Significance
Photoactivatable proteins are powerful tools for studying biological processes. Light-induced dimers are especially useful because they can be turned on and off with high spatial and temporal resolution in living systems, allowing for control of protein localization and activity. Here, we develop and apply methods for identifying mutations that improve the effectiveness of a light-induced dimer. The engineered switch is modular, can be used in most organisms, has more than 50-fold change in binding affinity upon light stimulation, and can be used to initiate signaling pathways in a specific region of a cell.
Keywords: optogenetic tool, PER-ARNT-SIM domain, computational library, phage display, Rosetta molecular modeling suite
Abstract
The discovery of light-inducible protein–protein interactions has allowed for the spatial and temporal control of a variety of biological processes. To be effective, a photodimerizer should have several characteristics: it should show a large change in binding affinity upon light stimulation, it should not cross-react with other molecules in the cell, and it should be easily used in a variety of organisms to recruit proteins of interest to each other. To create a switch that meets these criteria we have embedded the bacterial SsrA peptide in the C-terminal helix of a naturally occurring photoswitch, the light-oxygen-voltage 2 (LOV2) domain from Avena sativa. In the dark the SsrA peptide is sterically blocked from binding its natural binding partner, SspB. When activated with blue light, the C-terminal helix of the LOV2 domain undocks from the protein, allowing the SsrA peptide to bind SspB. Without optimization, the switch exhibited a twofold change in binding affinity for SspB with light stimulation. Here, we describe the use of computational protein design, phage display, and high-throughput binding assays to create an improved light inducible dimer (iLID) that changes its affinity for SspB by over 50-fold with light stimulation. A crystal structure of iLID shows a critical interaction between the surface of the LOV2 domain and a phenylalanine engineered to more tightly pin the SsrA peptide against the LOV2 domain in the dark. We demonstrate the functional utility of the switch through light-mediated subcellular localization in mammalian cell culture and reversible control of small GTPase signaling.
Inducible protein dimers are complexes that form when a specific stimulus is provided; for example, the protein FRB binds to the protein FKBP12 in the presence of rapamycin (1). Inducible dimers are powerful research tools because with genetic engineering they can be used to localize and activate proteins in living systems (2–5). For example, by fusing one half of an inducible dimer to a DNA binding domain and the other half to a transcription activation domain, transcription of target genes can be initiated by providing the stimulus that induces dimerization. Chemically induced dimers have been used to control a wide variety of biological processes but are limited by irreversibility and lack of spatial control within a cell. For this reason, there is strong interest in light-inducible dimers (LIDs) that can be activated in specific regions of a cell or an organism using light in a reversible manner.
Several LIDs are currently available and have been used to control signaling pathways in living cells. In almost all cases, the dimers are derived from naturally occurring photoactivable systems. The most widely used pair thus far is cryptochrome 2 (Cry2) and CIB1 from Arabidopsis thaliana. The Cry2/CIB1 pair shows blue light induced dimerization in both yeast and mammalian cell culture. Association occurs on a subsecond time scale and reversion within 10 min (4). The mechanism of light-activation is not fully understood and it has recently been shown that Cry2 oligomerizes into large clusters under blue light in addition to associating with CIB1. This could be a drawback for applications that require precise stoichiometry, but the oligomerization itself has been used for control of protein activation (6). Another dimerization pair is phytochrome B (PhyB) and PIF, also from A. thaliana. PhyB and PIF interact after irradiation with red light and dissociate with exposure to far-red light (3). This system requires a chromophore, phycocyanobilin that is not naturally present in many organisms, including mammals (7, 8). The tunable light-controlled interacting protein tags (TULIPs) make use of the blue light-sensing light-oxygen-voltage (LOV) domain and an engineered PDZ domain (9). Subcellular localization has been shown with TULIPs in both yeast and mammalian cells. However, the presence of a PDZ binding peptide and PDZ domain in the system could lead to cross-talk with endogenous signaling pathways. The FKF1/GIGANTEA (GI) heterodimer pair from A. thaliana also relies on a LOV domain for light-activated binding. This pair dimerizes in response to blue light in mammalian cells, associating within a few minutes of activation and dissociating on the order of a few hours (2). However, GI is a very large protein and the switching power of the pair is sensitive to expression levels.
An ideal LID would be small and modular, associate quickly in the presence of signal and quickly dissociate in its absence, and be fully orthogonal to the organisms in which they are to be used. To fulfill these criteria, we chose to use the LOV2 domain of phototropin 1 from Avena sativa (As) as the photoactive element of our LID. In its native setting, the AsLOV2 domain senses blue light and activates a C-terminal kinase domain. AsLOV2 can be genetically encoded and its chromophore, flavin mononucleotide, is abundant in most organisms. It is monomeric in dark state and remains so under activating blue light. The structure of AsLOV2 has been determined, making it amenable to structure-guided engineering. AsLOV2 is made up of a core per-arnt-sim (PAS) fold with flanking α-helicies on the N- and C-termini (10). Upon blue light irradiation, a conserved cysteine residue in the core of AsLOV2 becomes covalently bound to the flavin cofactor, structural rearrangement is passed along through the PAS fold, and ultimately the flanking helices unfold from the PAS core (11, 12). The covalent bond breaks with a half-life of 30–50 s, and the helices refold to their dark-state conformation (13, 14). To reengineer the AsLOV2 domain to be part of a light-inducible complex, we incorporated a naturally occurring binding element within the protein: seven residues of the Escherichia coli SsrA peptide that bind SspB, a 13-kD adaptor protein also from E. coli. Both SsrA and SspB have been structurally characterized and the SsrA peptide shares sequence identity with the Jα helix of AsLOV2 (15–17). We previously showed that incorporation of SsrA into the Jα helix of AsLOV2 led to steric occlusion of SspB binding in the dark and uncaging with blue light irradiation, yielding a light-inducible heterodimer pair (18). Our original light-inducible dimer (oLID), although containing some of the characteristics desirable for a versatile tool, did not show large changes in binding affinity with light stimulation. Original fusions of AsLOV2 and SsrA yielded a twofold change in affinity for SspB. This was improved to an eightfold switch by incorporating mutations known to stabilize the Jα helix in the dark, as well as including a C-terminal phenylalanine predicted by molecular modeling to further hold the Jα helix against the PAS domain in the dark. However, even in its final form, dimerization in the dark at moderate protein concentrations prevented large light-induced phenotypic changes in vivo.
To create a generalizable, versatile, and more powerful LID, we sought to improve oLID with a novel combination of computational library design and phage-display screening. Using the Rosetta macromolecular software suite, we ranked point mutations within AsLOV2 and created a library of mutations with the goal of improving the dynamic range of oLID, specifically reducing dark-state binding. We then used phage-display and ELISA-based binding assays to screen for mutations that both weakened dark-state binding and responded to light activation. Here, we describe two improved light-inducible (iLID) pairs that we identified with this approach: iLID nano, which switches from 130 nM under blue light to 4.7 µM in the dark, and iLID micro, which switches from 800 nM under blue light to 47 µM in the dark. iLID nano and iLID micro both colocalize under blue light within seconds, revert to dark state within minutes, and can be activated subcellularly in mammalian cell culture. To demonstrate functional utility in cells, we show light-dependent control of GTPase activity and the cytoskeleton through localization of the DH/PH domains of Intersectin (ITSN) and Tiam1.
Results
Computational Library Screened by Phage Display and Protein ELISA.
In oLID there was appreciable binding in the dark, ∼800 nM. Previous studies suggest that a major source of dark state “leakiness” comes from transient undocking and unfolding of AsLOV2’s Jα-helix, and mutations that increase the intrinsic stability of the Jα helix have proven successful in reducing dark-state activity of light-activated AsLOV2 switches (9, 19). We sought to extend this approach and use molecular modeling combined with high-throughput screening to find mutations throughout the domain that stabilize the docked state of the Jα-helix. The pmut_scan protocol in Rosetta (20) was used to model and score all possible point mutations within 6 Å of the Jα helix using the crystal structure of AsLOV2 in its dark state (PDB ID code 2V0U). All mutations that were predicted to be stabilizing or neutral (within 1 kcal/mole of the wild-type residue) were considered potential mutations that might stabilize the dark state and were used as the basis for creating a directed library. The final point-mutant library consisted of 743 mutations at 49 positions (SI Appendix, Table S4), and used the oLID sequence as the starting sequence (Fig. 1A).
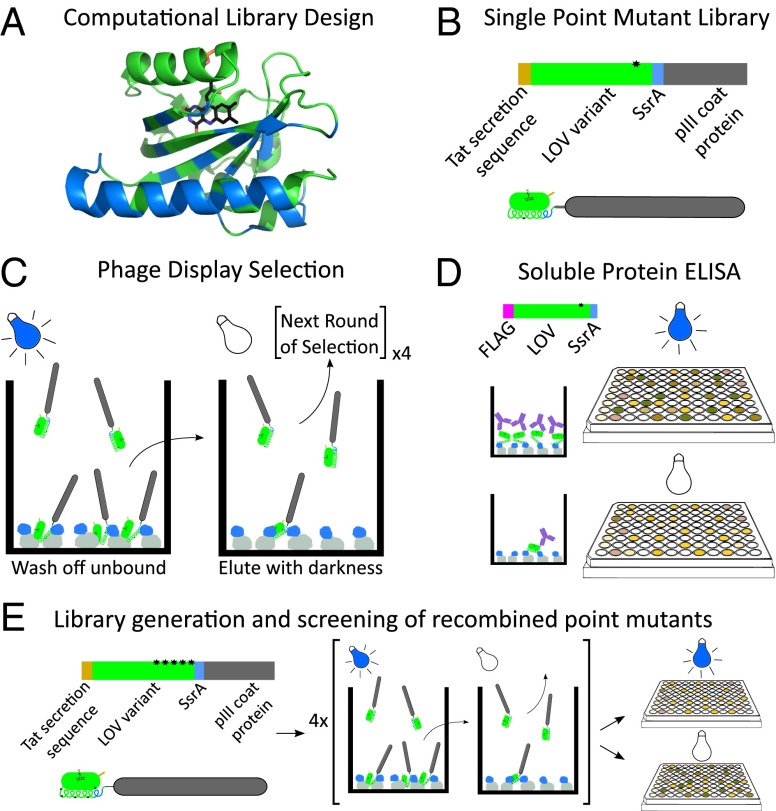
Fig. 1.
Schematic overview of selection and screening protocol for improved LOV variants. (A) Using Rosetta’s pmut_scan, we generated a library of point mutations at 49 positions within the AsLOV2 domain (blue residues). (B) Phage-display construct illustration. (C) The phage library was added to plates coated with MBP-SspB (gray-blue) in the presence of blue light and washed. Plates were moved to the dark and eluted phages were collected. (D) Top single-mutation sequences were recloned as flag tag fusions and individually expressed. Binding of soluble protein to MBP-SspB coated plates was measured after exposure to blue light and sequestration in the dark by ELISA. (E) Mutations with the highest dynamic range were recombined to generate a new library of LOV variants, which was screened using the procedure shown in C and D.
The oLID library was fused with the phage pIII coat protein and an N-terminal Tat (twin-arginine translocation) secretion sequence for display on the surface of phage (21). The Tat secretion pathway was used because it maintains proteins in the folded state during secretion, which allows them to remain bound to cofactors. In this case this was critical, as FMN binding is needed for LOV domain activity (Fig. 1B and SI Appendix, Fig. S10). We screened the library by rounds of positive and negative selection, binding under blue light to immobilized MBP-SspB and subsequent elution in the dark (Fig. 1C). After four rounds of lit and dark screening, individual sequences were tested by soluble protein ELISA (Fig. 1D). We ranked mutations by change in dynamic range, defined as ELISA signal under blue light/ELISA signal in darkness. Single point-mutant LOV variants had minimal improvement to overall switching. The top mutations were pooled and recombined with each other to yield a recombined library. Screening of the recombined library was repeated as before (Fig. 1E). The top 60 sequences had dynamic ranges ranging from 2.4 to 35.4 (SI Appendix, Fig. S1). Four sequences with the highest dynamic ranges were chosen for further characterization (SI Appendix, Fig. S2).
In Vitro Characterization.
We measured the binding affinity of our four top sequences for SspB in the dark and after irradiation with blue light using a competitive fluorescence polarization binding assay. All four had improved dynamic ranges compared with the parent clone oLID (Fig. 2 A and B) and switched roughly over the same affinity range, ∼100 nM to <5 μM (SI Appendix, Fig. S3). Clones D5 and F2 had the highest dynamic range of the clones tested: 43-fold and 36-fold changes, respectively. D5 switched from 77 nM under blue light to 3.4 μM in the dark and F2 switched from 132 nM under blue light to 4.7 μM in the dark. As dark-state activity can be the limiting factor in the usefulness of light-inducible tools, we chose to move forward with clone F2, naming it our improved light-inducible dimer, iLID (Fig. 2 C and E).
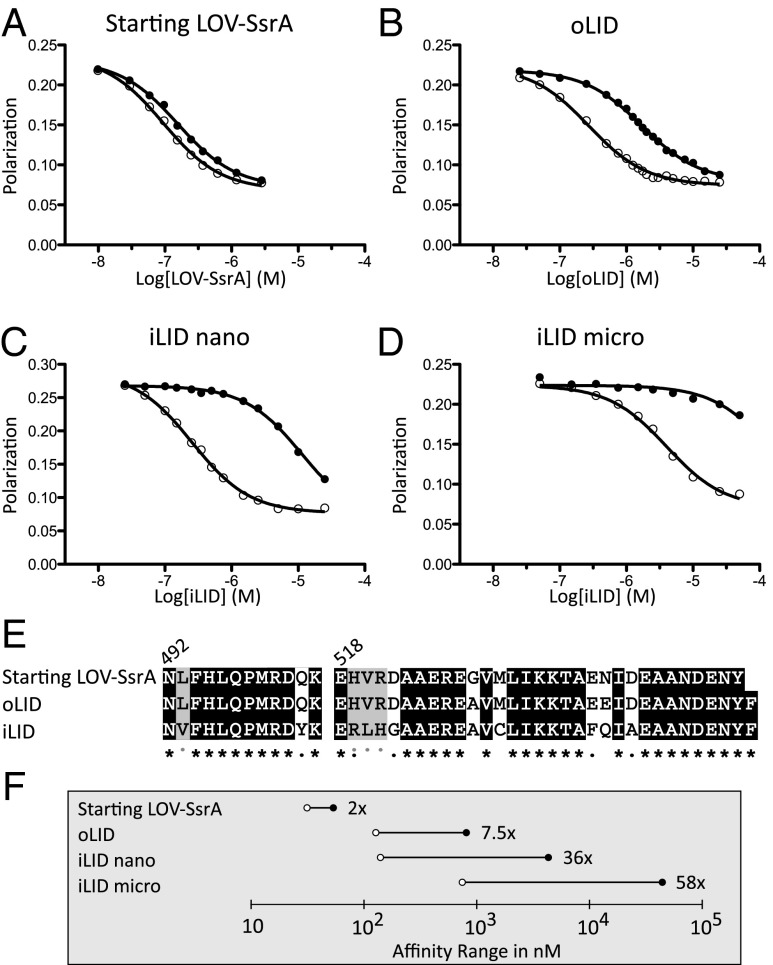
Fig. 2.
Characterization of iLID nano and iLID micro. Competitive fluorescence polarization binding assays measure affinities of heterodimerization under blue light (○) and in the dark (●). (A) Incorporation of SsrA into final turn of the AsLOV2 Jα helix. (B) Addition of helix-strengthening mutations (G528A, N538E) and C-terminal phenylalanine. (C) Top sequence from phage display and screening, iLID, has a 36-fold change in affinity for SspB because of light. (D) A mutation to SspB (R73Q) yields a heterodimer pair that switches over the micromolar range of affinities with a 58-fold change in affinity. (E) Sequence alignment of starting, oLID, and iLID. Black, unmutated; gray, similar amino acid mutation. Gray dots indicate iLID mutations that converged in all top four sequences. (F) Comparison of lit (○) and dark (●) affinities between the original heterodimer pairs and our two new pairs, iLID nano and iLID micro.
The concentration threshold required for activity varies for different signaling proteins within a cell. Having tools that switch over different ranges of affinities may allow a wider range of targets to be controlled. To create an alternate affinity range for iLID as well as retain our improved switching, we made a point mutant, R73Q to SspB, reducing the peptide/protein affinity to 900 nM (SI Appendix, Fig. S4). In the context of SspB R73Q, iLID has an affinity of 800 nM for SspB R73Q under blue light and a dark-state affinity of 47 μM, which is a 58-fold change in binding affinity (Fig. 2D). Thus, two sets of tools are available for light-inducible heterodimerization: iLID nano (makes use of wild-type SspB) and iLID micro (makes use of SspB R73Q). Comparisons of the affinity ranges of these tools to their predecessors can be seen in Fig. 2F.
Structural Characterization of iLID.
iLID contains 10 mutations compared with oLID. Four of the mutations are clustered in the hinge loop that connects the PAS domain to the Jα helix, four are located in the Jα helix, and two are in the PAS domain. To investigate how these mutations are improving the dynamic range of the switch, we solved the crystal structure of iLID in its dark state to a resolution of 2.05 Å (Fig. 3). In general, there are very few structural perturbations when comparing iLID to AsLOV2. Within the core PAS domain (residues 413–516), the Cα RMSD between iLID and AsLOV2 is 0.42 Å, and the hinge loop showed almost no movement in the protein backbone (0.23 Å Cα RMSD) (Fig. 3B). Several of the side-chains in the hinge region are forming interactions that may be important for stabilizing the dark state of the switch. R519 is packed against W491 from the PAS domain. The isoleucine replacing valine 520 is buried against side-chains from the Jα helix and the PAS domain and packs against a valine at position 493 that is a leucine in AsLOV2. The last residue in the hinge, position 522, is an aspartic acid in AsLOV2 and a glycine in iLID. This is the last residue before the Jα helix and has a positive ϕ angle: ϕ = 49.0°, ψ = −126°. Aspartic acids are observed in this region of the ramachandran plot, but glycine is by far the most common amino acid in this region of the plot. In a set of high-resolution crystal structure, 47% of the residues with ϕ near 50° and ψ near −130° are glycine, whereas only 7% are aspartic acids. This finding suggests that there may be some strain that is relieved when the aspartic acid is mutated to a glycine.
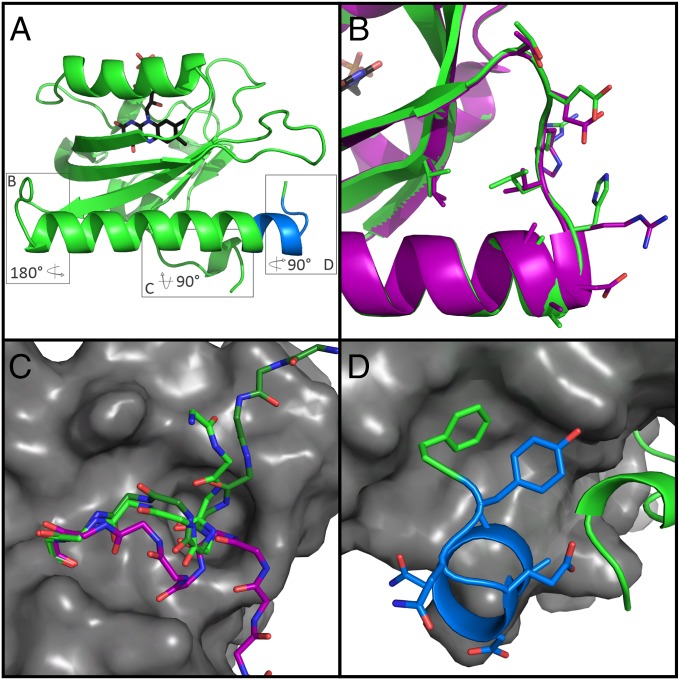
Fig. 3.
Structure of iLID yields insight to role of mutated residues. (A) Overall topology iLID (green) remains unchanged from AsLOV2. Boxes surround three areas of interest to be shown in B, C, and D. (B) Alignment with AsLOV2 (PDB ID code 2V0U, purple) reveals no major backbone rearrangement of the hinge region due to mutations. (C) Close up view of A'α helix from iLID (green), AsLOV2 (purple), and PA-Rac1 (PDB ID code 2WKP, dark green), show different orientation in caged LOV variants from uncaged. (D) SsrA epitope (blue) contains an extra helical turn before wrapping back and interacting with the PAS fold. Designed C-terminal phenylalanine (green) packs nicely into a hydrophobic pocket of the LOV domain.
The largest conformational change in the iLID structure was in the N-terminal residues and the A'α helix. The first two resolved residues, T406 and T407, point toward the Jα helix instead of the Dα helix, as in AsLOV2, and the A'α helix is shifted by 3 Å (Fig. 3C). A smaller ∼1 Å movement is seen in the last 12 residues of the Jα helix toward the A'α helix, allowing for tighter packing between the two. The slight unwinding of the A'α helix allows for the terminal nitrogen of the helix to be capped by E546 of the SsrA sequence. Additionally, the side-chain of E409 forms a hydrogen bond with the tyrosine from the SsrA sequence.
Finally, the structure of iLID revealed the location and conformation of the caged SsrA peptide. The designed C-terminal phenylalanine, F549, fits into a hydrophobic pocket on the surface of the PAS domain made up of I417, I428, F429, and Y508 (Fig. 3D). SsrA makes three hydrogen bonds with the rest of the LOV domain, all within the A'α helix. The carboxyl group of D545 interacts with the hydroxyl of T406, the carboxyl group of E546 with the backbone amide of L407, and the hydroxyl of Y548 with the carboxyl of E409. In comparison with AsLOV2, F549 occupies the same area as the C-terminal L546. Because of extra residues in iLID, the last turn of the Jα helix buckles out and wraps back in to make this placement of F549 possible. iLID atomic coordinates and structure factors have been deposited to the Protein Data Bank (PDB ID code 4WF0).
Reversion and Paired Down iLID.
Based on the structure of iLID, we chose six mutations to revert to determine their role or necessity in the improved switching of iLID (SI Appendix, Figs. S5 and S6, and Table S2). Reversion of R519 and D522 led to a lower dynamic range and weaker caging, which is apparent as tighter affinities under blue light and in the dark. Removal of F537 had a similar effect on dynamic range, but weakened blue-light and dark-state affinities for SspB to the micromolar range. The remaining three reversions, Y502Q, H521R, and C530M, had little effect on the dynamic range. The double-reversion mutant, Y502Q/H521R, binds about threefold tighter in both states. In addition to keeping the improved dynamic range, C530M has very little effect on the affinity range or reversion kinetics of the switch and can be removed without consequence (SI Appendix, Fig. S9).
Subcellular Recruitment.
Because oLID was previously untested in mammalian cell culture, we were interested in how well it would function and if the improvements of iLID, measured in vitro, would be apparent in cells. To test LID function in cells, we designed constructs by fusing the LIDs to the yellow fluorescent protein, Venus, and a well-characterized peptide sequence that would anchor the protein to specific subcellular localizations (CAAX, cell membrane; Mito, mitochondria). Furthermore, SspB (Nano) and SspB R73Q (Micro) were fused to the red fluorescent protein, TagRFP-T (22). IA32 mouse fibroblasts (23) transiently expressing the LID protein pairs were imaged using a confocal microscope and activated with a 488-nm laser. To assay whole-cell activation, cells expressing TagRFP-T-SspB and the Mito anchored LID pairs were imaged during a time course of intermittent whole cell 488-nm light stimulation. Cells expressing each pair of switches showed rapid recruitment of TagRFP-T to the mitochondria during stimulation followed by dissociation into the cytoplasm with the removal of stimulus (Fig. 4A, Upper, and Movie S1). Recruitment for each pair was quantified and compared between switches by measuring the ratio of mitochondrial/cytoplasmic TagRFP-T fluorescence intensity throughout time (Fig. 4B). To prevent bias, mitochondrial and cytoplasmic regions of interest (ROIs) were automatically identified for each frame using the Venus channel and the default Threshold function in FIJI software. The recruitment phase was fit to a single-phase exponential and we determined that the iLID/Nano and iLID/Micro pairs showed a 4.4- and 4.3-fold change in comparison with a 2.4-fold change seen for oLID/Nano in this assay; each with a half-life < 30 s. Furthermore, we hypothesized that the ratio of mitochondrial/cytoplasmic TagRFP-T fluorescence in the dark state would be representative of the dark-state binding affinities observed in vitro. As expected, the iLID/Nano dark state ratio is higher than the ratio for iLID/Micro. However, although the in vitro evidence would suggest that oLID/Nano would have the tightest binding affinity in the dark, their dark-state mitochondrial/cytoplasmic TagRFP-T fluorescence ratio is the lowest.
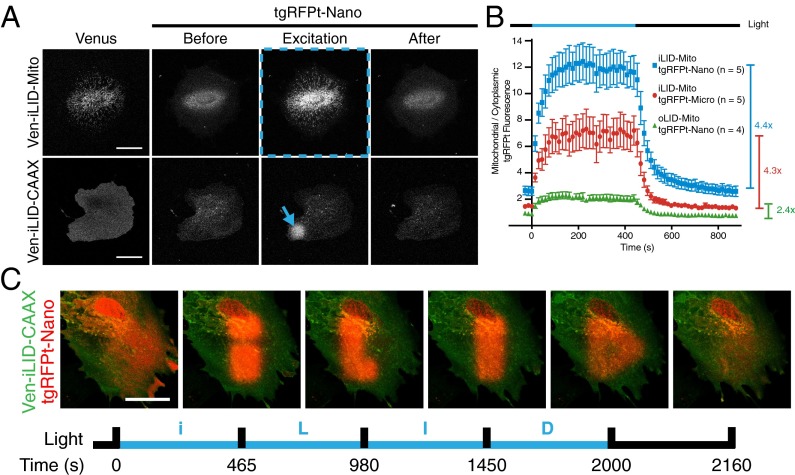
Fig. 4.
iLID provides improved local recruitment in cell culture. (A) IA32 fibroblasts expressing membrane (CAAX) and mitochondrial anchored Venus-iLID and cytoplasmic TagRFP-T-Nano. Localized activation (denoted by blue markings) caused relocalization of TagRFP-T-Nano. (Scale bar, 50 μm.) (B) A ratio of mitochondrial to cytoplasmic TagRFP-T signal intensity during a time course of whole-cell activation as shown in row 1 of A for each pair of mitochondria anchored switches. (C) Patterned activation of Venus-iLID-CAAX shows tight spatial and temporal control of TagRFP-T-Nano localization within a cell. (Scale bar, 50 μm.)
To test if the LID proteins could be used to recruit protein to a subcellular localization based on a ROI, the CAAX fused LID pairs were expressed and imaged in fibroblasts (Fig. 4A, Lower, and Movie S2). The optical sections obtained using the confocal were thick enough to capture light emitted from both apical and basal membrane bound proteins. This process allowed us to stimulate ROIs on these membranes and image the relocalization of TagRFP-T to these regions of the cell. Again, we imaged cells with intermittent stimulation at a ROI. During stimulation the TagRFP-T signal increased within the ROI but diminished outside the ROI. From this assay it is difficult to determine differences in dark-state binding because we do not know what portion of TagRFP-T-SspB is membrane bound versus cytoplasmic. However, by quantifying the TagRFP-T fluorescence intensity in a relatively small ROI (260 μm2), the cytoplasmic concentration of TagRFP-T-SspB should remain constant before and after 488-nm light stimulation. Therefore, by calculating pre:post stimulation ratio of TagRFP-T fluorescence intensity within the ROI, we determined the relative fold-change of binding in the light and dark. These values correlate with in vitro measured dynamic ranges (SI Appendix, Fig. S7). These experiments led us to realize that by anchoring the switching half of the dimer pair we obtained a relatively tight spatial resolution of activation. Combined with the minute time scales of association and dissociation, we were able to exemplify these attributes of the switch by sequentially writing the letters i, L, I, and D on the membrane of a cell (Fig. 4C and Movie S3).
Light-Induced GTPase Signaling Through Guanine Nucleotide Exchange Factor Localization.
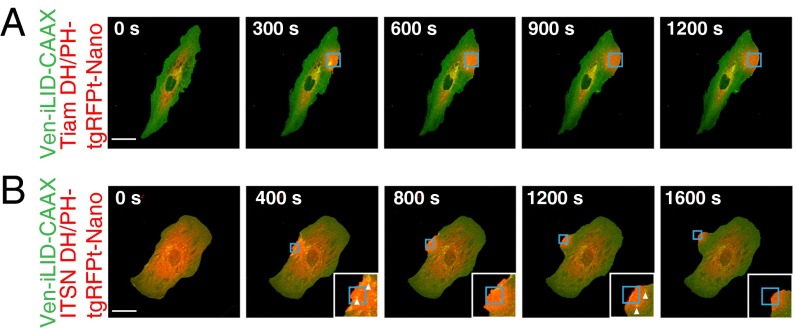
One area of interest for the use of LIDs is to control and study cell signaling. The Rho family of small GTPases (consisting of RhoA, Rac1, and Cdc42) are signaling proteins involved in many aspects of cell physiology, including actin cytoskeleton remodeling. In general, GTPase signaling is considered active when the GTPase is bound to GTP and inactive after the GTPase hydrolyzes GTP to GDP. Canonically, active Rac1 leads to branched actin formation and lamellipodial morphology, active Cdc42 leads to bundled actin and filopodial morphology, and active RhoA leads to bundled actin and actin stress fiber formation. Other groups have found that small GTPases can be controlled through the localization of guanine nucleotide exchange factor (GEF) Dbl homology (DH)/pleckstrin-homology(PH) domains to the membrane (3, 24). At the membrane the DH/PH domains are colocalized with GTPases, where they can aid in exchange of GDP for GTP, activating the GTPases. We hypothesized that the LID switches could be used to locally activate GTPase signaling, leading to a change in actin dynamics and a visible change in cell morphology (SI Appendix, Fig. S8A). Therefore, we fused the DH/PH domains of ITSN (Cdc42 GEF) and Tiam1 (Rac GEF) to the N terminus of the TagRFP-T-SspB constructs (SI Appendix, Fig. S8B). Fibroblasts were transiently transfected with each construct in combination with Venus-iLID-CAAX using a 1:1 ratio of DNA. Because each construct is under the control of a CMV promoter, we expect that on average each construct is expressed at equal levels. Two days later the cells were imaged and stimulated in a similar manner to the spot stimulation previously described. Here, stimulated areas were at the edge of the cell. As expected, the cells expressing the Tiam1 fusions, within minutes of stimulation, began to ruffle and formed a lamellipodia at the site of recruitment (Fig. 5A and Movie S4). Surprisingly, the cells expressing the ITSN fusions displayed a similar phenotype in the area of stimulation (Fig. 5B and Movie S5). Although we would expect a filopodial morphology to form, our results may be explained in two ways: cross-talk between GTPase pathways that leads to an increase in Rac GTPase signaling or a direct catalytic effect on a Rac GTPase, such as RhoG, from the ITSN DH/PH (25, 26). In addition to forming lamellipodia, most cells displayed increased appearance of vesicles in the stimulated area consistent with the role of Cdc42 in endocytosis (Fig. 5B, arrowheads, and Movie S6).
Fig. 5.
Spatial control of GEF DH/PH domains by iLID produces localized control of Rac and Cdc42 activity. IA32 cells expressing Venus-iLID-CAAX and (A) Tiam DH/PH-TagRFP-T-Nano or (B) ITSN DH/PH-TagRFP-T-Nano. iLID was activated in the regions highlighted in blue. White arrowheads mark vesicles. Images are representative (n > 5). (Scale bars, 50 μm.)
Discussion
A novel aspect of our study was the use of high-throughput screening and selection to improve the dynamic range of a photoactivatable protein switch. A specific challenge of engineering a switch is that the selection protocol should be sensitive to protein activity in both the on and off state. In our case, it was critical that the LOV domain be presented on the surface of the phage in a functional form. In phage display it is common to use signal sequences that direct a protein to the Sec-mediated translocation pathway, which transports proteins through membranes in an unfolded state. This approach was not appropriate for our application because we needed the LOV domain to remain folded during translocation so that it would remain bound to the flavin cofactor. Therefore, we made use of the Tat secretion pathway, which maintains proteins in a folded and cofactor-bound state during secretion. Once the LOV domain was on the surface of phage, we were able to find functional switches by selecting for binding under blue light, and then collecting phage that unbound when the light was turned off. The phage protocol was useful as a first filter in the search for better switches, but explicit screening in the light and dark with the ELISA-based binding assay allowed us to identify the top performing variants. The ELISA was particularly informative because the binding properties of single variants could be characterized side-by-side in the light and dark. This process allowed us to detect small improvements in switching, which allowed us to select variants to include in the recombination library.
In creating the directed library, we used molecular modeling to find mutations predicted to be compatible with the dark state of the switch. We used this approach because we were interested in stabilizing the closed state of the switch. Ideally, modeling would be used to find mutations that are compatible with the dark and lit state. This was not possible because there is not detailed structural information about what happens to the PAS domain when it is photoactivated. It is informative to see where the final mutations were located in iLID. The cluster of mutations in the hinge loop indicates that it has an important role in the docking and undocking of the Jα helix. In a multiple sequence alignment of the LOV domain, there is variability in the hinge loop, suggesting that LOV-domain homologs may have different switching properties.
Previous studies have highlighted the role of the A'α helix in stabilizing the docked state of the Jα helix in the dark and releasing it in the light (12). In the iLID structure the A'α helix rearranges to form several contacts with the Jα helix and the embedded SsrA sequence. Interestingly, the alternate conformation of the A'α helix is almost identical to that of the A'α helix of PA-Rac1, another photoactivatable switch based on the AsLOV2 domain (Fig. 3C). A shared feature of the iLID and PA-Rac1 structures is the capping of the A'α helix. In the AsLOV2 structure, the A'α helix is capped by a water molecule; however, in iLID the carboxyl group of E546 in SsrA caps the helix and in PA-Rac1 it is capped by the carbonyl group of N595 in Rac1. These capping interactions in Pa-Rac1 and iLID may enhance cooperativity between folding of the A'α helix and caging, and therefore lead to effective switching.
Our goal in creating iLIDs was to create a genetic tool for studying biochemical and cellular processes. We showed that both iLID pairs can be used in mammalian cell culture to form dimers and recruit fusion proteins to subcellular structures. Furthermore, the differing affinity ranges of iLID nano and iLID micro, measured in vitro, translate to different cell characteristics and can be used accordingly. We postulate that this inherent tuneability of the switch will lead to use in a broader spectrum of applications. Here, we were able to control local GTPase activity by driving GEF DH/PH domains to a localized spot on the membrane, and we expect that the small, modular nature of iLID nano/micro will open up control of a much wider variety of cellular targets.
Materials and Methods
Design and Construction of Computationally Directed Library.
Rosetta’s pmut_scan protocol was used to predict stability of all possible AsLOV2 point mutations. Stabilizing and mildly destabilizing mutations were filtered for proximity to the Jα helix and a library of single point mutations was generated using a comprehensive mutagenesis protocol (27). The recombined point-mutant library was generated using a gene-assembly protocol with oligonucleotides encoding for the top mutations suggested by ELISA ranking. See SI Appendix for details.
Phage Display Selection and Ranking by Soluble Protein ELISA.
The AsLOV2 library was cloned as a fusion with phage pIII coat protein and the Tat secretion sequence. Phage expressing the library was added to plates coated with MBP-SspB and placed under blue light, left for 1 h, washed, and moved to the dark. After 10 min in the dark, the eluted phage was collected, propagated, and quantified (28). Four rounds of positive and negative selection were performed before cloning into a modified pET-21b vector with an N-terminal FLAG tag. Overnight cultures were lysed and added to plates with immobilized MBP-SspB under blue light and in the dark. ELISA signal to anti-FLAG antibody was detected via ECL reagent. See SI Appendix for details.
Growth and Purification of Recombinant Proteins.
Purified proteins were prepared as previously published (18). See SI Appendix for details.
Fluorescence Polarization Binding Assays.
Binding assays were performed as previously described (18). See SI Appendix for details. All measured affinities can be found in SI Appendix, Tables S1–S3.
Crystallization and Structural Determination of iLID.
Initial crystals of iLID were grown using hanging drop vapor diffusion with 800 mM lithium chloride, 100 mM Tris⋅HCl pH 8.5, 32% (wt/vol) PEG 4000. Crystals were optimized with microseeding and grew to full size within 3 d. X-ray diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. The structure was determined using molecular replacement with PDB ID 2V0U. See SI Appendix for details.
Mammalian Cell Culture and Transfection.
IA32 mouse fibroblast cells were cultured at 37 °C and 5% (vol/vol) CO2 in DMEM supplemented with 10% FBS (HyClone), 100 U/mL penicillin, 100 μg/mL streptomycin, and 292 μg/mL l-glutamine. Cells were transiently transfected using NanoJuice (EMD Millipore) as recommended by the manufacturer.
Mammallian Cell Localization/GEF Microscopy and Image Analysis.
Fibroblasts were transfected with two vectors containing each component of the switch in equal amounts. Two days later, cells were imaged and photo-activated with an Olympus FluoView FV1000 scanning confocal inverted microscope equipped with a 1.30 N.A. 40× oil-immersion objective. Images were analyzed using FIJI software (29). See SI Appendix for details.
Cloning and Vector Information.
See SI Appendix, Methods for details and SI Appendix, Table S6 for a list of constructs and corresponding Addgene numbers.
Supplementary Material
Acknowledgments
This work was supported by the NIH (GM093208) and a grant from the Human Frontier Science Program. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4WF0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417910112/-/DCSupplemental.
References
- 1.Rivera VM, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2(9):1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 2.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein–protein interactions in live cells using light. Nat Biotechnol. 2009;27(10):941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 3.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461(7266):997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7(12):973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10(3):249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 7.Levskaya A, et al. Synthetic biology: Engineering Escherichia coli to see light. Nature. 2005;438(7067):441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 8.Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J Mol Biol. 2011;405(2):315–324. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strickland D, et al. TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9(4):379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46(49):14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 11.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301(5639):1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 12.Zayner JP, Antoniou C, Sosnick TR. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J Mol Biol. 2012;419(1-2):61–74. doi: 10.1016/j.jmb.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39(31):9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 14.Swartz TE, et al. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276(39):36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 15.Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a specificity factor for an AAA+ ATPase: Assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem Biol. 2002;9(11):1237–1245. doi: 10.1016/s1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- 16.Flynn JM, et al. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;98(19):10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levchenko I, Grant RA, Wah DA, Sauer RT, Baker TA. Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol Cell. 2003;12(2):365–372. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Lungu OI, et al. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol. 2012;19(4):507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strickland D, et al. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7(8):623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speck J, Arndt KM, Müller KM. Efficient phage display of intracellularly folded proteins mediated by the TAT pathway. Protein Eng Des Sel. 2011;24(6):473–484. doi: 10.1093/protein/gzr001. [DOI] [PubMed] [Google Scholar]
- 22.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5(6):545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134(5):828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2(6):415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 26.Jaiswal M, Dvorsky R, Ahmadian MR. Deciphering the molecular and functional basis of Dbl family proteins: A novel systematic approach toward classification of selective activation of the Rho family proteins. J Biol Chem. 2013;288(6):4486–4500. doi: 10.1074/jbc.M112.429746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firnberg E, Ostermeier M. PFunkel: Efficient, expansive, user-defined mutagenesis. PLoS ONE. 2012;7(12):e52031. doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat Protoc. 2007;2(6):1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- 29.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.