrations1. Currently, the most widespread approach taken to improve the detection limit is the use of PCR to amplify the DNA target to a detectable amount, greatly enriching the target to background DNA ratio (for reviews see2,3,4). Various quantitative PCR (qPCR) techniques can detect fewer than a hundred (and in some cases as little as5,6,7,8,9,10) copies of the target DNA in a given sample5. However, qPCR techniques require thermocycling and the use of high cost, complex instrumentation. Furthermore, non-specific primer binding3,5, and a high susceptibility to contamination4 are frequently problematic in PCR. Several other DNA amplification techniques have been developed to overcome PCR limitations and to amplify DNA using isothermal conditions, including transcription mediated amplification, strand displacement amplification, rolling circle amplification, etc.6,7,8. One of these techniques, loop-mediated isothermal amplification (LAMP), involves four custom-designed primers recognizing six distinct regions of the target DNA, and is based on autocycling strand displacing DNA synthesis performed by Bst DNA polymerase9,10. This setup ensures very high amplification specificity of LAMP9, and the sensitivity observed with some LAMP applications is similar to, or even higher, than that of qPCR10.
The main problem with DNA amplification techniques is a possible sequence-dependent bias, when certain sequences have much higher amplification efficiency than the other11. Sequence-dependent limitations can also complicate design and validation of the multiple primers necessary for DNA amplifications schemes, and for qPCR multiplexing. The desire to bypass target amplification has led to the development of alternative methods that are instead based on amplification of the signal generated by target-probe hybridization5,11. This approach relies on cleavage of the probe in a target-probe hybrid, where formation of the hybrid itself activates the cleavage reaction. In this way target integrity is preserved and the target is recycled, essentially serving as a catalyst for additional probe cleavage events (for a review see Ref. 7). A single target molecule has been shown to generate up to 100 cleavage events12. Probe cleavage can be achieved in several ways, for example it can result from formation of a ternary complex between two junction probes and a target11, or from activation of a DNAzyme self-cleaving domain contained within a single probe12,13. DNAzymes are catalytically active DNA molecules that are frequently used as reporters for target analyte detection based on nucleic acid hybridization13. Additionally, DNA-modifying enzymes, including endo- and exonucleases, and polymerases can also be used for target re-cycling and probe cleavage reactions7,14. Detection limits observed with these approaches may reach sub-picomolar range, i.e., 0.1 pM target detection was observed for the G-quadruplex DNAzyme approach7,15.
Further efforts have indicated that greater assay sensitivity can be achieved by amplification of detected signals using catalytic labels7. To develop isothermal ultrasensitive colorimetric detection systems, DNAzymes have been coupled with restriction endonucleases (i.e. FokI16) and nicking enzymes (Nt.BstNBI17) (for a review see Ref. 7). Coupling of polymerase with nicking enzyme reactions to generate DNAzymes resulted in a detection limit of 10 fM17. These coupled non-enzymatic and enzymatic cascades have advantages over PCR, since they are performed under isothermal conditions and do not involve complex instrumentation7. Despite utilizing rather complex setups, many of these assays can be engineered in portable formats for field tests or point-of-care applications (for a review see Ref. 7). However, most of these assays thus far have only been tested using pure samples composed of a target of interest in a simple buffer solution7,11. Thus, it is unclear whether this approach is valid for complex nucleic acid mixtures in which the target of interest is a relatively minor component, and/or biological samples containing a high background of proteins, such as human body tissues and fluids7,11.
Our previous work showed that the use of Type II restriction endonucleases (REases) in a DNA hybridization assay may prove effective with complex samples18. Our direct restriction assay (DRA) consists of two main steps: (i) hybridization of a target DNA to immobilized complementary oligonucleotide probes carrying the molecular marker horseradish peroxidase (HRP), and (ii) restriction cleavage with a target-specific REase, which releases the HRP marker into solution. The released HRP is then quantified colorimetrically18. The assay involves two independent bio-recognition steps, resulting in a greatly reduced incidence of false positives, and it is insensitive to addition of large excesses of heterologous genomic DNA18. The main caveat for use of the DRA is its relatively high limit of detection (approximately 1 nM)18.
The work described in this report was focused on development and testing of a novel ultrasensitive technique for signal amplification that could replace DRA. The ultimate goals for assay development were: (i) a detection limit in the attomolar range, comparable to qPCR; (ii) high specificity for use in complex mixtures containing both target and foreign DNA; and (iii) simplicity, flexibility, and low cost. The result was development of the ‘Restriction Cascade Exponential Amplification’ (RCEA) assay, based on a cascade of enzymatic reactions catalyzed by REases19,20. The initial step involves hybridization of a DNA target with a specific oligonucleotide probe carrying an immobilized enzymatic marker, a REase for amplification (Ramp) (Fig. 1). Ramps are genetically engineered for attachment to oligonucleotide probes, and Ramp-probe conjugates are immobilized on a solid surface (Fig. 1a). After the DNA target hybridization, the probe-target hybrids are specifically cleaved by a free recognition REase (Rrec). As a result, immobilized Ramp is released into the reaction solution, thus generating the enzymatic ‘signal’ (Fig. 1b). This signal is amplified in a special chamber that contains an excess of immobilized Ramp attached to a solid surface through a double-stranded (dsDNA) linker. The linker sequence contains the corresponding Ramp recognition site, however, the immobilized Ramp is incapable of cleaving their own or neighboring linkers. Linker cleavage is initiated by the addition of reaction solution containing the released Ramp signal. Every cleavage step doubles the amount of free Ramp in the reaction solution, resulting in an exponential cascade of restriction events (Fig. 1c). In addition to Ramp, each linker is also conjugated to HRP, which, upon release into the reaction solution with cleavage is used for colorimetric quantification (Fig. 1d). This RCEA assay, designed using mutant BamHI and EcoRI REases, achieved a limit of detection of 10 aM target concentration, making its sensitivity comparable to that of qPCR. This report describes the RCEA scheme, experimental setup, and assay performance in detail.
Figure 1. General schematic of the restriction cascade exponential amplification (RCEA) assay.
(a) An oligonucleotide probe specific for a target of interest is conjugated to a REase for amplification (Ramp), and attached to a solid substrate using biotin. A test sample containing the target of interest is added. (b) The target in the test sample hybridizes to the probe, and the hybrid is specifically cleaved by a recognition REase (Rrec) that is present in solution. Ramp is subsequently released into the solution. (c) The reaction solution is transferred to the amplification chamber, which contains an excess of immobilized Ramp conjugate. The Ramp conjugate is attached to a solid surface through an oligonucleotide linker that contains the Ramp recognition sequence. The linker is double-stranded, and the complement to the Ramp conjugate strand is conjugated with horseradish peroxidase (HRP). All Ramp molecules in the amplification reaction are immobilized and thus incapable of cleaving their own or neighboring linkers. Addition of the free Ramp generated in (b) activates linker cleavage, releasing additional Ramp, which in turn cleaves new linkers. Each step of this exponential cascade of cleavage reactions doubles the amount of free Ramp in the reaction solution. (d) The linker cleavage releases HRP, which is quantified colorimetrically. (e) Each initial target-probe hybridization event produces an exponentially amplified number of HRP molecules, with the value dependent on the assay time.
Results
Generation of genetically engineered REases for conjugation
Most REases contain cysteines that can be used for ligand conjugation through -SH groups. However, these residues may be buried inside the 3D enzyme structure, and their modifications may destroy the catalytic activity. Previous studies of the REase EcoRI showed that modifying the enzyme typically led to total inactivation (for a review see Ref. 21). Our initial attempts to generate enzymatically active conjugates using commercially available WT REases resulted in failure (data not shown). Thus, enzymes had to be engineered for ligand attachment by identifying some surface “non-essential” amino acid residues that can be mutated into a cysteine21. The genetically engineered K249C EcoRI mutant has been previously produced for biotinylation, and shown to retain enzymatic activity and specificity of binding to its target sequence GAATTC21. Another mutant, S17C BamHI, containing a single cysteine residue for ligand attachment was generated in this work. First, three cysteines present in the WT protein were mutated into serines (C34S/C54S/C64S), and then the surface-exposed non-essential Ser17 was mutated into a cysteine. The resultant quadruple S17C mutant was compared to the WT BamHI enzyme that was expressed and purified under the same conditions. The mutant retained the specific activity, however, it was approximately 6.2% of the WT (4.8 × 104 versus 7.6 × 105 Units/mg protein for the S17C and WT, respectively). A second BamHI mutant T41C in the same C34S/C54S/C64S background was also constructed. However, it was not used for the assay development, since it showed lower specific activity than the S17C enzyme (data not shown).
Mutant REase conjugation to oligonucleotides, conjugate immobilization and testing
Both mutant REases, S17C BamHI and K249C EcoRI, were conjugated by chemical binding of the -SH groups of the engineered surface cysteines with maleimide groups introduced to the 3′ end of the oligonucleotide probes and linkers. The conjugate testing and RCEA assay development were done using a probe-target oligonucleotide pair, MCA-BG-Bio and AMC-BG-40 (Table 1), designed and validated in our previous work with the mecA gene of MRSA18. The 40-mer target-probe hybrid contained a BglII restriction sequence (AGATCT) in the center.
Table 1. The oligonucleotide probe, targets and linkers used in the current study Capital letters show sequences that are cognate between the probe and a target oligonucleotide, with the restriction site shown in bold. 5′ and 3′ mod indicate the modifications of oligonucleotide ends for surface attachment and enzyme conjugation.
| Usage | Oligo name | Oligo sequence | 5′ mod | 3′ mod | Conjugate | REase | Length |
|---|---|---|---|---|---|---|---|
| Probe, recognition, testing | MCA-BG-Bio | CAATTAAGTTTGCATAAGATCTATAAATATCTTCTTTATG | Biotin | Amino | BamHI, EcoRI | BglII | 40 |
| Target, recognition, testing | AMC-BG-40 | CATAAAGAAGATATTTATAGATCTTATGCAAACTTAATTG | no | no | no | BglII | 40 |
| HRP linker, conjugate testing | L1-RB-Bio-for-HRP | CATTGCTAGTTGTTTGGATCCCGGTAGTGCGAAATG | Biotin | Thiol | HRP | BamHI | 36 |
| HRP linker, conjugate testing | L2-RE-Bio-for-HRP | GCTTTCTTAAACAAAGGAATTCAAATCACATTAAGA | Biotin | Thiol | HRP | EcoRI | 36 |
| 2nd strand, conjugate testing | ASL1-RB | CATTTCGCACTACCGGGATCCAAACAACTAGCAATG | no | no | no | BamHI | 36 |
| 2nd strand, conjugate testing | ASL2-RE | TCTTAATGTGATTTGAATTCCTTTGTTTAAGAAAGC | no | no | no | EcoRI | 36 |
| Mutant target | AMC-12/40 | atgcctactacgtacTATAGATCTTATaacggacctccat | no | no | no | BglII | 40 |
| Ramp linker, amplification | L1-RB-Bio-for-Ramp | CATTGCTAGTTGTTTGGATCCCGGTAGTGCGAAATG | Biotin | Amino | BamHI | BamHI | 36 |
| Ramp linker, amplification | L2-RE-Bio-for-Ramp | GCTTTCTTAAACAAAGGAATTCAAATCACATTAAGA | Biotin | Amino | EcoRI | EcoRI | 36 |
| HRP linker, amplification | ASL1-RB-for-HRP | CATTTCGCACTACCGGGATCCAAACAACTAGCAATG | Thiol | no | HRP | BamHI | 36 |
| HRP linker, amplification | ASL2-RE-for-HRP | TCTTAATGTGATTTGAATTCCTTTGTTTAAGAAAGC | Thiol | no | HRP | EcoRI | 36 |
The first step of conjugate testing was to ensure that the oligonucleotides were indeed attached to the enzymes, and that the immobilized REases stayed on the surface under the applied assay conditions (Fig. 2). The mutant REase conjugates with the oligonucleotide probes were immobilized onto streptavidin-agarose beads through biotins of the probes. The extensive washing was applied to remove all non-conjugated enzymes. Then positive controls containing the target AMC-BG-40 at 1 μM concentration, or negative controls containing no target were added together with the free BglII REase in the restriction buffer (Fig. 2a, b). After 1 hour incubation, the reaction solutions were removed from the beads and applied to the corresponding HRP reporter systems to quantify enzymatic activity of the released mutant REases (Fig. 2c, d).
Figure 2. General schematic of the assay used for REase conjugate testing.
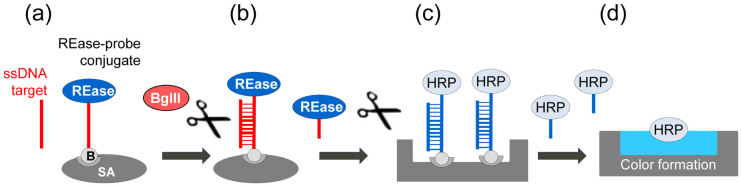
(a) An oligonucleotide probe specific for a target of interest is conjugated to a mutant REase (either the S17C BamHI or K249C EcoRI), and attached to a solid surface using biotin. An oligonucleotide target complementary to the probe is added, and it hybridizes to the probe. (b) The hybrid is specifically cleaved by a free REase BglII that is added to the reaction solution. The mutant REase is subsequently released into the solution. (c) The reaction solution is transferred to a chamber containing an HRP conjugate immobilized through a dsDNA linker containing the recognition sequence of the mutant REase. The addition of the released REase generated in (b) catalyzes the linker cleavage and HRP release. (d) The released HRP is quantified colorimetrically.
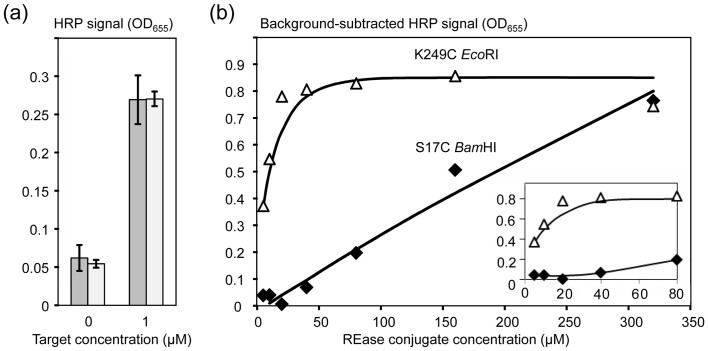
As shown in Figure 3a, for both S17C BamHI (dark grey bars) and K249C EcoRI (light grey bars), the HRP signal generated with the target exceeded the mean negative control approximately 5-fold. Apparently, both conjugates were able to (i) bind to the agarose beads through their oligonucleotide parts, and (ii) stay enzymatically active.
Figure 3. Testing of the REase-oligonucleotide conjugates, S17C BamHI-MCA-BG-Bio and K249C EcoRI-MCA-BG-Bio.
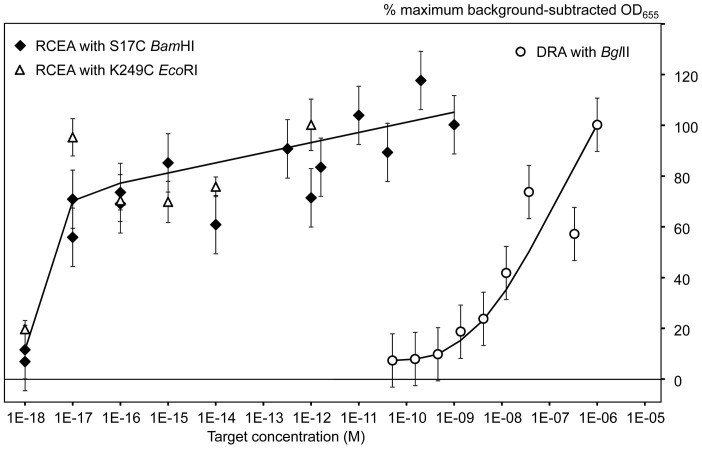
(a) The REase conjugates were tested for the presence of oligonucleotide parts and immobilization as described by the schematic in Figure 2. The X-axis shows the target AMC-BG-40 concentration (the negative controls had no target added); and the Y-axis shows the HRP reporter signals quantified colorimetrically as OD655. Dark and light grey bars show the data obtained for S17C BamHI and K249C EcoRI, respectively. The tests were performed in triplicate for calculations of the mean values and standard deviations (error bars). (b) Titration of free REase conjugates using the HRP reporter systems. The X-axis shows the conjugate concentrations. Negative controls were prepared by addition of the restriction buffer without conjugates and used for background subtraction. The Y-axis shows the background-subtracted HRP signals (OD655) generated using the two mutant enzyme conjugates, S17C BamHI and K249C EcoRI. The inset shows the data corresponding to the REase conjugate concentrations between 0 and 80 μM.
Interestingly, the negative controls showed very variable, albeit always relatively low HRP signals indicating some release of immobilized REases. Since no enzymatic activity on ssDNA targets has been reported for BglII (see the REBASE entry at http://rebase.neb.com/cgi-bin/hybcombolist?BglII), there was apparently some non-enzymatic release of the REase from ssDNA probes. This could be caused by carrier (agarose bead) degradation, and/or mechanical breakage of the probes. To improve the assay performance we separated the two processes, by first performing the target hybridization to the probes for 1 h, and then adding the free BglII for a short 20-min cleavage reaction. Such assay setup was used with the highly enzymatically active K249C EcoRI REase, and it resulted in the reduced background values.
The second step of conjugate testing was performed for the direct comparison of enzymatic activity of S17C BamHI and K249C EcoRI conjugates, and for conjugate titration. Conjugate dilution series were prepared and applied for the HRP-based enzymatic activity tests directly, without prior surface immobilization. Negative controls were prepared with no conjugate addition, and used for background subtraction. The resultant titration curves are shown in Figure 3b. The signal saturation was observed at 50 and 320 μM concentrations for the S17C BamHI and K249C EcoRI conjugates, respectively (Fig. 3b). As expected, the K249C EcoRI conjugate was at least 6.4X more active than the S17C BamHI conjugate. Thus, when designing the assays with the EcoRI mutant, we reduced the conjugate amounts 10 to 100X in comparison with the BamHI system to achieve similar enzymatic activities.
RCEA assay design
The general scheme of the RCEA assay is shown in Figure 1. For the first recognition step, the oligonucleotide probe MCA-BG-Bio was conjugated to Ramp, either S17C BamHI or K249C EcoRI. The recognition chamber contained the Ramp-probe conjugate attached to agarose beads through its oligonucleotide part. Next, a sample containing the target AMC-BG-40 was hybridized to the immobilized probes (Fig. 1a). The resultant dsDNA probe-target hybrids contained the specific recognition site of the Rrec BglII (AGATCT). Addition of this enzyme resulted in cleavage and release of Ramp into the reaction solution at a rate of approximately one free Ramp per target DNA molecule (Fig. 1b). The RCEA recognition stage was designed to use the optimum REase amount and reaction time to achieve complete digestion of the target-probe hybrids (as recommended by the enzyme manufacturer).
The reaction solution from the recognition step was next transferred to a chamber containing an excess of another bead-immobilized conjugate of the same Ramp with a dsDNA oligonucleotide linker for the amplification step (Fig. 1c). The first strand of the linker was attached to the surface through the 5′ end, and to the Ramp through the 3′ end. The complementary second strand was conjugated to HRP through the 5′ end (Fig. 1c). The Ramp/HRP dsDNA linker conjugates contained the sequence of the corresponding ‘self’ recognition site (GGATCC or GAATTC, for the S17C BamHI or K249C EcoRI, respectively). The restriction reactions started upon addition of the free Ramp released during the recognition stage. The released Ramp triggered the dsDNA linker cleavage, releasing additional Ramp, which in turn cleaved more new linkers. Each step of this exponential cascade of cleavage reactions doubled the amount of free Ramp in the reaction solution (Fig. 1c). The linker cleavage also released HRP, which then was quantified colorimetrically (Fig. 1d). Generally, each initial target-probe hybridization event produced an exponentially amplified number of HRP molecules (Fig. 1e).
The amplification step setup was tested to ensure that the immobilized REases were incapable of cleavage of their own and neighbor's linkers in the absence of free REase. During the immobilization process, the high salt content (200 mM NaCl) and the absence of magnesium in the applied RE-store buffer prevented REase cleavage. After completion of immobilization, agarose beads were incubated in the restriction buffer (1X NEBuffer 3) for 75 min. Then the reaction solution was separated from the beads and applied for the colorimetric HRP assay. The resultant HRP signals were identical to the negative control, an aliquot of the restriction buffer with no beads added. Evidently, once attached to the surface, the immobilized REases could not cleave the dsDNA linkers, and the restriction reaction cascade could only start upon addition of the free (non-immobilized) REase.
Calibration curves and the attomolar detection limit of RCEA assays
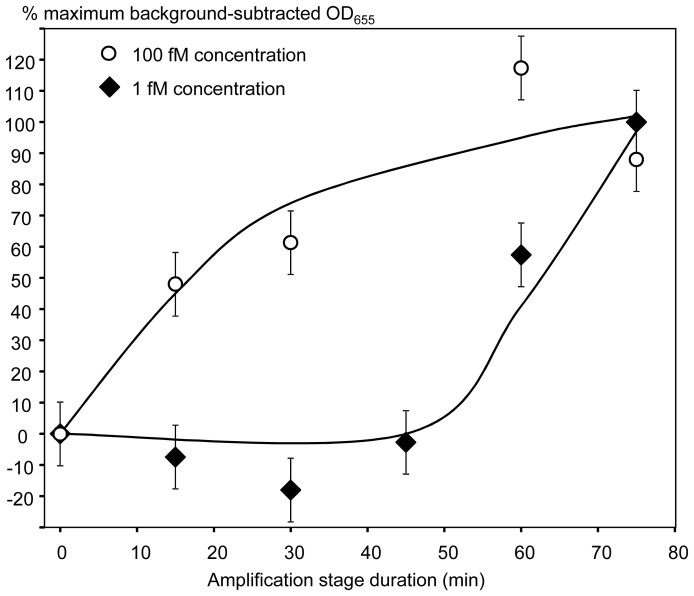
The RCEA assay scheme was tested using serial dilutions of the target AMC-BG-40, with the negative control of no target added. The mean negative control value was used to generate background-subtracted signal values for the test dilutions. The RCEA assays were performed using both Ramp, and then the assay signals were expressed as the percentages of the maximum ones that were measured at 1 nM or 1 pM target concentrations for the S17C BamHI or K249C EcoRI, respectively. To generate calibration curves, the background-subtracted and normalized signals were plotted versus the target concentration (Fig. 4).
Figure 4. The RCEA limit of detection evaluated using the oligonucleotide target AMC-BG-40.
The X-axis shows the target concentrations (M), and the Y-axis shows the background-subtracted HRP signal values (with the background calculated as the mean signal generated for zero target concentrations). For normalization and comparison of sample series, the HRP signal values were expressed as the percentages of the maximum background-subtracted OD655, corresponding to each series. Open circles show the data generated using the Direct Restriction Assay (DRA) with no amplification. Other two series were generated using the RCEA assays with the mutant S17C BamHI (closed diamonds) and K249C EcoRI (open triangles) as amplification REases. Error bars show standard deviations.
The RCEA calibration curves were analyzed in parallel with the one generated using the direct restriction assay (DRA). The DRA was performed using the AMC-BG-40 target dilutions according to the previously described protocol18 with a small modification of using streptavidin beads instead of ELISA plate wells. As before, the DRA calibration curve analysis showed the detection limit of 1 nM (Fig. 4)18. The RCEA calibration curves for both Ramp showed logarithmic signal dependence on the target concentration in the range from 10 aM to 1 nM (Fig. 4). The RCEA assay signal was above the background even at the lowest target concentration of 1 aM (10−18 M), the value was approximately 15% of the maximum, however, the error bars overlapped with those of the negative controls (Fig. 4). Thus, we evaluated the RCEA detection limit as 10−17 M, or 10 aM concentration of the oligonucleotide target. At this concentration, the assay signals were 60 to 100% of the maximum, well above the background (Fig. 4). Thus, the RCEA assay gained approximately 8 orders of magnitude improvement of the detection limit over the DRA.
Our data indicated that the RCEA assay produced an exponentially amplified number of HRP molecules for each DNA target added. The degree of signal amplification depended on (i) the REase enzyme turnover rate, (ii) the amplification stage duration, and (iii) the efficiency of mass transfer in the system (Fig. 1e). The RCEA assay employs a double-phase system, thus, the reciprocal mass transfer between the solid and liquid phases is determined by multiple factors including the surface to volume ratio and suspension mixing speed. Further assay improvement will be done in the future. However, the first test of the developed RCEA system resulted in an extremely low detection limit of approximately 200 target molecules per sample, thus, approaching the detection limit of qPCR (5–100 molecules per sample5).
Development of a real-time RCEA format
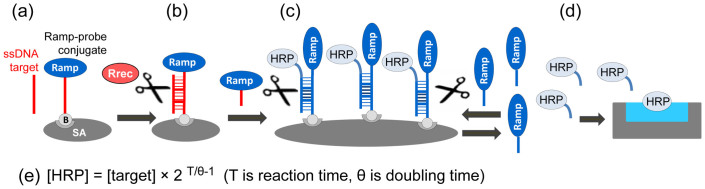
Two concentrations of the oligonucleotide target AMC-BG-40, 1 and 100 fM were used to analyze the dependence between the assay signal and the duration of the amplification stage for the Ramp S17C BamHI (Fig. 5). For each time point the signals were background-subtracted using the mean background value calculated from the corresponding negative control with no target added. Then the data were expressed as the percentages of the maximum signal achieved for the 100 fM target concentration using the standard assay time of 75 min. As expected for the high 100 fM concentration, the RCEA signal reached 50% of the maximum very quickly, at the first time point of 15 min (Fig. 5). In contrast, for the low concentration of 1 fM, the increase of signal above background was observed only after 60 min incubation (Fig. 5). Thus, the time required for the RCEA assay to generate a signal exceeding the background was dependent on the initial target concentration.
Figure 5. Dependence between the assay signal and the duration of the amplification stage analyzed using the S17C BamHI system.
The X-axis shows the duration of the RCEA amplification stage (min). The Y-axis shows the background-subtracted HRP signal values (with the background calculated for each time point as the mean signal generated for zero target concentrations). For normalization and comparison of sample series, the HRP signal values were expressed as the percentages of the maximum background-corrected OD655 obtained for 1 fM concentration after 75 min incubation. Two different target AMC-BG-40 concentrations, 100 fM (open circles) and 1 fM (closed diamonds), were used for the assays.
RCEA Assay specificity
A partially complementary target AMC-12/40 composed of (i) a fully cognate 12-mer portion containing the 6-mer BglII restriction site and 3 adjacent nucleotides from each end, and (ii) non-cognate ends of 14 nucleotides each (Table 1) was applied to the RCEA assay in parallel with the fully cognate target AMC-BG-40. Our previous study using the DRA showed that such targets produced zero signals, since a minimum of 16-bp target-probe duplex length was required for significant BglII cleavage18. The fully and partially cognate targets were assayed at 1 pM concentrations using the Ramp K249C EcoRI system (Fig. 6). The setup of the first recognition step was slightly changed to separate the target hybridization from the Rrec (BglII) cleavage (see Materials and Methods). The assay signals were background-subtracted using the negative control of no oligonucleotides added, and normalized as the percentages of the AMC-BG-40 signal. As expected based on the DRA study, the partially cognate target did not produce a positive signal, moreover, the signal was substantially, almost 40%, below the background (Fig. 6). As described above, the negative controls produced low variable signal values, probably due to some accidental Ramp release during the recognition step. The partially cognate target (i) had a sequence too short for successful REase cleavage, and (ii) could still hybridize to the probes. Apparently, this target could somehow stabilize the probe-Ramp conjugates and reduce the accidental REase release in comparison with the negative controls with no targets added.
Figure 6. Effects of the target sequence alteration and addition of foreign DNA on performance of the RCEA assay employing the K249C EcoRI system.
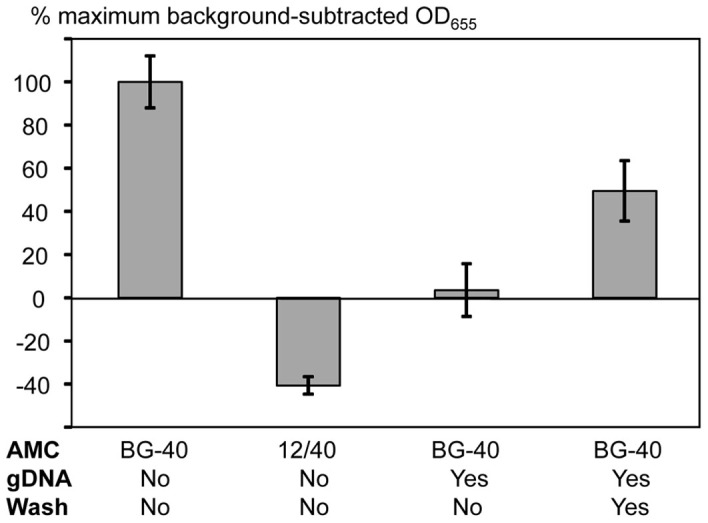
The X-axis shows the experimental conditions: AMC, the target type (BG-40 is fully cognate; 12/40 contains 12 cognate nucleotides centered around the restriction site and non-cognate ends, Table 1); gDNA, presence or absence of mouse genomic DNA (80 ng per reaction); Wash, whether the beads carrying immobilized Ramp conjugates were washed after target hybridization prior to addition of the Rrec BglII. The Y-axis shows the background-subtracted HRP signal values (with the background calculated as the mean signal generated for zero target concentrations). For normalization and comparison of sample series, the HRP signal values were expressed as the percentages of the maximum background-subtracted OD655 obtained for the fully cognate target (the left-most bar of the graph). For all test samples, the target concentrations were 1 pM.
RCEA sensitivity to foreign DNA addition
Previously, we have shown that the DRA can detect target DNA in the presence of non-cognate, heterologous mouse genomic DNA18. The applied DRA setup involved two separate steps, first the target probe hybridization in the presence of foreign DNA, then washing of the dsDNA probe-target hybrids, and finally, the Rrec addition for the specific hybrid cleavage18. We applied the same approach for the recognition step of RCEA assay to study the influence of foreign DNA. Mouse genomic DNA (80 ng per reaction) was added to the AMC-BG-40 target dilution of 1 pM, and the mixture was hybridized to the immobilized Ramp conjugates without adding Rrec. After hybridization completion, the reaction solution was removed, and the restriction buffer with the Rrec BglII was added to the beads either without or after washing. If no washing was performed, the assay signal stayed at the background level (Fig. 6). Apparently, the excess of foreign DNA (undoubtedly, containing multiple restriction sites) interacted with both REases, the free Rrec and released Ramp, efficiently removing them from the assay signal generation process. In contrast, if the beads carrying target-probe hybrids were washed 4 times (as described in Materials and Methods) prior to the Rrec BglII addition, the assay signal was restored to approximately 50% of the one observed with no foreign DNA added (Fig. 6).
Discussion
The RCEA technology provides a cost-effective alternative to PCR-based assays with the selective and sensitive detection of DNA targets by signal amplification. The RCEA assay could detect attomolar concentrations, or approximately 200 target molecules per sample, approaching the sensitivity achieved with qPCR-based techniques.
The time required for the RCEA assay to generate a signal exceeding the background was dependent on the initial target concentration. Comparable to qPCR22, accumulation of signal from different initial target concentrations produced distinct curves in the assay (Fig. 5). This remarkable similarity to qPCR suggests that the RCEA assay could be developed in a real-time format (qRCEA) for precise quantification of initial target concentrations. Initially, this type of format would include periodic sampling of the amplification reaction mixture and measurement of the HRP signal in order to plot signal accumulation over time. In the future we plan to replace the periodic sampling with a real-time detection format based on continuous electrochemical monitoring of the HRP content in the reaction solution. Development of a ‘qRCEA’ assay would furthermore provide a way to evaluate the efficiency of RCEA detection for different DNA target sequences.
Specificity achieved in the RCEA assay was comparable to that observed previously in the DRA, which is also based on the use of REase enzymes. The DRA study18 concluded that no REase-generated signal was observed if point mutations occurred in the restriction site, or a cognate target-probe length of 12 or fewer nucleotides was used to detect the DNA target. In fact, the target-probe length of 16 nucleotides was determined to be the minimum required to generate a robust signal using DRA. Double, and especially triple, point mutations in the probe located outside of the restriction site usually reduced the DRA signal to 50% of that observed for the unaltered probe18. Similar constraints are anticipated to be important for the design of RCEA assays, for example, the RCEA probes need to be selected to minimize the secondary structure formation18.
Specificity of DNA assays can be further improved by the use of thermophilic DNA-modifying enzymes at high temperatures, as shown for the nicking enzyme-mediated amplification9. Previously, thermophilic REases have been used in conjunction with PCR to block the synthesis of WT amplicons in favor of the amplification of mutant PCR products23. We speculate that the use of thermophilic REases with optimum temperatures of 50–65°C may further improve the RCEA assay stringency and thus, facilitate detection of mutations and allelic variation in target sequences outside of the REase recognition sites.
Both DRA and RCEA involve two independent biorecognition steps, which likely contributed, in both assays, to the absence of false positives (due to non-specific binding), and specific detection of target sequences in the presence of foreign genomic DNA. The RCEA assays designed for analysis of complex nucleic acid mixtures should include multiple washings during the recognition step between the target-probe hybridization and the addition of free Rrec. The separation of target hybridization and cleavage reactions may also help in designing particular hybridization stringency (i.e. salt concentrations) to distinguish fully from partially cognate DNA target-probe pairs without disrupting the REase cleavage reaction. Such adaptation of the RCEA assay for an excess of foreign DNA relies on a simple assay scheme involving one target-specific probe. In contrast, the high specificity of some other DNA assays requires complex setups, i.e. the LAMP amplification involves four different primers that must bind six distinct regions on the target DNA9.
Conclusion
The novel isothermal RCEA assay provided for a detection limit in the attomolar range, comparable to the sensitivity achieved with qPCR-based assays. The assay functions without fluorescent reagents or complex instrumentation. In the future it can be developed in a wide variety of portable and field-deployable formats, including qRCEA based on real-time electrochemical detection. Further development may also help to drastically reduce the assay time by improving the mass transfer conditions. RCEA can be adapted easily for use with new DNA targets through design of corresponding probes for the recognition step. The amplification step can be common for different DNA targets of interest, and it can be designed as described in this work using the mutant REase enzymes.
Methods
Oligonucleotide probes for recognition and amplification chambers
Table 1 provides a list of applied oligonucleotides purchased from Eurofins MWG/Operon (Huntsville, AL). The hybridization probe for Ramp immobilization in the recognition chamber, 40-mer oligonucleotide MCA-BG-Bio, corresponded to the conserved mecA gene sequences flanking the BglII restriction site AGATCT18. The oligonucleotide linkers for Ramp and HRP immobilization in the amplification chamber consisted of: (i) the first strand (L1-RB-Bio-for-Ramp or L2-RE-Bio-for-Ramp, used for S17C BamHI or K249C EcoRI attachment, respectively), and (ii) the complementary second strand (ASL1-RB-for-HRP or ASL2-RE-for-HRP) used for HRP attachment.
Preparation of the mutant BamHI REase for oligonucleotide conjugation
Site-directed mutagenesis was first used to prepare a triple cysteine-less BamHI endonuclease mutant allele, in which the Cys34, Cys54, and Cys64 residues of the wild-type (WT) bamHIR gene were replaced with Ser by using mutagenic oligonucleotides and USER Friendly Cloning Kit (New England Biolabs, Ipswich, MA) (Rebecca Kucera, unpublished results). Both WT bamHIR gene and C34S/C54S/C64S allele (5′-NcoI-3′-blunt fragment) were subcloned into pTYB2 (5′-NcoI-3′-SmaI cut) in fusion with the intein and CBD tag coding sequence. The constructs were transferred into pre-modified E. coli competent cells T7 Express lacIq [pACYC184-bamHIM] as described24,25,26. Then, the surface Ser17 of the C34S/C54S/C64S allele was changed into Cys by site-directed mutagenesis. The desired mutations were confirmed by DNA sequencing of the entire allele. The resultant BamHI endonuclease gene encoded C34S/C54S/C64S/S17C mutant protein that carried a single surface Cys exposed into solution for ligand conjugation (above it is designated as ‘S17C BamHI’). Cultures carrying the WT and the S17C mutant constructs (1 to 2 L) were grown in the LB media plus ampicillin (100 μg/ml) at 37°C to the mid-log phase. Then, the recombinant fusion protein production was induced by adding IPTG to 0.25 mM final concentration and cultivation at 30°C overnight.
The WT and S17C BamHI proteins were purified by affinity chromatography through chitin columns (New England Biolabs) following with the DTT-assisted intein cleavage (in the cleavage buffer containing 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 50 mM DTT) at 4°C overnight, as previously described26. After the cleavage, one extra Gly residue was retained at the C-terminus of the recombinant protein. The enzymatically active fractions were further purified by chromatography through HiTrap heparin columns (5 ml) using an AKTA purification system (GE Healthcare Life Sciences, Marlborough, MA). Eluted fractions were assayed for REase activity with phage λ DNA. Pooled active fractions were concentrated and dialyzed against a buffer containing 20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.1 mM EDTA, 50% w/v glycerol. The yield of the mutant S17C was approximately 1 mg of purified protein per L of culture. The purified enzyme was stored at −20°C.
REase conjugation to oligonucleotide probes and linkers to prepare for immobilization
REases and oligonucleotides were conjugated at approximately 1:1 ratio. The oligonucleotides for REase conjugation (Table 1) carried amino groups at the 3′ end. The groups were activated by reaction with succinimide of the sulfosuccinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (Sulfo-SMCC) reagent (Thermo Fisher Scientific Inc., Rockford, IL) to introduce maleimide groups18. The reaction was carried out in 1X phosphate buffered saline (PBS) (diluted from 10X stock solution (Thermo Fisher Scientific)). First, 100 μL of 10 μM oligonucleotide solution in PBS was treated with 5 μL of 10 mg/mL (23 mM) Sulfo-SMCC solution in dimethylformamide (DMF). The reaction was incubated at room temperature for 2 h. For each reaction, two Micro Bio-Spin columns with Bio-Gel P-6 (P-6 column) (Bio-Rad, Hercules, CA) were washed 3 times with 0.5 mL of a REase storage buffer (‘RE-store’) containing 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 0.1 mM EDTA, 10% w/v glycerol. Then the activated oligonucleotide solution was sequentially applied to the columns to remove the excess Sulfo-SMCC.
The S17C BamHI mutant was prepared as 40 μM stock solution in the buffer containing 20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.1 mM EDTA, 50% w/v glycerol. For conjugation, 20 μL of the stock was added to 80 μL of the activated oligonucleotide solution for the final oligonucleotide concentration of 8 μM. The K249C mutant of EcoRI was kindly provided by Dr. L. Jen-Jacobson21 as 4 μM stock solution in a storage buffer containing 20 mM sodium phosphate buffer, pH 7.3, 600 mM NaCl, 1 mM EDTA, 1 mM NaN3, 5% v/v DMSO, 10% w/v glycerol. For conjugation, 100 μL of the K249C stock was added to 40 μL of the activated oligonucleotide solution (for the final concentration of 2.9 μM). Both mixtures of mutant REases and activated oligonucleotides were incubated for 2 h at room temperature, and then stored at 4°C and −20°C for short-term and long-term storage, respectively. The preparations were used in all assays directly, with no conjugate purification. The REase conjugate concentration values were estimated based on their oligonucleotide parts, since only full conjugates containing oligonucleotides were retained on the surface after immobilization.
HRP conjugate preparation, HRP detection and quantification of assay signals
HRP-oligonucleotide conjugates for immobilization were prepared as previously described18 using the oligonucleotides shown in Table 1. The conjugates were immobilized through biotin-streptavidin interactions on the surface of either streptavidin-precoated 96-microwell plates, or the streptavidin agarose bead suspension (both from Thermo Fisher Scientific). HRP was released from the surface attachment as the result of REase cleavage of probes and linkers. Reaction mixtures containing released HRP were transferred to wells of a 96-well ELISA microplate (Thermo Fisher Scientific). Then 100 μL of the BioFX TMB One Component HRP Microwell Substrate (SurModics, Eden Prairie, MN) was added into each well. The HRP-generated signal was quantified after 2–5 min incubation at room temperature by the blue color formation that was measured colorimetrically at the wavelength of 655 nM (OD655), using an iMark Microplate Reader (Bio-Rad).
Manipulations with streptavidin agarose beads (common for all bead-based conjugate immobilization protocols)
To prepare for the enzyme conjugate immobilization, an aliquot (typically, 60 μL) of the streptavidin agarose bead suspension was placed into a spin column (Pierce Spin Columns, screw cap with Luer lock, Thermo Fisher Scientific) and centrifuged at 1,500 g for 15 sec to remove the manufacturer's buffer. To block non-specific binding, the agarose beads were re-suspended in 150 μL of 0.5% bovine serum albumin (BSA, Thermo Fisher Scientific) solution in the RE-store buffer (‘RE-store-BSA’). The column was incubated for 30 min at room temperature in a Labquake Shaker Rotisserie (Thermo Fisher Scientific) set for 8 rpm. After incubation, the columns were centrifuged at 1,500 g for 15 sec to remove the flow-through.
The prepared bead suspensions were used for conjugate immobilization in the RE-store-BSA buffer. The immobilization reaction mixtures were incubated for 1 h at room temperature in the rotisserie. Unless specified otherwise, the same washing procedure was performed after completion of immobilization. The columns were centrifuged at 1,500 g for 15 sec to remove the flow-through, and washed sequentially with: (i) RE-store-BSA buffer, 6 times, 200 μL each, (ii) RE-store buffer, 10 times, 200 μL each, and (iii) 1X NEBuffer 3 (New England Biolabs) (100 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT), 3 times, 200 μL each. Each washing was done by re-suspending the agarose suspension in the buffer, and centrifugation at 1,500 g for 15 sec to remove the flow-through. The prepared bead suspensions with immobilized enzyme conjugates were stored on ice, and aliquoted into micro-spin columns for subsequent assays (Pierce Micro-Spin Columns, Thermo Fisher Scientific).
All REase cleavage reactions (including the RCEA recognition and amplification stages) were carried out in 1X NEBuffer 3 by using incubations at 37°C for 1–1.5 h with rotation (8 rpm). After completion, the flow-through reaction solutions were collected by centrifugation at 3,000 g for 2 min.
Testing of REase conjugates for the presence of oligonucleotides and immobilization
Four initial aliquots (45 μL each) of the prepared streptavidin agarose bead suspension were res-suspended in 140 μL of the RE-store-BSA buffer each. Two aliquots were used to immobilize the REase-oligonucleotide conjugates by adding either 4 μL of 8 μM solution of S17C BamHI-MCA-BG-Bio, or 11 μL of 2.9 μM of K249C EcoRI-MCA-BG-Bio in the RE-store buffer. Other two aliquots were used to prepare reporter systems of the immobilized HRP conjugates by adding either (i) 5 μL of 5 μM solution of HRP-L1-RB-Bio plus 5 μL of 10 μM solution of ASL1-RB, or (ii) 5 μL of 5 μM solution of HRP-L2-RE-Bio plus 5 μL of 10 μM solution of ASL2-RE (Table 1), for testing of the S17C BamHI and K249C EcoRI, respectively. After incubation for 1 h at room temperature, the agarose suspensions with immobilized REases were washed as described above, re-suspended in 158 μL of 1X NEBuffer 3, and divided into 2 aliquots of 77 μL each. The bead suspensions with the immobilized HRP conjugates were washed and re-suspended in 60 μL of 1X NEBuffer 3, then divided into 6 aliquots of 10 μL each.
As shown in the Figure 2, the procedure started with the addition of 4 μL of the REase BglII (10 units/μL) to each aliquot containing the beads with immobilized REase conjugates. For each conjugate, one aliquot was used to add 9 μL of 10 μM solution of the AMC-BG-40 target oligonucleotide (for the final concentration of 1 μM), and another - as a negative control by adding 9 μL of 1X NEBuffer 3 with no targets. The resultant reactions were incubated at 37°C for 1 h. The flow-through was collected by centrifugation at 1,500 g for 2 min, divided into three aliquots and added to the prepared 10 μL aliquots containing either HRP-L1-RB-Bio or HRP-L2-RE-Bio, for the S17C BamHI or K249C EcoRI, respectively. The reactions were incubated at 37°C for 1 h, and the flow-through reaction solutions were collected by centrifugation at 1,500 g for 2 min, and applied directly for the HRP detection as described above.
Enzymatic activity assays and titration of REase-oligonucleotide conjugates
An HRP reporter system was prepared by treating streptavidin pre-coated wells with 50 μL per well of PBS solution containing HRP conjugates and the corresponding 2nd strand oligonucleotides. Two reporters were prepared: (i) 50 nM solution of HRP-L1-RB-Bio plus 100 nM solution of ASL1-RB, or (ii) 50 nM solution of HRP-L2-RE-Bio plus 100 nM solution of ASL2-RE (see Table 1 for sequences), for testing of the S17C BamHI and K249C EcoRI, respectively. The plate was incubated for 2 h at room temperature, followed by washing 4 times with PBS supplemented with 0.5% (v/v) Tween-20 (PBST). Next the wells were incubated with 1:100 dilution of saturated biotin solution in PBS for 15 min at room temperature to block the free streptavidin. Then the plate was washed 3 times with PBST and 4 times with PBS.
The mutant REase conjugates with the oligonucleotide probe MCA-BG-Bio (Table 1), S17C BamHI-MCA-BG-Bio and K249C EcoRI-MCA-BG-Bio, were used to prepare sample series in 1X NEBuffer 3. Each series started with a 320 μM/μL conjugate solution, then it was diluted by a factor of 2 for each consecutive sample, with the last sample of 5 μM/μL. Negative controls were prepared from 1X NEBuffer 3 with no conjugate added. The dilution samples were added to the corresponding wells with the immobilized HRP conjugates (HRP-L1-RB-Bio or HRP-L2-RE-Bio, for S17C BamHI-MCA-BG-Bio or K249C EcoRI-MCA-BG-Bio, respectively). The microwell plate was incubated at 37°C for 1 h. Then the reaction solutions were collected and applied for the HRP detection as described above.
Preparation of immobilized REase-probe conjugates for the RCEA recognition stage
The agarose bead suspension was prepared as described above and re-suspended in the RE-store-BSA buffer, 200 μL for the initial bead aliquot of 60 μL. Then the bead suspension was supplemented with 6 μL of either 400 nM solution of the S17C BamHI-MCA-BG-Bio (2.4 pmol/initial aliquot) or 4 nM solution of the K249C EcoRI-MCA-BG-Bio (24 fmol/initial aliquot). The agarose suspensions were incubated for 1.5 h at room temperature, then the reaction solution was removed by centrifugation, and the beads were washed as described above. Finally, the beads with immobilized REases were re-suspended in 216 μL of 1X NEBuffer 3, divided into 8 aliquots (27 μL each) and placed on ice prior to the recognition reaction.
Preparation of immobilized REase/HRP dsDNA linker conjugates for the RCEA amplification stage
The agarose bead suspension was re-suspended in the RE-store-BSA buffer, 200 μL for the initial bead aliquot of 60 μL. To immobilize the REase- and HRP-linker conjugates, the bead solution was supplemented with either (i) 6 μL of 8 μM solution of the S17C BamHI-L1-RB-Bio (48 pmol/initial aliquot) plus 12 μL of 5 μM solution of the HRP-ASL1-RB (60 pmol/initial aliquot) conjugates or (ii) 1.5 μL of 2.9 μM solution of the K249C EcoRI-L2-RE-Bio (4.35 pmol/initial aliquot) and 1.5 μL of 5 μM solution HRP-ASL2-RE (7.5 pmol/initial aliquot) conjugates. The agarose suspensions were incubated for 1.5 h at room temperature, then the reaction solution was removed by centrifugation, and the beads were washed as described above. Finally, the beads with immobilized conjugates were re-suspended in 80 μL of 1X NEBuffer 3, divided into 8 aliquots (10 μL each) and placed on ice prior to the recognition reaction.
RCEA assay protocol: the recognition stage
For the S17C BamHI conjugate, the recognition reaction was set up using a 27 μL aliquot of the agarose bead suspension with the immobilized BamHI-MCA-BG-Bio. The reaction solution was supplemented with 0.75 U/μL of the recognition REase (Rrec) specific for the DNA target, BglII (New England Biolabs), and 3 μL of a target (i.e., AMC-BG-40) diluted in 1X NEBuffer 3 at a desired concentration. In all cases, negative controls were prepared using 3 μL 1X NEBuffer 3 with no target added. The recognition reaction was then incubated at 37°C for 1.5 h with rotation (8 rpm), and the flow-through reaction solutions were collected by centrifugation.
For the K249C EcoRI conjugate, the recognition reaction was set up using a 27 μL aliquot of the agarose bead suspension with the immobilized EcoRI-MCA-BG-Bio. The reaction was performed in two steps, first a desired target analyte dilution in 1X NEBuffer 3 (or a negative control) was added for hybridization to the immobilized probe conjugates. The recognition reaction was incubated at 37°C for 1 h with rotation (8 rpm). Second, the REase cleavage was performed by adding the Rrec BglII to the same reaction to achieve 0.75 U/μL concentration. The reaction mixture was incubated at 37°C for 20 min. Then the flow-through reaction solutions were collected by centrifugation.
The recognition reaction protocol was separated into two steps for several experiments with the Ramp K249C EcoRI. The target dilutions were prepared in the RE-store buffer with addition of mouse genomic DNA to achieve 2.7 ng/μL concentration (80 ng/reaction). At the first step, the target mixture was hybridized to the probes without the addition of Rrec, The reaction solution was then removed, and the beads were washed with (i) RE-store buffer (3 times, 200 μL each), and (ii) 1X NEBuffer 3 (1 time, 200 μL). At the second step, the beads were re-suspended in 27 μL of 1X NEBuffer 3 and subjected to BglII cleavage as described above.
RCEA assay protocol: the amplification stage
Both recognition and amplification stages were designed to use the same immobilized REase (i.e. the recognition reaction employing the BamHI-MCA-BG-Bio was applied for the amplification stage with the BamHI-L1-RB-Bio/HRP-ASL1-RB conjugates). The flow-through solution collected at the completion of the recognition stage (approximately 30 μL per reaction) was mixed with a 10 μL aliquot of the corresponding immobilized REase/HRP-dsDNA linker conjugate prepared as described above. The resultant suspension was then incubated at 37°C for 75 min with rotation (8 rpm). For the experiments on the time dependence of the assay signal on the duration of the amplification stage, the incubation times varied from 0 to 75 min with 15 min increments. After incubation, the reaction was centrifuged to collect the flow-through solution for the HRP detection performed as described above.
Data analysis
The OD655 measurements of HRP signal were subjected to background subtraction using the mean background values calculated from 4 replicate negative controls. Test samples were measured in triplicate, to calculate mean and standard deviation values. The data were additionally normalized to construct calibration curves for the direct comparison of different assay schemes. For each assay, we specified the maximum background-corrected signal as 100%, and expressed all other values in the series as the percentages of the maximum.
Author Contributions
A.L.G., M.W.S., and N.S.Y. conceived and designed the study, performed the experiments, and wrote the manuscript. H.M.S., I.A.S. and K.S. contributed to the study design, data analysis and the manuscript writing and editing. L.J.-J. provided the mutant EcoRI and consulted the other authors on the experimental protocols. I.A.M. constructed the single Cys BamHI mutant; X.F. and S.Y.X. purified the mutant enzyme. S.Y.X. contributed to the writing of the BamHI mutagenesis/purification portion of the manuscript and editing of the entire manuscript.
Acknowledgments
We thank Rebecca Kucera for providing the triple Cys-less BamHI mutant plasmid, and Siu-Hong Chan for help with the protein purification. This publication was made possible by NPRP 5-593-3-149 grant from the Qatar National Research Fund (a member of the Qatar Foundation, http://www.qnrf.org/). The statements made herein are solely the responsibility of the authors. Support was also provided by National Science Foundation (http://www.nsf.gov/), grant OCE 0424602. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
M.W.S., N.S.Y., and K.S. are named inventors of several patents related to the RCEA technology described in this manuscript and owned by Cascade Biosystems, Inc. Other authors declare no potential conflict of interest. S.Y.X. is a member of the Editorial Board of Scientific Reports.
References
- Epstein J. R., Biran I. & Walt D. R. Fluorescence-based nucleic acid detection and microarrays. Analyt. Chim. Acta 469, 3–36 (2002). [Google Scholar]
- Wolk D., Mitchell S. & Patel R. Principles of molecular microbiology testing methods. Infect. disease clinic. North America 15, 1157–1204 (2001). [DOI] [PubMed] [Google Scholar]
- Yang S. & Rothman R. E. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 4, 337–348 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barken K. B., Haagensen J. A. J. & Tolker-Nielsen T. Advances in nucleic acid-based diagnostics of bacterial infections. Clinic. Chim. Acta 384, 1–11 (2007). [DOI] [PubMed] [Google Scholar]
- Rosi N. L. & Mirkin C. A. Nanostructures in biodiagnostics. Chem. Rev. 105, 1547–1562 (2005). [DOI] [PubMed] [Google Scholar]
- Gill P. & Ghaemi A. Nucleic acid isothermal amplification technologies - a review. Nucleosid. Nucleotid. Nuc. Acids 27, 224–243 (2008). [DOI] [PubMed] [Google Scholar]
- Wang F., Lu C.-H. & Willner I. From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem. Rev. 114, 2881–2941 (2014). [DOI] [PubMed] [Google Scholar]
- Nycz C. M., Dean C. H., Haaland P. D., Spargo C. A. & Walker G. T. Quantitative reverse transcription strand displacement amplification: quantitation of nucleic acids using an isothermal amplification technique. Analyt. Biochem. 259, 226–234 (1998). [DOI] [PubMed] [Google Scholar]
- Tanner N. A. & Evans T. C. Loop-mediated isothermal amplification for detection of nucleic acids. Cur. Prot. Mol. Biol. 10.1002/0471142727.mb1514s105 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang X., Lowe S. B. & Gooding J. J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectr. 61, 491–499 (2014). [DOI] [PubMed] [Google Scholar]
- Nakayama S., Yan L. & Sintim H. O. Junction probes - sequence specific detection of nucleic acids via template enhanced hybridization processes. J. Amer. Chem. Soc. 130, 12560–12571 (2008). [DOI] [PubMed] [Google Scholar]
- Abe H. & Kool E. T. Destabilizing universal linkers for signal amplification in self-ligating probes for RNA. J. Amer. Chem. Soc. 126, 13980–13986 (2004). [DOI] [PubMed] [Google Scholar]
- Sando S., Sasaki T., Kanatani K. & Aoyama Y. Amplified nucleic acid sensing using programmed self-cleaving DNAzyme. J. Amer. Chem. Soc. 125, 15720–15721 (2003). [DOI] [PubMed] [Google Scholar]
- Ji H., Yan F., Lei J. & Ju H. Ultrasensitive electrochemical detection of nucleic acids by template enhanced hybridization followed with rolling circle amplification. Analyt. Chem. 84, 7166–7171 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou Z., Du Y., Zhang L. & Dong S. A label-free, G-quadruplex DNAzyme-based fluorescent probe for signal-amplified DNA detection and turn-on assay of endonuclease. Biosens. Bioelectr. 34, 100–105 (2012). [DOI] [PubMed] [Google Scholar]
- Weizmann Y., Cheglakov Z. & Willner I. A Fok I/DNA machine that duplicates its analyte gene sequence. J. Amer. Chem. Soc. 130, 17224–17225 (2008). [DOI] [PubMed] [Google Scholar]
- Weizmann Y. et al. A virus spotlighted by an autonomous DNA machine. Angewandte Chem. 118, 7544–7548 (2006). [DOI] [PubMed] [Google Scholar]
- Smith M. W. et al. A new restriction endonuclease-based method for highly-specific detection of DNA targets from methicillin-resistant Staphylococcus aureus. PLOS One 9 10.1371/journal.pone.0097826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. D., Yazvenko N. & Smit M. Detecting nucleic acid. U.S. Patent, USPTO: US8278048 B2, Cascade Biosystems, Inc., USA (2012).
- Smith K. D., Yazvenko N. & Smit M. Methods and materials for detecting viral or microbial infections. U.S. Patent, USPTO: US8597886 B2, Cascade Biosystems, Inc., USA (2013).
- Dylla-Spears R., Townsend J. E., Sohn L. L., Jen-Jacobson L. & Muller S. J. Fluorescent marker for direct detection of specific dsDNA sequences. Analyt. Chem. 81, 10049–10054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- Wei A. et al. Efficient isolation of targeted Caenorhabditis elegans deletion strains using highly thermostable restriction endonucleases and PCR. Nuc. Acid. Res. 30, e110 10.1093/nar/gnf109 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E. et al. Overexpression, purification and crystallization of BamHI endonuclease. Nuc. Acid. Res. 19, 1825–1829 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.-Y. & Schildkraut I. Isolation of BamHI variants with reduced cleavage activities. J. Biol. Chem. 266, 4425–4429 (1991). [PubMed] [Google Scholar]
- Chong S. et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192, 271–281 (1997). [DOI] [PubMed] [Google Scholar]