Abstract
Segmentation of the vertebrate hindbrain is an evolutionarily conserved process. Here, we identify the transcription factor early growth response 4 (egr4) as a novel regulator of posterior hindbrain development in Xenopus. egr4 is specifically and transiently expressed in rhombomeres 5 and 6 (r5/r6), and Egr4 knockdown causes a loss of mafb/kreisler and krox20/egr2 expression in r5/r6 and r5, respectively. This phenotype can be fully rescued by injection of frog or mouse Egr4 mRNA. Moreover Egr4-depleted embryos exhibit a specific loss of the neural crest stream adjacent to r5, and have inner ear defects. While the homeodomain protein vHnf1/Hnf1b directly activates Mafb and Krox20 expression in the mouse hindbrain to specify r5, we show that in Xenopus this process is indirect through the activation of Egr4. We provide evidence that rearrangements in the regulatory sequences around egr4 and mafb genes may account for this difference.
Development of the vertebrate hindbrain along the anteroposterior axis is an evolutionarily conserved process. It involves the transient formation of seven to eight transversal swellings known as rhombomeres (r). These territories are cell lineage restricted compartments and constitute developmental units for cranial motor nerves and neural crest cells. In most vertebrates, cranial neural crest cells originate from r2, r4 and r6 and migrate as three streams that populate the 1st, 2nd and 3rd branchial arches, respectively. Neural crest cells give rise to the ganglia of cranial nerves V, VII/VIII and IX, and to craniofacial skeletal elements unique for each branchial arch. Fewer neural crest cells are produced from r3 and r5; these cells typically migrate rostrally and caudally to join the adjacent streams1,2,3. This conserved pattern of neural crest cell migration directly depends on the proper segmentation of the hindbrain. Caudal hindbrain patterning is also critical for the specification and development of the inner ear which arises from the ectoderm adjacent to r4/r54.
The gene regulatory network underlying hindbrain segmentation involves a complex interplay of transcription factors and signaling molecules leading to the regional expression of different combination of genes, which imparts a unique molecular identity to each rhombomere5. Several of these genes encode transcription factors, such as Krox20/Egr2, MafB/Kreisler/Valentino, Hnf1b/vHnf1 and members of the Hox family. In the posterior hindbrain, the zinc finger transcription factor Krox20 is expressed in the prospective r3 and r56,7. The basic leucine zipper Mafb is expressed in r5 and r6, and controls the specification of these two rhombomeres, and activates Krox20 in r58,9. The homeodomain transcription factor vHNF1 (variant hepatocyte nuclear factor 1) cooperates with FGF to promote caudal hindbrain identity, by activating Mafb and Krox20 expression in r5/r6 and r5, respectively10,11. Hoxb1 is expressed in the neural plate with an anterior limit at the prospective r3/r4 boundary, and later is only maintained in r4, where it acts with other Hox factors to specify r4 identity12. The regulatory cascade underlying rhombomere specification is highly conserved among vertebrates, and only minor variations in this blueprint have been reported.
Here we describe the function of the transcription factor early growth response 4 (Egr4), a novel regulator of hindbrain development in Xenopus. Egr4 is transiently expressed in r5/r6 at neurula stages. Using loss- and gain-of-function approaches we demonstrate that Egr4 is required downstream of Hnf1b to activate mafb and krox20 expression and promote r5 identity in the posterior hindbrain. This is the first report describing a role for Egr4 in the vertebrate nervous system.
Results
Xenopus laevis egr4 is a target of Pax3 and Zic1
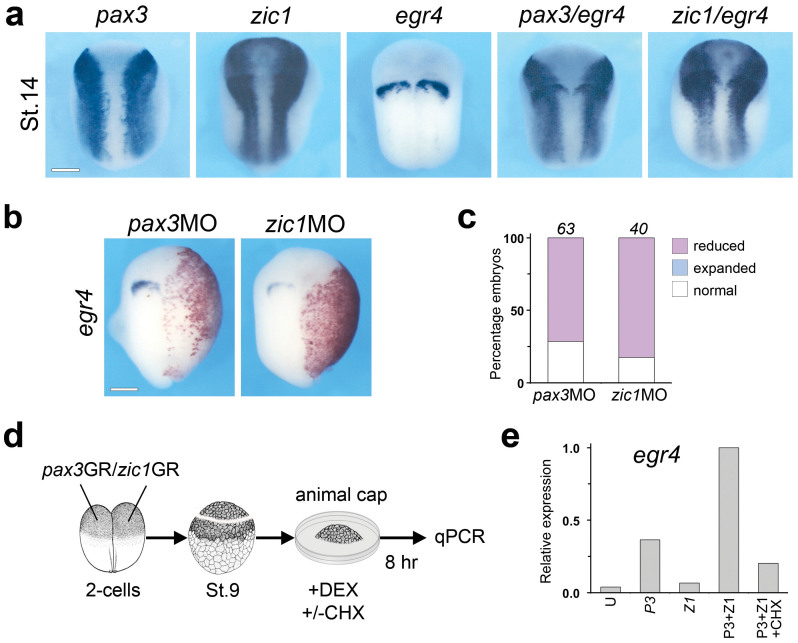
Egr4, a member of the early growth response (EGR) family of zinc-finger transcription factors, was identified in a microarray screen for targets of Pax3 and Zic113, two transcription factors that are necessary and sufficient to specify the neural plate border in Xenopus14,15,16. Xenopus laevis egr4 possesses an open reading frame encoding 486 amino acids. At the amino acid level, Xenopus laevis Egr4 shares 40% identity with human EGR4 (accession # NP_0011956), 44% identity with mouse Egr4 (accession # NP_065621), 50% identity with chicken Egr4 (accession # SP_420888) and 89% identity with Xenopus tropicalis Egr4 (accession # XP_004911346). By whole mount in situ hybridization egr4 is detected in a single stripe across the neural plate, extending into the neural plate border where egr4 overlaps with pax3 and zic1 expression domains (Fig 1a). To confirm that egr4 is a target of Pax3 and Zic1, we performed perturbation experiments in the embryo using pax3- and zic1-specific morpholino antisense oligonucleotides (MOs)14,15,16. Pax3 or Zic1 knockdown resulted in a reduction or loss of egr4 expression in most embryos (Fig 1b–c), suggesting that both factors are independently required for egr4 expression. In animal cap explants from embryos injected at the 2-cell stage (Fig 1d), expression of pax3GR and/or zic1GR (the hormone-inducible versions of pax3 and zic1, fused to the human glucocorticoid receptor ligand-binding domain) showed that Pax3 and Zic1 are sufficient to synergistically activate egr4 (Fig 1e). Moreover, egr4 expression in pax3/zic1-injected explants was dramatically reduced when the protein synthesis inhibitor cyclohexamide (CHX) was added to the culture medium, indicating that the induction of egr4 by Pax3 and Zic1 is indirect (Fig 1e).
Figure 1. egr4 is a downstream target of Pax3 and Zic1.
(a) Whole-mount in situ hybridization for egr4, pax3 and zic1. The DIG-labeled probes were used alone or in combination (pax3/egr4 and zic1/egr4). Dorsal views, anterior to top. Scale bar, 300 μm. (b) Injection in one blastomere at the 2-cell stage of MOs to block Pax3 (pax3MO; 40 ng) or Zic1 (zic1MO; 40 ng) function resulted in a reduction of egr4 expression. The injected side is to the right as indicated by the presence of the lineage tracer (Red-Gal). Dorsal views, anterior to top. Scale bar, 300 μm. (c) The graph indicates the percentage of embryos with normal (white) or reduced/lost (red) egr4 expression. The number of embryos analyzed is indicated on top of each bar. (d) mRNA encoding pax3GR and zic1GR (100 pg each), alone or in combination were injected into both blastomeres in the animal pole at the 2-cell stage. At the blastula stage (stage 9), animal cap explants were dissected and cultured for 8 hours in the presence of dexamethasone (DEX). In some samples, cyclohexamide (CHX) was used to block protein synthesis. (e) egr4 expression in pax3GR and zic1GR injected animal cap explants analyzed by qPCR.
Egr4 is specifically and transiently expressed in the posterior hindbrain
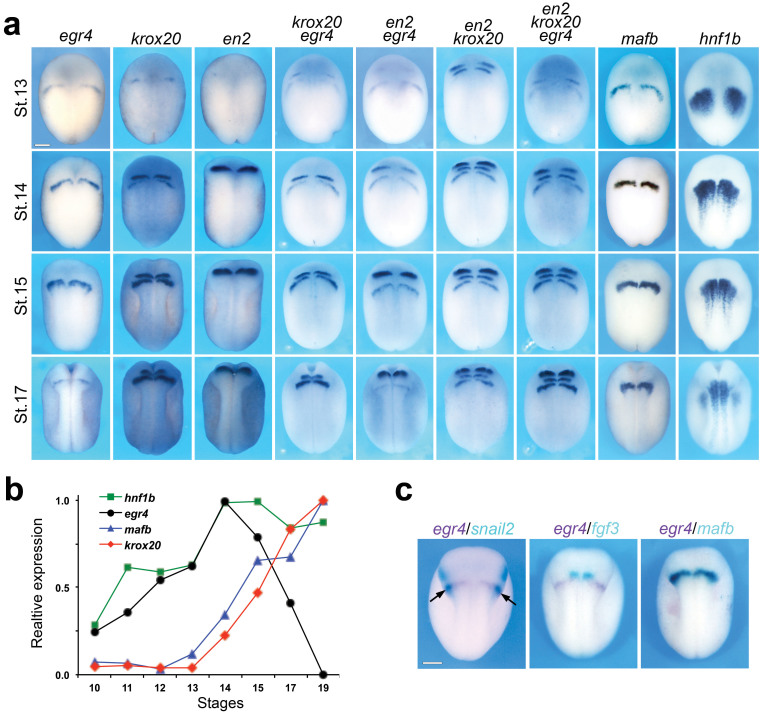
In order to more precisely map the expression domain of egr4, we performed in situ hybridization on embryos at different stages using digoxigenin-labeled RNA probes for a number of genes expressed in similar region of the ectoderm, including en2 (midbrain-hindbrain boundary17), krox20/egr2 (hindbrain r3/r518), fgf3 (hindbrain r419) mafb/kreisler (hindbrain r5/r620), hnf1b/vHnf1b (hindbrain r5/r6/r721) and snail2 (neural crest22). The probes were used alone or in combination as indicated (Fig 2a). egr4 expression is initiated at stage 13 as a single domain across the neural pate. This expression progressively increases to reach a maximum around stage 14–15, and then slowly decreases to become undetectable around stage 19/20 (Fig 2a; not shown). This expression profile was independently confirmed by qPCR (Fig 2b). Spatially, there is no overlap between egr4 and en2 expression domain, however, egr4 is co-expressed with krox20 in r5 (Fig 2a). Temporally, the escalation of egr4 expression precedes krox20 and mafb expression as shown by qPCR (Fig 2b). We also performed two-color in situ hybridization using snail2, fgf3 and mafb in combination with egr4 (Fig 2c). The most lateral domain of egr4 overlaps with snail2 demonstrating that egr4 is expressed in a subpopulation of neural crest progenitors. In addition, egr4 expression domain in the hindbrain lays immediately posterior to fgf3 expression domain in r4, and overlaps completely with mafb in r5/r6 (Fig 2c). Taken together, these results indicate that egr4 is specifically and transiently expressed in r5/r6, and its expression escalates ahead of the expression of krox20 and mafb expression in this region.
Figure 2. egr4 is specifically and transiently expressed in r5 and r6.
(a) Analysis of the spatiotemporal expression of egr4 as compared to genes expressed in similar region of the ectoderm, including en2 (midbrain-hindbrain boundary), krox20/egr2 (hindbrain r3/r5), mafb/kreisler (hindbrain r5/r6) and hnf1b/vHnf1b (hindbrain r5/r6/r7). The probes were used alone or in combination as indicated. Scale bar, 300 μm. (b) Temporal dynamics of the expression of egr4, krox20, mafb, and hnf1b analyzed by qPCR in whole embryos at the indicated stages. The values were normalized to odc1. (c) Two-color in situ hybridization for egr4/snail2, egr4/fgf3, and egr4/mafb at the neurula stage. The expression of egr4 at the neural plate border overlaps with the neural crest marker, snail2 (arrows). egr4 is detected immediately adjacent and posterior to fgf3 expression domain in r4. egr4 and mafb have overlapping expression domains in r5/r6. Scale bar, 300 μm.
Egr4 is required for mafb and krox20 expression in the posterior hindbrain
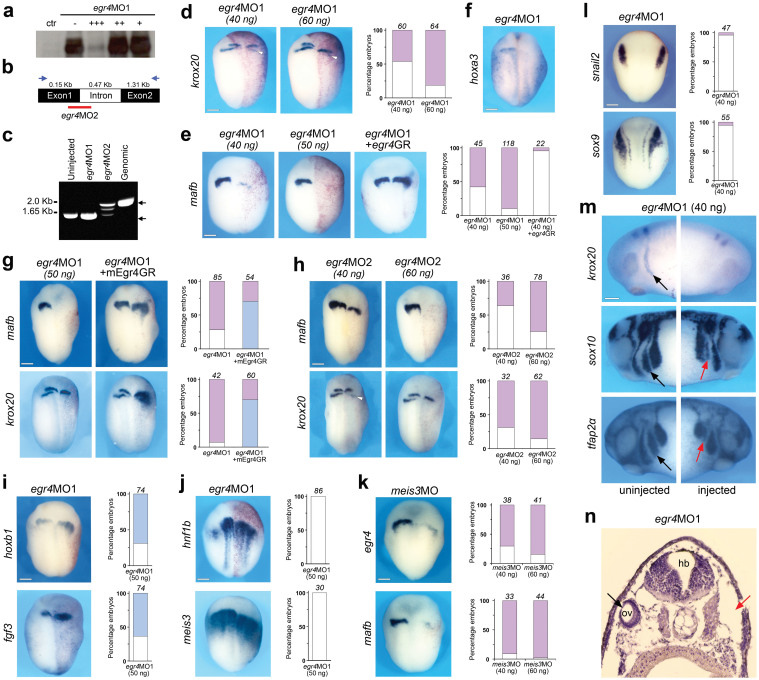
To determine whether Egr4 function is required for posterior hindbrain development we performed Egr4 knockdown in developing embryos using MOs. The first MO (erg4MO1) was designed to specifically interfere with translation of egr4 mRNA. This was confirmed in an in vitro transcription/translation assay in which erg4MO1 blocked Egr4 protein production (Fig 3a; Supplementary Figure 1). Unilateral injection of egr4MO1 at the 2-cell stage led to a reduction of krox20 expression in r5 at stage 15, without affecting krox20 expression in r3; however, r3 appeared shifted posteriorly in some embryos (Fig. 3d). egr4 knockdown also strongly inhibited the expression of mafb in r5/r6 (Fig. 3e). In morphant embryos the loss of mafb expression in r5/r6 was associated with a reduction of hoxa3 expression, a direct target of Mafb23 (Fig 3f). The loss of mafb and krox20 in morphant embryos could be rescued by injection of mRNAs encoding the hormone inducible version of Xenopus egr4 (egr4GR) or mouse Egr4 (mEgr4GR) (Fig. 3e, g). To confirm the specificity of the knockdown phenotype, we used a second MO (egr4MO2) that specifically interfered with egr4 pre-mRNA splicing by targeting the splice donor site of exon 1 (Fig 3b), resulting in the production of a transcript of higher size, due to intron retention (Fig 3c). The phenotype of egr4MO2-injected embryos was indistinguishable from the phenotype generated by injection of the translation blocking MO, with strong inhibition of mafb and krox20 expression in r5/r6 and r5, respectively (Fig 3h). Furthermore, coinjection of the two MOs at low doses showed an additive effect on the repression of mafb expression (Supplementary Figure 2). The similarity of the two MO phenotypes, and the ability to rescue krox20 and mafb expression in morphant embryos vouch for a specific requirement of Egr4 in posterior hindbrain development.
Figure 3. Egr4 is required for caudal hindbrain development.
(a) Increasing amounts of egr4MO1 10 ng (+), 100 ng (++), and 1000 ng (+++) blocks translation directed by egr4 mRNA in an in vitro coupled transcription/translation reaction. Full-length blot is presented in Supplementary Figure 1. (b) Schematic representation of the egr4 locus. The PCR primers used for the analysis of spliced transcripts are indicated (blue arrows). The position of the splice (egr4MO2) blocking MO is indicated. (c) In egr4MO2-injected embryos a larger egr4 transcript is detected (~2.0 kb) due to intron retention. For all samples, the RT-PCR was performed under the same experimental conditions. (d–f) Unilateral injection of egr4MO1 at the 2-cell stage caused a reduction of expression of krox20 in r5 (d), mafb in r5/r6 (e) and hoxa3 (f). Note the residual krox20 in r5 (arrowheads). Injection of Xenopus egr4GR (e) or mouse Egr4GR (g) mRNAs (100 pg) can efficiently rescue mafb or krox20 expression in egr4-depleted embryos. (h) Unilateral injection of egr4MO2 results in a specific reduction/loss of expression of krox20 in r5 and mafb in r5/r6. Note the residual krox20 in r5 (arrowhead; left panel). (i–j) Unilateral injection of egr4MO1 (50 ng) at the 2-cell stage caused a posterior expansion of hoxb1 and fgf3 (i), while hnf1b and meis3 were largely unaffected (j). (k) Unilateral injection of meis3MO (40 ng) at the 2-cell stage caused a reduction of egr4 and mafb expression. (l) Egr4 knockdown (egr4MO1; 40 ng) has no effect on snail2 and sox9 expression at the neural plate border. In all panels (d–l), the graphs indicate the percentage of embryos with normal (white), reduced/lost (red) or expanded/ectopic (blue) gene expression. The number of embryos analyzed is indicated on top of each bar. Dorsal views, anterior to top. (m) At stage 25 egr4MO1-injected embryos (40 ng) show a specific loss of anterior branchial neural crest stream that normally travels to the third branchial arch (red arrows). Lateral views, anterior to left (uninjected side; black arrows) or to right (injected side; red arrows), dorsal to top. Panels (d–m) scale bars, 300 μm. (n) Transverse section through an embryo injected with 40ng of egr4MO1. On the injected side (red arrow) the otic vesicle failed to form. The otic vesicle on the control side is indicated (black arrow). hb: hindbrain; ov: otic vesicle.
To evaluate the consequences of the loss of krox20 and mafb on other regions of the hindbrain, we analyzed the expression of hoxb1, fgf3 and hnf1b in morphant embryos. We found that the expression domain of hoxb1 and fgf3, normally confined to r4, was expanded posteriorly (Fig 3i), consistent with a loss of r5 identity. The expression of hnf1b was largely unaffected, although slightly shifted posteriorly (Fig 3j), suggesting that Egr4 is functioning downstream of hnf1b. The transcription factor Meis3 is expressed in r2-r424 and has been shown to be acting downstream of Pax3 and Zic125. meis3 expression level was unchanged in egr4-depleted embryos, however there was a posterior shift of its anterior boundary of expression, similar to the one observed for hnf1b (Fig 3j). We also analyzed the regulation of egr4 expression by Meis3. Meis3 MO-mediated knockdown resulted in a severe reduction of egr4 and mafb expression in r5/r6 (Fig 3k), suggesting that Meis3 is acting upstream of Egr4 and Mafb. This result is consistent with a previous study showing that Meis3 regulate cell fates in the hindbrain in a non-cell autonomous manner24.
Egr4-depleted embryos have neural crest and inner ear defects
Because egr4 expression at the neural plate border overlaps with the neural crest territory (Fig 2c), we analyzed the expression of two early neural crest transcriptional regulators, snail2 and sox9, in morphant embryos. We found that both genes were largely unaffected in Egr4-depleted embryos (Fig 3l), suggesting that egr4 knockdown did not interfere with neural crest specification. In Xenopus, neural crest cells at hindbrain level migrate towards the branchial arches into four streams, the mandibular, hyoid, anterior and posterior branchial neural crest26. The stream of neural crest expressing egr4, completely overlaps with krox20-positive neural crest cells (Fig 2a), which corresponds to the anterior branchial stream that travels caudal to the otic vesicle (centered onto r4 in Xenopus27) into the third branchial arch28. In egr4-depleted embryos this stream of neural crest cells was specifically reduced or lost, as revealed by the expression of krox20, sox10 and tfap2α at the tailbud stage (Fig 3m). Interestingly, even in morphant embryos retaining some krox20 expression in r5, the stream of neural crest cells adjacent to this axial level was often missing, suggesting that both domains are regulated by different levels of egr4. The Mafb mouse mutant Kreisler, and its counterpart in fish valentino, which exhibit mis-patterning of hindbrain caudal to r3, have severe inner ear defects23,29,30. Since egr4-depleted embryos fail to express mafb, we analyzed otic vesicle formation in egr4 morphant embryos by histology. We found that a majority of these embryos had reduced or missing otic vesicles on the injected side (Fig 3n). We posit that the neural crest and the inner ear phenotypes observed in egr4-depleted embryos could be the result of defects in posterior hindbrain patterning.
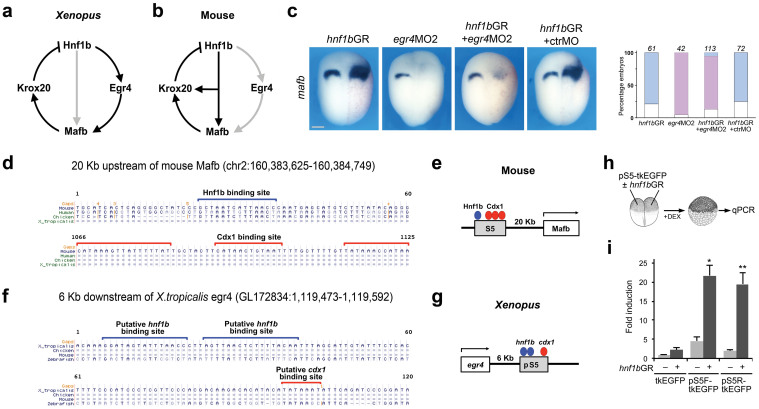
Hnf1b activates egr4 expression in r5/r6
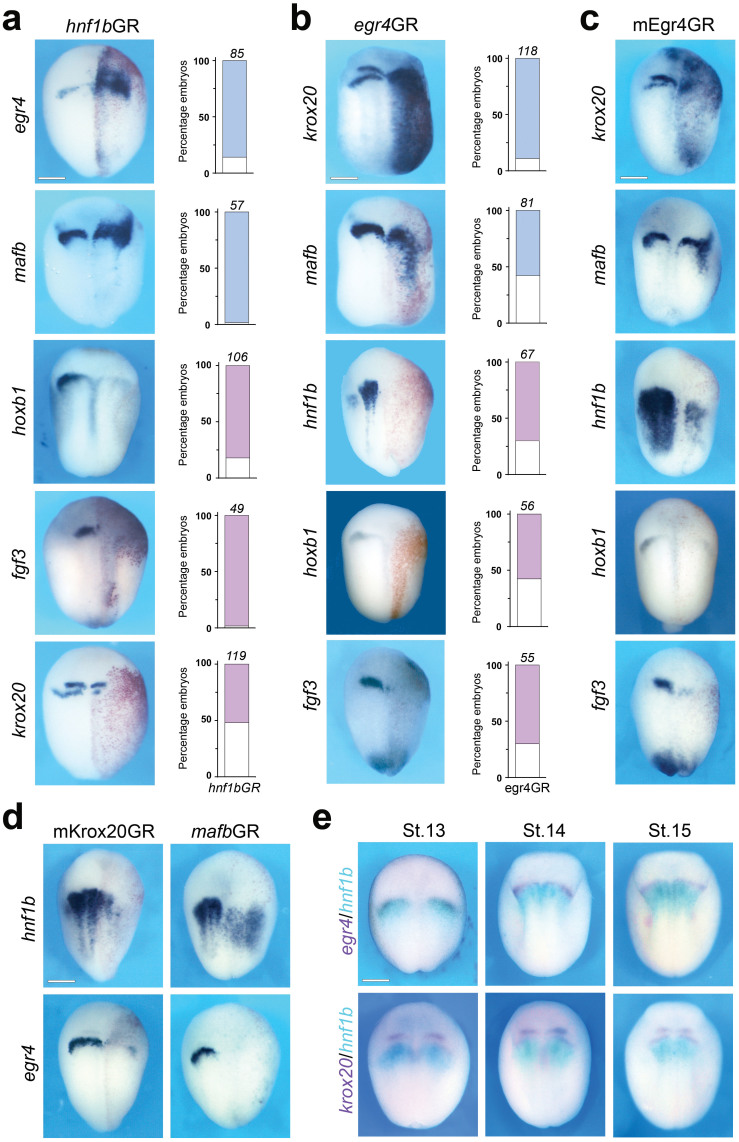
Hnf1b/vHnf1 is a major regulator of posterior hindbrain development where it participates in the formation of the boundary between r4 and r5 and establishes r5 and r6 identities. More specifically, Hnf1b represses r4 identity in r5/r6, and synergizes with r4-derived Fgf3 signal to drive mafb expression in r5/r6, and cooperate with Mafb for the direct transcriptional activation of krox20 in r510,11. To determine the position of Egr4 in this regulatory cascade we performed gain-of-function experiments by injection of mRNAs encoding a hormone inducible version of hnf1b (hnf1bGR). Unilateral injection of hnf1bGR mRNAs at the 2-cell stage caused an anterior expansion of egr4 and mafb expression domains, associated with a down-regulation of hoxb1 and fgf3 (Fig 4a), indicative of a loss of r4 identity. The expression of krox20 was reduced in these embryos (Fig 4a), since r4-derived Fgf signals are required to maintain krox20 expression in r531,32. These results suggest that egr4 is activated by Hnf1b to establish r5 identity. Consistent with this view, egr4 gain-of-function by injection of Xenopus egr4 (egr4GR) or mouse Egr4 (mEgr4GR) mRNAs caused a dramatic expansion of krox20 and mafb expression domains, often extending to the entire injected side (Fig 4b–c). The expression of hnf1b, hoxb1, and fgf3 was strongly repressed in these embryos (Fig 4b–c).
Figure 4. Cross-regulation of Hnf1b and Egr4.
(a) Injection of 100 pg hnf1bGR mRNA expands egr4 and mafb expression anteriorly, while repressing hoxb1 and fgf3 expression. (a) Injection of 100 pg egr4GR mRNA expands krox20 and mafb expression and represses hnf1b, hoxb1 and fgf3 expression. The graphs indicate the percentage of embryos with normal (white), reduced/lost (red) or expanded/ectopic (blue) gene expression. The number of embryos analyzed is indicated on top of each bar. (c) Similar results were obtained by injection of 100 pg mouse Egr4 (mEgr4GR) mRNA. (d) Embryos injected with 100 pg of mouse Krox20 (mKrox20GR) or Xenopus mafb (mafbGR) mRNAs exhibit a dramatic reduction of hnf1b and egr4 expression. (e) Two-color in situ hybriidization for egr4/hnf1b and krox20/hnf1b. Overtime, the anterior limit of hnf1b retracts posteriorly and segregates from the r5 egr4 and krox20 expression domains. Dorsal views, anterior to top. In all panels scale bars, 300 μm.
The repression of hnf1b in Egr4GR-injected embryos suggested the existence of a negative feedback loop in the regulation of hnf1b expression in r5/r6 that may involve Egr4 directly or other factors activated by Egr4. To test this possibility, embryos were injected with mRNAs encoding a hormone inducible version of mouse Krox20 (mKrox20GR) or Xenopus mafb (mafbGR), and in both cases we observed a strong inhibition of hnfb1 expression as well as a loss of egr4 on the injected side (Fig 4d), indicating that Krox20 and Mafb are the likely factors repressing hnf1b expression in r5/r6. In most vertebrates, Hnf1b is initially expressed in the posterior hindbrain with an anterior limit at the r4/r5 boundary33,34,35. Later, as Krox20 is activated in r5 this anterior limit progressively retracts posteriorly, and hnf1b is eventually excluded from r5 and r621,33,34,35. We performed two-color in situ hybridization to assess the spatio-temporal expression of egr4 and hnf1b. At stage 13, egr4 overlaps with hnf1b expression domain anteriorly, and as development proceeds, the anterior limit of hnf1b is shifted posteriorly and segregates from egr4 expression domain (Fig 4e).
Egr4 directly regulates mafb expression
Our results suggest that Hnf1b induces mafb and krox20 expression indirectly through the activation of Egr4 (Fig 5a). This is in contrast to what has been described in the mouse in which Hnf1b has been shown to directly activate Mafb expression34,36 (Fig 5b). To determine whether the activation of mafb by Hnf1b is primarily mediated through Egr4 in Xenopus, we evaluated the ability of hnf1bGR to activate mafb in the absence of Egr4 function. Co-injection of egr4MO2 with hnf1bGR mRNA completely blocked endogenous as well as ectopic mafb expression by Hnf1b (Fig. 5c). These results support the view that Hnf1b activates mafb via Egr4 in Xenopus.
Figure 5. Hnf1b directly activates egr4 expression.
(a) In Xenopus Hnf1b induces mafb and krox20 indirectly through the activation of egr4. (b) In the mouse Hnf1b directly activates mafb and krox20 expression. (c) The expansion of mafb in embryos injected with hnf1bGR mRNA (10 pg) is completely blocked by co-injection of egr4MO2, but not by a control morpholino (ctrMO). Scale bar, 300 μm. The graph indicates the percentage of embryos with normal (white), reduced/lost (red) or expanded (blue) mafb expression. The number of embryos analyzed is indicated on top of each bar. (d) A conserved region upstream of the Mafb gene containing and Hnf1 binding site has been characterized as an r5/r6-specific enhancer (S5). (f) A putative S5 enhancer (pS5) downstream of Xenopus tropicalis egr4 containing two putative Hnf1 binding sites was identified using rVISTA 2.0. (e, g) Schematic representation of the S5 and pS5 enhancer around Mafb (mouse) and egr4 (Xenopus) genes, respectively. (h) Experimental procedure to test the activity of the pS5 in Xenopus embryo. (i) Fold EGFP induction analyzed by qPCR. Values are normalized to ef1α. Graph represents mean ± S.E. of 3 independent experiments. *, P<0.05; **, P<0.01; versus embryos injected with each vector alone. Student's t-test.
In the mouse, Hnf1b directly binds with an evolutionally conserved region of DNA upstream of the Mafb gene, known as the S5 enhancer34,36. This region also contains three Cdx1 binding sites. In the hindbrain Cdx1 has an anterior limit of expression at the prospective r6/r7 boundary, and Cdx1 represses Mafb expression in this region, leading to the characteristic r5/r6 expression of Mafb36. The S5 enhancer has been mapped to a region 20 Kb upstream of the mouse Mafb gene. Sequence alignments indicate that this regulatory module is well conserved in human and chicken. A similar region could not be found around Xenopus tropicalis mafb gene using the UCSC Gene browser (Fig 5d,e). However, a region with similar characteristics, containing two putative Hnf1b binding sites and a single putative Cdx1 binding site, was identified 6Kb downstream of Xenopus tropicalis egr4 gene (Fig 5f,g), here referred as a putative S5 enhancer (pS5). To determine whether this regulatory module can drive egr4 expression, pS5 was cloned in forward (pS5F) and reverse (pS5R) orientation into ptkEGFP vector containing a basal thymidine kinase promoter driving enhanced GFP expression37. Both constructs, pS5F-ptkEGFP and pS5R-ptkEGFP, were co-injected with hnf1bGR mRNA at the 2-cell stage and immediately treated with dexamethasone. Embryos were cultured for 6 hours and analyzed by qPCR (Fig 5h). We observed a significant up-regulation of EGFP transcripts in embryos co-injected with hnf1bGR mRNA and pS5F-ptkEGFP (4.6 fold) or pS5R-ptkEGFP (10.2 fold) as compared to embryos injected with each vector alone (Fig 5i). These results indicate that Hnf1b may directly activate egr4 expression via the pS5 enhancer.
Discussion
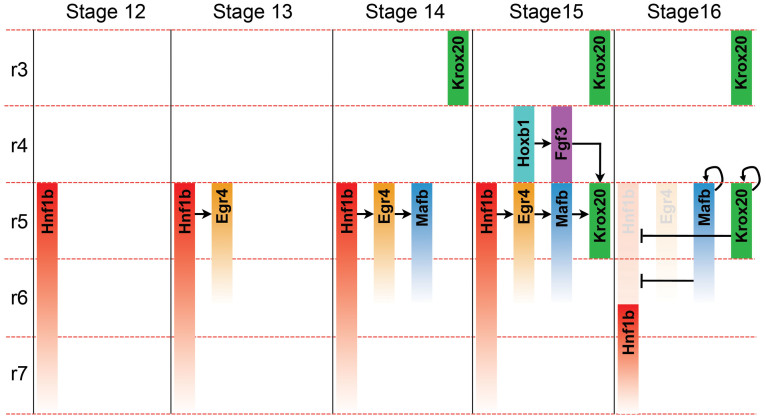
Our findings reveal a novel function for Egr4 in posterior hindbrain development. Egr4 is specifically and transiently activated in r5/r6 by Hnf1b, and is required for mafb and krox20 expression in r5/r6 and r5, respectively (Fig 6). In addition, Egr4 knockdown leads to a specific loss of the anterior branchial neural crest stream that normally travels to the third branchial arch, and inner ear defects. While a recent study has implicated Egr4 in the differentiation of the brain primordia during planarian regeneration38, our study is the first report describing a role for Egr4 in the vertebrate nervous system.
Figure 6. Model for the temporal regulation of r5 identity by Hnf1b, Egr4, Mafb and Krox20 in Xenopus posterior hindbrain.
Using a combination of gain- and loss-of-function approaches we provide evidence that Egr4 is part of a conserved regulatory cascade, involving Hnf1b, Mafb and Krox20, essential to specify r5 identity in the hindbrain. Hnf1b/vHnf1 is a major regulator of posterior hindbrain development. It is believed to repress r4 identity in r5/r6, and to synergize with r4-derived FGF signals to directly drive mafb expression in r5/r6. Hnf1b eventually cooperates with Mafb for the direct transcriptional activation of krox20 in r510,11. Our gain-of-function studies indicate that Hnf1b can promote egr4 and mafb expression. Moreover, ectopic expression of egr4 is sufficient to activate mafb and krox20 in the embryo, suggesting that Hnf1b may control the expression of these factors indirectly in Xenopus, through the activation of Egr4. Consistent with this possibility, hnf1bGR is unable to rescue mafb expression in Egr4-depleted embryos. This is in contrast to what has been reported in other species in which Hnf1b directly activates Mafb and Krox20 expression11,21,36,39. Once activated by Hnf1b in r5/r6, Egr4 induces the expression of mafb1 and krox20, which in turns act as negative regulators of Hnf1b to restrict its expression to a level posterior to r6. Accordingly, ectopic expression of Krox20 or Mafb represses hnf1b expression in the caudal hindbrain. The inhibitory activity of Krox20 in restricting Hnf1b expression posteriorly has been well described in other species33,34,35. This negative feedback loop may also account for the transient expression of egr4 in r5/r6 in Xenopus, which appears to require continued Hnf1b input for its maintenance. The expression of Krox20 and Mafb1 is maintained primarily through an autoregulatory mechanism36,39,40 (Fig 6).
Egr4 belongs to the early growth response factor family of transcription factors. Members of this family were initially identified as genes rapidly induced by growth factors. A defining feature of Egr proteins is a highly conserved DNA-binding domain composed of three zinc finger motifs that bind a 9-bp response element to initiate transcriptional response41. Egr2/Krox20, the most extensively studied member of this family, is essential for the development of r3/r5 by directly activating Hoxa2 and Hoxb2 in r3/r5 and Hoxb3 in r542,43,44. Egr2/Krox20 mutant mouse embryos also have peripheral nerve myelination defects45. Egr1/Krox24 plays a critical role in regulating the luteinizing hormone (LH) β gene expression; Egr1-deficient mice have low levels of LHβ in the pituitary and low levels of LH in serum, and this appears to primarily affect female fertility46,47. Egr3 is required for muscle spindle morphogenesis and normal proprioception in mice48.
Egr4 is detected at low levels in testicular germ cells and Egr4-deficient mice are characterized by a defect in germ cell maturation, leading to male infertility49. Egr4 is also expressed at high level in postnatal rat brain50, and at E13.5 in the mouse spinal cord51. Beside this report, there is no detailed description of Egr4 developmental expression in the mouse. Egr4 mutant mouse embryos exhibit no obvious phenotypic abnormalities in the nervous system. However, embryos have not been examined specifically for defects in the developing brain. It is possible that the effects may be transient or subtle. Alternatively, Egr4 may not be an essential component of the regulatory cascade controlling r5 identity in mammals. Interestingly, mouse Egr4 can fully compensate for the loss of Egr4 in frog embryos, and in overexpression studies its function is indistinguishable from that of Xenopus Egr4. Therefore, the difference in the regulation of r5 identity between frog and mouse cannot be explained by intrinsic functional differences in the Egr4 proteins.
In the mouse, Hnf1b regulates Mafb expression through the S5 enhancer36, a region highly conserved in chicken and human. This enhancer could not be identified around Xenopus tropicalis mafb gene. However, we found that a regulatory module similar to the S5 enhancer, containing two Hnf1b binding sites, was present downstream of the egr4 gene in Xenopus tropicalis, a region also conserved in zebrafish. Using a reporter assay in the embryo we were able to show that Hnf1b can activate this putative enhancer. We propose that rearrangements in the regulatory regions around egr4 and mafb genes may account for the evolutionary changes in the importance of Egr4 in establishing r5 identity in the hindbrain of amniotes versus anamniotes.
Experimental Procedures
Plasmid constructs and morpholino antisense oligonucleotides
The full-length Xenopus laevis egr4 was obtained by RACE-PCR using SMARTerTM RACE cDNA amplification Kit (Clontech, Mountain View CA) according to the manufacturer's instructions. Briefly, a first-strand cDNA was synthesized from 1 μg of total RNA isolated from neurula stage embryos. Two sets of gene-specific primers (GSP), 5′-RACE-egr4 and 3′-RACE-egr4 (5′-AGCAAGGGCTGTGGCGACTTGACTGT CT-3′ and 5′-AGCCGAAGCGTCAAAGTGGGAAACGCGA-3′) were designed based on EST sequences (Unigene ID Xl.53808). The RACE-PCR conditions were as follows: 5 cycles at 94°C for 30 sec and at 72°C for 3 min; 5 cycles at 94°C for 30 sec, at 70°C for 30 sec, and 72°C for 3 min; 27 cycles at 94°C for 30 sec, at 68°C for 30 sec, and 72°C for 3 min. 5′-RACE and 3′-RACE PCR products were cloned into pGEM-T Easy vector (Promega, Madison WI) and sequenced. The sequence of Xenopus egr4 has been deposited into GeneBank (Accession # KF957598). The ORF of Xenopus laevis hnf1b and mafb were amplified by PCR from neurula stage embryo cDNA using primers based on published sequences (GeneBank accession # NM_001089811 and NM_001090383). Mouse Egr4 was purchased from Open Biosystems and mouse Krox20 was a gift of Drs. Schneider-Maunoury and Gilardi-Hebenstreit52. Hormone-inducible constructs were generated by sub-cloning the coding region of each gene into pCS2+GR53. These constructs are referred as egr4GR, hnf1bGR, mafbGR, and mKrox20GR. Pax3 (pax3MO15;), Zic1 (zic1MO16;), Meis3 (meis3MO24;) and Egr4 (egr4MO1; CCTGGCAGGAGAGGTCCATCGT TAT, egr4MO2; GTCCTTACCTG ACCTGAGCTGAGT) MOs were purchased from GeneTools. The specificity of the egr4MO1 was tested in an in vitro transcription/translation coupled rabbit reticulocyte lysate assay (Promega, Madison WI) as previously described54. Synthetic mRNAs encoding pax3GR, zic1GR, egr4GR, hnf1bGR, mafbGR, mEgr4GR, mKrox20GR and β-galactosidase were synthesized in vitro using the Message Machine kit (Ambion-Life Technologies, Grand Island NY).
Embryos, injections and explants culture
Xenopus laevis embryos were staged as previously described55 and raised in 0.1X NAM (Normal Amphibian Medium56;). This study was performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocol (# 120311) was approved by the Institutional Animal Care and Use Committee of New York University. Embryos were injected in one blastomere at the 2-cell stage and analyzed by in situ hybridization at stage 15. MOs (40–50 ng) and synthetic mRNA (100 pg) were injected together with 500 pg of β-galactosidase mRNA as a lineage tracer. Embryos injected with mRNA encoding the hormone inducible version of Xenopus egr4 (egr4GR), hnf1b (hnf1bGR) and mafb (mafbGR), and mouse Egr4 (mEgr4GR) and Krox20 (mKrox20) embryos were treated at stage 12.5 with 10 μM dexamethasone (DEX; Sigma-Aldrich, St Louis MO) and collected at stage 15/17. For animal cap explant experiments, both blastomeres at the 2-cell stage were injected in the animal pole region 100 pg of mRNAs encoding pax3GR and zic1GR, explants were dissected at the late blastula stage and immediately cultured for several hours in NAM 0.5X plus 10 μM of dexamethasone. In some experiments, 10 μg/ml cycloheximide (CHX; Sigma-Aldrich, St Louis MO) was added to the culture medium to block protein synthesis, in which case explants were pretreated with cycloheximide for 30 min before dexamethasone treatment53. The explants were subsequently analyzed by quantitative PCR (qPCR).
Lineage tracing, in situ hybridization and histology
Xenopus embryos were fixed in MEMFA and processed for Red-Gal (Research Organics, Clevelan OH) staining to visualize the lineage tracer (β-galactosidase) prior to in situ hybridization. Whole-mount in situ hybridization was performed as previously described57. Digoxygenin (DIG)- and fluorescein isothiocyanate (FITC)-labeled antisense RNA probes (Roche Diagnostics, Indianapolis, IN) were synthesized using template cDNA encoding Zic114, Pax314, En217, Krox2028, Mafb58, Hnf1b59, Meis324, Fgf360, Hoxb161, Hoxa362, Snail222, Sox963, Sox1064 and Tfap2a65. For double in situ hybridization DIG- and FITC-labeled probes were hybridized simultaneously and sequentially detected using anti-FITC and anti-DIG alkaline phosphatase conjugated antibodies. FITC-labeled probe was visualized first using 4-toluidine salt (BCIP; Roche Diagnostics, Indianapolis, IN) and after inactivation of the anti-FITC antibody, the color reaction for the DIG-labeled probe was performed using Magenta Phosphate (5-bromo-6chloro-3indoxyl phosphate; Biosynth, Itasca IL). For histology, fixed embryos were embedded into Paraplast+, 12 µm sections cut on a rotary microtome (Olympus, Center Valley PA) and counterstained with Hematoxylin.
RNA preparation and qPCR
Total RNA from whole embryos was extracted using RNeasy Micro Kit (Qiagen, Valencia CA) according to the manufacturer's instructions. During the extraction procedure, the samples were treated with DNase I to eliminate possible genomic DNA contamination. The amount of isolated RNA was quantified using a Nanodrop spectrophotometer (Nanodrop Technologies; Wilmington, DE). For mRNA extraction from animal cap explants, pools of 8 explants were homogenized and mRNA was isolated using Dynabeads® mRNA DIRECTTM Micro Kit (Invitrogen, Grand Island NY). cDNA synthesis from total RNA (whole embryos) and mRNA (animal cap explants) were performed using Superscript VILO cDNA Synthesis Kit (Invitrogen, Grand Island, NY) according to the manufacturer's instructions. Quantitative PCR was performed on an Eco Real-Time PCR System (Illumina, San Diego CA) using the primers shown in Table 1 and the Power SYBR Green PCR Master Mix (Invitrogen, Grand Island NY). The reaction mixture consisted of 10 µl of Power SYBR Green PCR Master Mix, 200 nM primers, and 2 µl of cDNA in a total volume of 10 µl. The PCR conditions were as follows: incubation at 50°C for 2 min; activation at 95°C for 10 min; 40 cycles at 95°C for 10 s and at 60°C for 30 s; melt curve at 95°C for 15 s, 55°C for 15 s, and at 95°C for 15 s. The ΔΔCT method was used to quantify the qPCR results. Each reaction included a standard curve of serial dilution points (in 10-fold increments) of test cDNA. In each case, ornithine decarboxylase 1 (odc1) or elongation factor 1α (ef1α) was used for normalization.
Table 1. Primers for qPCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| hnf1b | 5′-GTTCTGTTCCCAGTTGTTTGTT-3′ | 5′-GGATATACCACGTGAGCTTCTG-3′ |
| egr4 | 5′-AACGCGAGACCGTGTTATT-3′ | 5′-GGAACAGAAAGTGCGTGTAAATAG-3′ |
| krox20 | 5′-TCTCCTACTCGTCCAACTACC-3′ | 5′-CGCTCACTAGATTGAAGATCCC-3′ |
| mafb | 5′-GACGCAGTAGAAGCCCTTATT-3′ | 5′-GATGCTGGTTCTGGTGATGA-3′ |
| odc1 | 5′-ACATGGCATTCTCCCTGAAG-3′ | 5′-TGGTCCCAAGGCTAAAGTTG-3′ |
| ef1α | 5′-ACCCTCCTCTTGGTCGTTTT-3′ | 5′-TTTGGTTTTCGCTGCTTTCT-3′ |
| egfp | 5′-GAACCGCATCGAGCTGAA-3′ | 5′-TGCTTGTCGGCCATGATATAG-3′ |
Isolation of conserved regions
Using the UCSC Gene browser, we identified regions in Xenopus tropicalis genome containing two putative Hnf1b binding sites and a cdx1 binding site in the vicinity of Egr4 coding region (pS5). Using PCR, we amplified this putative regulatory region from Xenopus tropicalis genomic DNA. The pS5 module was cloned in forward and reverse orientation upstream of ptkEGFP vector that contains the Herpes simplex virus thymidine kinase basal promoter driving enhanced GFP expression37. These constructs are referred as pS5F-ptkEGFP and pS5R-ptkEGFP, respectively.
Author Contributions
C.-J.B. and J.-P.S.-J. designed the experiments, prepared the figures and wrote the manuscript. C.-J.B., J.J. and J.-P.S-J. performed the experiments and analyzed the data.
Supplementary Material
Supplementary Figures
Acknowledgments
We are grateful to Dr. Jane McCutcheon for comments on the manuscript and to Mr. Arun Devotta for technical assistance. We thank Dr. Marianne Bronner, Dr. Pascale Giraldi-Hebenstreit, Dr. Sylvie Schneider-Monoury, Dr. Hajime Ogido and Dr. Jing Yang for reagents. We thank members of the Saint-Jeannet and Jeong laboratories for discussions. This work was supported by a grant from the National Institutes of Health to J.-P. S.-J.(R01-DE014212).
Funding Work was funded by a National Institutes of Health (NIDCR) grant [R01-DE014212] to J-P. S-J.
References
- Trainor P. A. & Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci 1, 116–124 (2000). [DOI] [PubMed] [Google Scholar]
- Santagati F. & Rijli F. M. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci 4, 806–818 (2003). [DOI] [PubMed] [Google Scholar]
- Le Douarin N. M. K. C. The neural crest. 2nd edn, (Cambridge University Press, 1999). [Google Scholar]
- Schneider-Maunoury S. & Pujades C. Hindbrain signals in otic regionalization: walk on the wild side. Int J Dev Biol 51, 495–506 (2007). [DOI] [PubMed] [Google Scholar]
- Gavalas A. & Krumlauf R. Retinoid signalling and hindbrain patterning. Curr Opin Genet Dev 10, 380–386 (2000). [DOI] [PubMed] [Google Scholar]
- Irving C., Nieto M. A., DasGupta R., Charnay P. & Wilkinson D. G. Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Dev Biol 173, 26–38 (1996). [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G., Bhatt S., Chavrier P., Bravo R. & Charnay P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 337, 461–464 (1989). [DOI] [PubMed] [Google Scholar]
- Cordes S. P. & Barsh G. S. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79, 1025–1034 (1994). [DOI] [PubMed] [Google Scholar]
- Prince V. E., Moens C. B., Kimmel C. B. & Ho R. K. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development 125, 393–406 (1998). [DOI] [PubMed] [Google Scholar]
- Wiellette E. L. & Sive H. vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development 130, 3821–3829 (2003). [DOI] [PubMed] [Google Scholar]
- Hernandez R. E., Rikhof H. A., Bachmann R. & Moens C. B. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development 131, 4511–4520 (2004). [DOI] [PubMed] [Google Scholar]
- Studer M. et al. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125, 1025–1036 (1998). [DOI] [PubMed] [Google Scholar]
- Bae C. J. et al. Identification of Pax3 and Zic1 targets in the developing neural crest. Dev Biol 386, 473–483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C. S. & Saint-Jeannet J. P. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell 18, 2192–2202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq A. H., Wang E. & Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell 8, 167–178 (2005). [DOI] [PubMed] [Google Scholar]
- Sato T., Sasai N. & Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development 132, 2355–2363 (2005). [DOI] [PubMed] [Google Scholar]
- Brivanlou A. H. & Harland R. M. Expression of an engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development 106, 611–617 (1989). [DOI] [PubMed] [Google Scholar]
- Schneider-Maunoury S. et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell 75, 1199–1214 (1993). [DOI] [PubMed] [Google Scholar]
- Maves L., Jackman W. & Kimmel C. B. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development 129, 3825–3837 (2002). [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Martin G. R., Cordes S. P., Halamek L. P. & Barsh G. S. Altered rhombomere-specific gene expression and hyoid bone differentiation in the mouse segmentation mutant, kreisler (kr). Development 117, 925–936 (1993). [DOI] [PubMed] [Google Scholar]
- Sun Z. & Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev 15, 3217–3229 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., Morgan R. & Sargent M. G. Induction of the prospective neural crest of Xenopus. Development 121, 767–777 (1995). [DOI] [PubMed] [Google Scholar]
- Manzanares M. et al. The role of kreisler in segmentation during hindbrain development. Dev Biol 211, 220–237 (1999). [DOI] [PubMed] [Google Scholar]
- Dibner C., Elias S. & Frank D. XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development 128, 3415–3426 (2001). [DOI] [PubMed] [Google Scholar]
- Gutkovich Y. E. et al. Xenopus Meis3 protein lies at a nexus downstream to Zic1 and Pax3 proteins, regulating multiple cell-fates during early nervous system development. Dev Biol 338, 50–62 (2010). [DOI] [PubMed] [Google Scholar]
- Sadaghiani B. & Thiebaud C. H. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol 124, 91–110 (1987). [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. & Jessell T. M. Retinoic acid modifies the pattern of cell differentiation in the central nervous system of neurula stage Xenopus embryos. Development 112, 945–958 (1991). [DOI] [PubMed] [Google Scholar]
- Bradley L. C., Snape A., Bhatt S. & Wilkinson D. G. The structure and expression of the Xenopus Krox-20 gene: conserved and divergent patterns of expression in rhombomeres and neural crest. Mech Dev 40, 73–84 (1993). [DOI] [PubMed] [Google Scholar]
- Deol M. S. The Abnormalities of the Inner Ear in Kreisler Mice. J Embryol Exp Morphol 12, 475–490 (1964). [PubMed] [Google Scholar]
- Moens C. B., Yan Y. L., Appel B., Force A. G. & Kimmel C. B. valentino: a zebrafish gene required for normal hindbrain segmentation. Development 122, 3981–3990 (1996). [DOI] [PubMed] [Google Scholar]
- Marin F. & Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development 127, 4925–4935 (2000). [DOI] [PubMed] [Google Scholar]
- Weisinger K., Kayam G., Missulawin-Drillman T. & Sela-Donenfeld D. Analysis of expression and function of FGF-MAPK signaling components in the hindbrain reveals a central role for FGF3 in the regulation of Krox20, mediated by Pea3. Dev Biol 344, 881–895 (2010). [DOI] [PubMed] [Google Scholar]
- Aragon F. et al. vHnf1 regulates specification of caudal rhombomere identity in the chick hindbrain. Dev Dyn 234, 567–576 (2005). [DOI] [PubMed] [Google Scholar]
- Kim F. A. et al. The vHNF1 homeodomain protein establishes early rhombomere identity by direct regulation of Kreisler expression. Mech Dev 122, 1300–1309 (2005). [DOI] [PubMed] [Google Scholar]
- Lecaudey V., Anselme I., Rosa F. & Schneider-Maunoury S. The zebrafish Iroquois gene iro7 positions the r4/r5 boundary and controls neurogenesis in the rostral hindbrain. Development 131, 3121–3131 (2004). [DOI] [PubMed] [Google Scholar]
- Sturgeon K. et al. Cdx1 refines positional identity of the vertebrate hindbrain by directly repressing Mafb expression. Development 138, 65–74 (2011). [DOI] [PubMed] [Google Scholar]
- Barembaum M. & Bronner M. E. Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Dev Biol 382, 567–575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraguas S., Barberan S., Iglesias M., Rodriguez-Esteban G. & Cebria F. egr-4, a target of EGFR signaling, is required for the formation of the brain primordia and head regeneration in planarians. Development 141, 1835–1847 (2014). [DOI] [PubMed] [Google Scholar]
- Chomette D., Frain M., Cereghini S., Charnay P. & Ghislain J. Krox20 hindbrain cis-regulatory landscape: interplay between multiple long-range initiation and autoregulatory elements. Development 133, 1253–1262 (2006). [DOI] [PubMed] [Google Scholar]
- Ghislain J., Desmarquet-Trin-Dinh C., Gilardi-Hebenstreit P., Charnay P. & Frain M. Neural crest patterning: autoregulatory and crest-specific elements co-operate for Krox20 transcriptional control. Development 130, 941–953 (2003). [DOI] [PubMed] [Google Scholar]
- O'Donovan K. J., Tourtellotte W. G., Millbrandt J. & Baraban J. M. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci 22, 167–173 (1999). [DOI] [PubMed] [Google Scholar]
- Sham M. H. et al. The zinc finger gene Krox20 regulates HoxB2 (Hox2.8) during hindbrain segmentation. Cell 72, 183–196 (1993). [DOI] [PubMed] [Google Scholar]
- Nonchev S. et al. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 122, 543–554 (1996). [DOI] [PubMed] [Google Scholar]
- Manzanares M. et al. Krox20 and kreisler co-operate in the transcriptional control of segmental expression of Hoxb3 in the developing hindbrain. EMBO J 21, 365–376 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P. et al. Krox-20 controls myelination in the peripheral nervous system. Nature 371, 796–799 (1994). [DOI] [PubMed] [Google Scholar]
- Lee S. L. et al. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 273, 1219–1221 (1996). [DOI] [PubMed] [Google Scholar]
- Topilko P. et al. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol 12, 107–122 (1998). [DOI] [PubMed] [Google Scholar]
- Tourtellotte W. G. & Milbrandt J. Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat Genet 20, 87–91 (1998). [DOI] [PubMed] [Google Scholar]
- Tourtellotte W. G., Nagarajan R., Auyeung A., Mueller C. & Milbrandt J. Infertility associated with incomplete spermatogenic arrest and oligozoospermia in Egr4-deficient mice. Development 126, 5061–5071 (1999). [DOI] [PubMed] [Google Scholar]
- Crosby S. D. et al. Neural-specific expression, genomic structure, and chromosomal localization of the gene encoding the zinc-finger transcription factor NGFI-C. Proc Natl Acad Sci U S A 89, 6663 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. et al. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci 29, 11399–11408 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi P., Schneider-Maunoury S. & Charnay P. Krox-20: a candidate gene for the regulation of pattern formation in the hindbrain. Biochimie 73, 85–91 (1991). [DOI] [PubMed] [Google Scholar]
- Kolm P. J. & Sive H. L. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol 171, 267–272 (1995). [DOI] [PubMed] [Google Scholar]
- Park B. Y., Hong C. S., Weaver J. R., Rosocha E. M. & Saint-Jeannet J. P. Xaml1/Runx1 is required for the specification of Rohon-Beard sensory neurons in Xenopus. Dev Biol 362, 65–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P. D. & Faber J. Normal table of Xenopus laevis (Daudin). (North Holland Publishing Company, 1967). [Google Scholar]
- Slack J. M. & Forman D. An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. J Embryol Exp Morphol 56, 283–299 (1980). [PubMed] [Google Scholar]
- Harland R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 36, 685–695 (1991). [DOI] [PubMed] [Google Scholar]
- Ishibashi S. & Yasuda K. Distinct roles of maf genes during Xenopus lens development. Mech Dev 101, 155–166 (2001). [DOI] [PubMed] [Google Scholar]
- Demartis A., Maffei M., Vignali R., Barsacchi G. & De Simone V. Cloning and developmental expression of LFB3/HNF1 beta transcription factor in Xenopus laevis. Mech Dev 47, 19–28 (1994). [DOI] [PubMed] [Google Scholar]
- Lombardo A., Isaacs H. V. & Slack J. M. Expression and functions of FGF-3 in Xenopus development. Int J Dev Biol 42, 1101–1107 (1998). [PubMed] [Google Scholar]
- Godsave S. et al. Expression patterns of Hoxb genes in the Xenopus embryo suggest roles in anteroposterior specification of the hindbrain and in dorsoventral patterning of the mesoderm. Dev Biol 166, 465–476 (1994). [DOI] [PubMed] [Google Scholar]
- Lee Y. H. et al. Early development of the thymus in Xenopus laevis. Dev Dyn 242, 164–178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokony R. F., Aoki Y., Saint-Germain N., Magner-Fink E. & Saint-Jeannet J. P. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development 129, 421–432 (2002). [DOI] [PubMed] [Google Scholar]
- Aoki Y. et al. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol 259, 19–33 (2003). [DOI] [PubMed] [Google Scholar]
- Luo T., Lee Y. H., Saint-Jeannet J. P. & Sargent T. D. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc Natl Acad Sci U S A 100, 532–537 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures