Significance
TM7 is one of the most enigmatic bacterial phyla among the uncultivated candidate phyla referred to as “microbial dark matter,” and it has potential pathogenic associations. We revealed molecular insights into its uncultivability and pathogenicity, as well its unique epibiotic and parasitic lifestyle phases. These novel discoveries shed significant light on the biological, ecological, and medical importance of TM7, as well as providing useful information for culturing other TM7 and currently uncultivable bacteria that may evade standard cultivation approaches.
Keywords: TM7, human-associated, epibiont, oral microbiome, interspecies interaction
Abstract
The candidate phylum TM7 is globally distributed and often associated with human inflammatory mucosal diseases. Despite its prevalence, the TM7 phylum remains recalcitrant to cultivation, making it one of the most enigmatic phyla known. In this study, we cultivated a TM7 phylotype (TM7x) from the human oral cavity. This extremely small coccus (200–300 nm) has a distinctive lifestyle not previously observed in human-associated microbes. It is an obligate epibiont of an Actinomyces odontolyticus strain (XH001) yet also has a parasitic phase, thereby killing its host. This first completed genome (705 kb) for a human-associated TM7 phylotype revealed a complete lack of amino acid biosynthetic capacity. Comparative genomics analyses with uncultivated environmental TM7 assemblies show remarkable conserved gene synteny and only minimal gene loss/gain that may have occurred as TM7x adapted to conditions within the human host. Transcriptomic and metabolomic profiles provided the first indications, to our knowledge, that there is signaling interaction between TM7x and XH001. Furthermore, the induction of TNF-α production in macrophages by XH001 was repressed in the presence of TM7x, suggesting its potential immune suppression ability. Overall, our data provide intriguing insights into the uncultivability, pathogenicity, and unique lifestyle of this previously uncharacterized oral TM7 phylotype.
DNA-based culture-independent methods have revealed a comprehensive inventory of microorganisms from environments and human bodies, the majority of which are still categorized as uncultivated phylotypes (1–4). Cultivation and functional analyses of these “yet-to-be cultivated” microbial organisms have been and will continue to be one of the major frontiers in microbiology research.
Among all of the candidate divisions (5), TM7 has been one of the most challenging bacterial phyla. Since its initial discovery by culture-independent sequence analysis more than 20 y ago (6), TM7 representatives have been identified in a variety of natural habitats, such as soil, seawater, deep-sea sediments, hot springs, and termite guts, as well as in different human body sites, including the gastrointestinal tract, skin, and genital tract (2, 7–15), and at least five genera of TM7 have been found in the oral cavity (2, 9) alone (Fig. S1). Recent investigations of ancient dental calculus showed that TM7 has been part of the human microbiome in hunter-gatherers since before the introduction of processed sugar during the Industrial Revolution (16). Moreover, TM7 has been implicated in association with host inflammatory mucosal diseases (9, 10, 17). It is particularly prevalent in the oral cavity, although commonly at low abundance, generally around 1% of the whole oral microbial population based on culture-independent molecular analysis (9, 18). However, an increase in the abundance (as high as 21% of the whole oral bacterial community in some studies) of TM7 members was detected in patients with various types of periodontitis (13, 15). Furthermore, certain oral TM7 phylotypes, such as the oral clones I025 and EW086 (National Center for Biotechology Information GenBank nucleotide database accession nos. AF125206 and AY134895, respectively), are more prevalent in diseased samples, and some of these phylotypes can even be detected on or within the host crevicular epithelial cells (14). Based on these findings, the association of TM7 with periodontitis has been suggested. The partial and highly fragmented genome from the oral TM7 phylotype TM7a (19) provided a glimpse into its pathogenic potential, revealing the presence of genes encoding several putative virulence factors, such as cytotoxic necrotizing factor 1, hemolysin toxin protein, and type III secretion protein. However, due to their seemingly recalcitrant nature toward cultivation, only one single TM7 phylotype has been reportedly cultivated, with no detailed genomic information (20). Thus, knowledge regarding TM7 genotypes and physiological properties, as well as their potential role in the pathogenesis of oral mucosal disease, is very limited.
In a recent study, we developed a new oral culture medium (SHI medium) that supported the growth of many uncultivated bacteria within a multispecies community, including several phylotypes of TM7 (21, 22). In this work, by using targeted enrichment approaches on SHI medium-cultivated saliva samples, we were able to obtain a stable coculture of an oral TM7 phylotype (TM7x) attached to the surface of a previously uncultivated Actinomyces odontolyticus strain (XH001). The coculture enabled complete sequencing of the highly reduced circular 705-kb genome for this human-associated TM7 phylotype. Our genomic, transcriptomic, and metabolomics profiles provide intriguing insights into the unculturability, pathogenicity, and unique lifestyle of this previously uncharacterized TM7 phylotype as a possible parasitic epibiont of XH001.
Results and Discussion
TM7x Shows a Unique Lifestyle as a Parasitic Epibiont of Oral XH001.
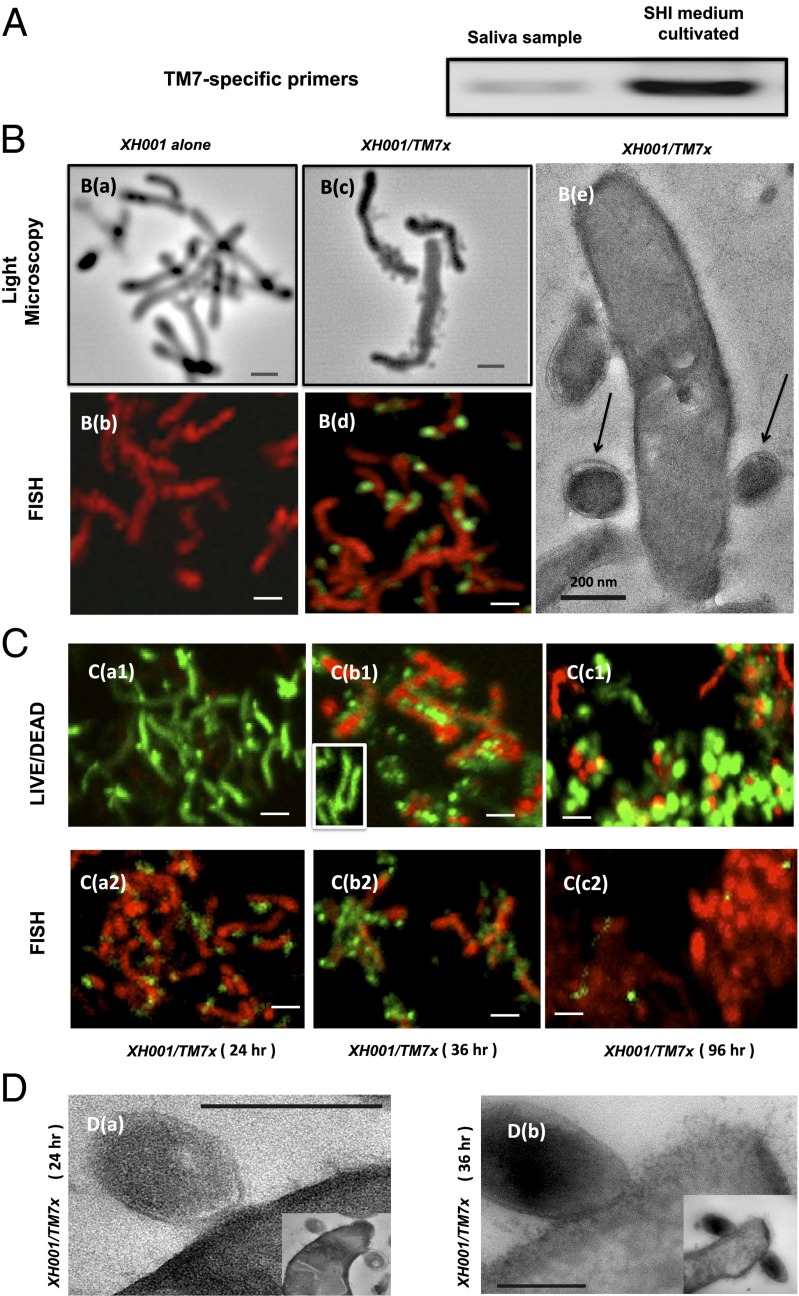
Analysis of 16S rRNA gene profiles revealed that some of these TM7 phylotypes contained an atypical base substitution (equivalent to Escherichia coli position 912: C to U), which is known to confer streptomycin resistance (12). Consistent with the prediction, addition of streptomycin to the in vitro oral community indeed resulted in enrichment of one specific TM7 phylotype (named TM7x) within the multispecies community (Fig. 1 A and B).
Fig. 1.
Cultivation and coisolation of TM7x with its host species Actinomyces spp. XH001. (A) PCR using a phylum-specific primer reveals the presence of TM7 within SHI medium-cultivated saliva samples. (B) Light microscopy and FISH images of XH100 monoculture (a and b) and XH100/TM7 coculture (c and d). (B, c) Cells of XH100 with several cells of TM7x attached to them. (B, d) Confocal laser scanning micrograph after hybridization with the cyanine5-labeled TM7567 (TM7x) and hexachloro-fluorescein–labeled universal eubacterial probe EUB338. XH001 appears red, whereas TM7 appears green. (B, e) TEM image of TM7x cell (indicated by arrows) attached to an XH001 cell. (Scale bars: 200 nm.) (C) Live/dead staining (a1, b1, and c1), and FISH (a2, b2, and c2) images. (C, a1 and a2) XH001/TM7x coculture 24 h after inoculation, with approximately two TM7x cells per XH001 cell. (C, b1 and b2) XH001/TM7x coculture 36 h after inoculation, with approximately six TM7x cells per XH001 cell. (C, b1, Inset) Live/dead staining of monoculture of XH001 36 h after inoculation. (C, c1 and c2) XH001/TM7x coculture 96 h after inoculation. For live/dead staining, live cells appear green and dead cells appear red; whereas for FISH, XH001 appears red and TM7x appears green. (D) TEM images showing the cell membrane of XH001 at/near the TM7x attachment site 24 h (a) and 36 h (b) after inoculation. (Insets) Original images from which D (a and b) are derived. (Scale bars: B, a–d and C, 1 μm; D, 200 nm.)
Next, we plated serial-diluted TM7x-enriched cultures on SHI medium agar plates in an attempt to isolate individual TM7x colonies, and found that TM7x-containing colonies only grew when a previously uncultivated A. odontolyticus strain (designated XH001 in this study) was also part of the colony. Co-occurrence of the two species was identified in multiple saliva samples from different human subjects; thus, this association is independent of the sample source or the original oral community composition. This intriguing finding prompted further investigation of the relationship between TM7x and XH001. Microscopic examination of independently isolated cocultures of TM7x and XH001 showed that TM7x cells are spherical, with a diameter of 200–300 nm, and that they are physically bound to rod-shaped XH001 cells (Fig. 1B). Various physical and chemical treatments were used to disrupt the attachment between TM7x and XH001 in an attempt to isolate TM7x and XH001 separately, including repeatedly passing the coculture through a 28-gauge needle and filtering the mixture through a 0.22-μm filter to separate TM7x from XH001. Although XH001 was able to form pure single colonies on SHI medium agar plates after physical disruption, no TM7x colonies were obtained. Addition of spent coculture medium or heat-killed XH001 also did not yield independent growth of TM7x. Although TM7x/XH001 cells coexisted well under nutritionally replete environments (Fig. 1 C, a), a different picture emerged under extended starvation conditions: After 36 h or more of coculture without addition of nutrients, the majority of XH001 cells associated with TM7x lost their viability even though XH001 cells alone persisted under the same starvation condition (Fig. 1 C, b). In contrast, the TM7x cells remained vital and multiplied under the same condition (Fig. 1 C, b). More interestingly, a subpopulation of XH001 cells developed exospore-like structures, which coincided with a drastic reduction of physically associated TM7x (Fig. 1 C, c). Transmission EM (TEM) images revealed a severely disrupted and most likely compromised cell membrane of XH001 (Fig. 1 D, b), which correlates with the live/dead staining results when XH001 was physically associated with TM7x under the starvation condition (Fig. 1 C, b). The apparently obligate surface attachment to XH001 indicates an epibiotic (ectosymbiontic) lifestyle for TM7x. Given the fact that TM7x cells can only survive in the presence of XH001 and have a negative impact on the viability of XH001 under extended starvation conditions, it is reasonable to consider that the type of relationship between TM7x and XH001 is parasitic overall rather than mutualistic or commensal.
It is worth mentioning that multiple TM7 phylotypes were observed in our original saliva samples based on PCR denaturing gradient gel electrophoresis analysis. Also, although 16S rRNA gene analysis suggested that most TM7 phylotypes are resistant to streptomycin, only TM7x was successfully enriched after streptomycin selection. One possible explanation could be that, like TM7x, which requires host species XH001 for supporting its growth, other TM7 phylotypes might also need specific oral partner species to achieve optimal growth. Whereas XH001 happened to be highly resistant to streptomycin as well, the partner species for other TM7 phylotypes might not be streptomycin-resistant, resulting in no apparent enrichment of other TM7 phylotypes under streptomycin selection.
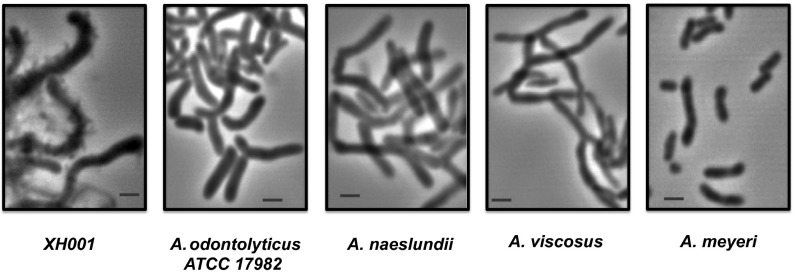
To determine the host specificity of TM7x further, associated TM7x cells were separated from their host XH001 by repeatedly passing the coculture through a 28-gauge needle and filtering the mixture through a 0.22-μm filter to collect TM7x cells. Isolated TM7x cells were mixed with different Actinomyces species, including A. odontolyticus strains XH001 and American Type Culture Collection 17982, Actinomyces naeslundii, Actinomyces viscosus, and Actinomyces meyeri. Cocultures were incubated anaerobically at 37 °C, and samples were monitored under microscopy periodically. As shown in Fig. 2, during a 72-h observation period, only XH001 was able to establish a physical association with TM7x, forming “grape on a vine” structures as observed for the TM7x/XH001 coculture in Fig. 1. Our result suggested that TM7x is likely to have a narrow host range. Although the host range has not yet been fully established and awaits further investigation, it is tempting to suspect that TM7x and its preferred host, XH001, might have undergone coevolution during their establishment within the oral cavity. In this regard, one interesting finding was that polyamine metabolism is unique in TM7x compared with environmental TM7 representatives. A spermidine synthase [Enzyme Commission (EC) 2.5.1.16] and an S-adenosylmethionine decarboxylase proenzyme (EC 4.1.1.50) are present. These genes are evolutionarily most closely related to members of A. odontolyticus species, including XH001, and may represent lateral gene transfer (LGT) between TM7 and XH001 resulting from their close interaction during coevolution. In nearly all cases, and more commonly in multicellular eukaryotes, recruitment of foreign genes and novel metabolic capabilities is highly favored by symbiotic associations (23).
Fig. 2.
Host specificity of TM7x. Microscopic images of different Actinomyces species infected with isolated TM7x. Cultures were monitored under a microscope periodically up to 72 h, and random photographs were taken. Representative images are shown. (Scale bars: 1 μm.)
A similar symbiotic relationship has been reported in Archaea, where a nanosized hyperthermophilic archaeon, Nanoarchaeum equitans, grows attached to the surface of a specific archaeal host, a member of the genus Ignicoccus (24). N. equitans is an obligate symbiont and is unable to grow when separated from its host, and too high a burden of N. equitans cells inhibits Ignicoccus growth, suggesting a parasitic behavior (25). To the best of our knowledge, what we are reporting is the first example of parasitic ectosymbiosis between two different species of bacteria. Unlike the N. equitans/Ignicoccus pair, far more dynamic interactions can be observed between TM7x/XH001 with different phases, including coexistence and induction of lysis, as well as exospore formation in XH001 by TM7x. These intriguing interactions are currently under further investigation.
TM7x Represses XH001-Induced TNF-α mRNA in Macrophages.
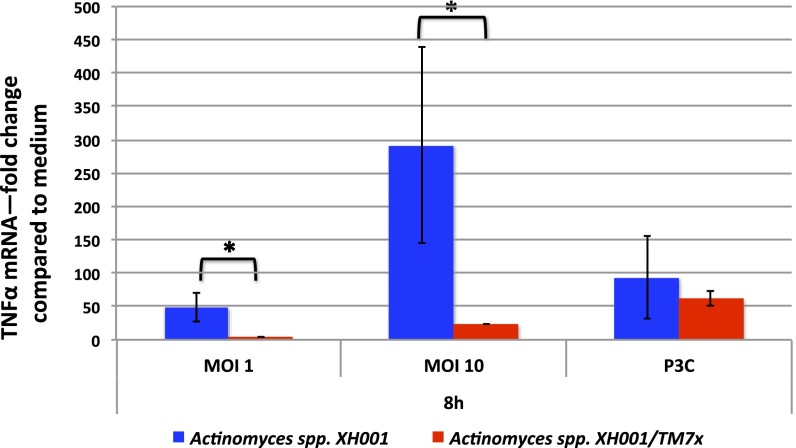
A number of 16S rRNA gene sequencing-based studies have implicated TM7 as a potential pathogen because it can be detected more frequently in human body sites with inflammatory mucosal diseases, such as vaginosis, inflammatory bowel disease, and periodontitis (9, 10, 17) (Fig. S1). Similarly, strains of A. odontolyticus related to XH001 have been associated with pathogenesis (26). The TM7x/XH001 coculture obtained in this study allowed researchers to examine the virulence potential of TM7x for the first time to our knowledge. For these studies, both XH001 alone and a coculture of TM7x/XH001 cells were used to infect J2 immortalized bone marrow macrophages (BMMs) and examined for their ability to modulate the expression of key cytokine-encoding genes. As shown in Fig. 3, XH001 is a strong inducer of TNF-α gene expression, consistent with observations for other Actinomyces species (27). Interestingly, when TM7x was physically attached to XH001, induction of TNF-α gene expression was greatly reduced, suggesting that TM7x can either prevent detection of XH001 by macrophages or possibly suppress TNF-α gene expression in macrophages. One of the crucial roles of macrophages in the host/defense system is to recognize pathogens and induce inflammation, a process characterized by the production of many inflammatory cytokines, including TNF-α (28). This experiment provided the first evidence, to our knowledge, that TM7x could indeed modulate the immune response. The data also suggest that the interactions between TM7x and XH001 are more complex than a simple metabolic dependency.
Fig. 3.
Induction of TNF-α production in macrophages by Actinomyces spp. XH001 monoculture and Actinomyces spp. XH001/TM7x coculture. Macrophages were treated with XH001 alone, XH001/TM7x coculture with a different multiplicity of infection (MOI), or Pam3CSK4 (P3C) as a positive control for 8 h. TNF-α mRNA was quantified using quantitative PCR. Fold induction was normalized to medium control. Each assay was performed in triplicate. Average values ± SD are shown. A Student t test (unpaired, two-tailed) was used for statistical analysis. An asterisk indicates a significant difference between the two values (P < 0.05).
Conserved Gene Synteny but Further Genome Reduction Is Evident in TM7x Compared with Environmental TM7 Genomes.
The enigmatic nature of the TM7 phylum has made it a major target for genomic capture. The first partial TM7 genomes included an oral phylotype (19), as well as a soil-derived TM7 phylotype (18). However, due to the lack of in vitro cultures, these early efforts relied on assembling the genome from single-cell amplifications, which resulted in valuable but highly fragmented and partial genomes (Table S1). Recently, sophisticated binning of metagenomic assemblies derived from complex aquifer (29) and sludge bioreactor communities (30) has produced two additional environmental TM7 genomic datasets (Table S1). The in vitro-grown coculture of TM7x and XH001 achieved in this study provided DNA of sufficient quality and quantity to obtain the first complete circular genome, to our knowledge, of this human-associated TM7 (Tables S1 and S2). TM7x has a reduced genome size (705,138 bp), with 699 coding sequences and 43 RNAs. The presence of 96 (of 111) single-copy marker genes nearly universally found in free-living bacteria (Table S1) and used for genome completeness estimation in other potentially symbiotic candidate phyla, such as TM6 (31), is in overall agreement with other published TM7 genomes (30). The coculture cells also allowed the generation of the first transcriptomic map, to our knowledge, for this TM7x genome using RNA sequencing (RNASeq) (Fig. S2), which enabled prediction of transcriptomic start sites and putative intergenic small RNAs (Table S2). It also served to validate the transcription of predicted ORFs, with the majority of the coding regions being transcribed over a wide dynamic range under coculture conditions with XH001 (94% of genes covered more than 10-fold over 85% of their length) (Fig. S2).
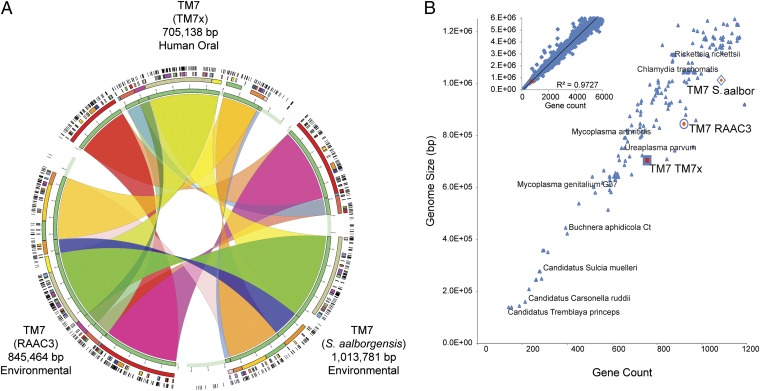
Comparative genomics revealed remarkably highly conserved gene synteny that has been maintained between TM7x and the aquifer and sludge bioreactor-associated TM7 (Fig. 4). A large fraction of the TM7x genome is syntenic to these two genomes (alignment results in 95% of the TM7x genome being in syntenic blocks) at the protein level, and nearly two-thirds of the TM7x proteins show high sequence similarity (i.e., average amino acid similarity in alignments is 72%, with an expected value cutoff of 1e−8, and matches covering >80% of the length of the TM7x genes) (Fig. S3). Further genome reduction in this human-associated strain, compared with the environmental TM7 genomes, is evident, and the strain-specific variation clusters mostly within a single genomic region (Fig. 4). These findings beg future investigation into the evolution of this phylum and the small number of genes gained or lost that possibly occurred during adaptation to the human host environment.
Fig. 4.
Comparative genomic analyses. (A) Genome synteny maintained between highly reduced genomes of human oral and environmentally derived TM7 phylum members. The RAAC3 and Saccharimonas aalborgensis genomes were assembled from environmental metagenomic reads into single contigs. The closed TM7x genome was assembled from an Actinomyces sp. (XH001) and TM7x symbiotic coculture. Related syntenic blocks between genomes are joined with colored ribbons revealing the larger number of syntenic regions and the maintained order of genes within each genome. From the inner ring to the outer ring, the colored blocks display the shared syntenic regions at increasing resolution. (B) Relationships between the microbial genome size of all finished genomes available in the Integrated Microbial Genomes Database (n = 2,086) and the number of predicted gene sequences. Smaller finished genomes are shown in the main panel. The three complete genomes representing members of the candidate phylum TM7 (TM7x, RAAC3, and S. aalborgensis) are marked with a color. (Inset) All microbial genomes that have less than 6 Mbp are shown, with the TM7x genome highlighted with a red marker.
Genome Properties of TM7x Are Consistent with the Observed Lifestyle as an Obligate Symbiont.
The combination of genomic and transcriptomic data allowed further analyses of how these genome features relate to the observed parasitic epibiont phenotypes described above. TM7x, in particular, has a high coding density genome (Fig. S4) and ranks among the smallest bacteria found in nature or the human body in both genome size and gene count to date (Fig. 3B). These facts, as well as analysis of its functional repertoire in relation to other known parasitic and mutualistic bacteria (Fig. S5), are consistent with its observed lifestyle as an obligate parasite instead of a free-living bacterium. Similar to other TM7 genomes, TM7x lacks genes necessary for de novo biosynthesis of any essential amino acid. Interestingly, these missing key biosynthetic pathways are complete in the genome of XH001, which could potentially account for TM7x metabolic dependency on XH001. It has been shown that for many reduced genomes, the pathways lost most frequently are those pathways with more enzymatic steps and higher energy requirements, including amino acid biosynthesis pathways (23, 32). The insect endosymbiont Buchnera spp. (33) and the parasitic N. equitans (25) encode no known amino acid transporters in their genomes. Buchnera relies on the host to provide nonessential amino acids, presumably through host transporters, and in return, the bacterium provides essential amino acids to the host. Bacterial endosymbionts that have lost genes for biosynthesis of amino acids have documented domain-level LGT to the host genome to provide these missing amino acids. N. equitans is an obligate epibiotic parasite that cannot import substrates from the environment, and therefore must stay attached to its host archaeon Ignicoccus to survive (26). Presumably, N. equitans acquires pathways from its host that are not encoded in its tiny genome, which include biosynthesis of amino acids, nucleotides, or cofactors (25). In the case of Ignicoccus, there are no apparent detrimental effects of the parasite N. equitans during normal growth, yet in the association between TM7x and XH001, TM7x clearly degrades and induces exospore formation in its host. In contrast, a motile obligate epibiotic bacterial predator, Micavibrio aeruginosavorus, missing only the biosynthetic capacity for seven amino acids and no apparent transporters, forms the basis for explaining its obligate parasitic lifestyle (34). Another predator, Bdellovibrio bacteriovorus, is capable of synthesizing 11 amino acids but has a large repertoire of 113 transporters for transporting amino acids, peptides, or amines (35). As a phylum, TM7 appears somewhat different from these types of interactions because they may not synthesize any essential amino acids. A transporter for Arg (Arg/ornithine antiporter) completes the full Arg deiminase pathway. An amino acid ABC transporter for charged and polar amino acids is present (His/Glu/Gln/Arg/opine family). However, it is still unclear how most other amino acids are acquired, although gaining essential amino acids through the several identified peptidases is likely. Furthermore, the TM7x genome contains a notably high number of genes encoding proteins with transmembrane domains, possibly including uncharacterized transporters (Table S3) to obtain nutrients from XH001. However, it is interesting to note that the genome contains a relatively low percentage of genes encoding proteins with signal peptides (Fig. S4).
The complete genomic information of TM7x also provided clues to the possible molecular basis for its underlying pathogenic nature. We found that despite its relatively small genome, TM7x contains many pathogenesis-related virulence gene homologs, including two virulence islands encoding separate type IV secretion systems and membrane-associated virulence-related proteins, such as OmpA (36) and LemA (37), as well as choline-binding proteins (38) (Table S2). It is particularly interesting to point out that the TM7x genome contains various ORFs encoding predicted proteins with toxin-antitoxin (TA) domains, such as VapC, VapB, and xenobiotic response element (39), as well as an abortive infection protein homolog known to promote cell death and limit phage replication within a bacterial population (40). These proteins could potentially play roles for TM7x to maintain its parasitic status against XH001.
Transcriptomic and Metabolic Responses of XH001 to the Presence of TM7x.
In an effort to investigate XH001/host cellular response and/or adaptations to the presence of TM7x, we conducted comparative transcriptomic and metabolomic studies of TM7x-associated XH001 and XH001 monocultures alone. Analyzing and comparing whole-genome expression (via RNASeq) and secreted molecule profiles [via liquid chromatography (LC) MS (41)] could also reveal gene functions and gene products that are associated with XH001 and are potentially important for the successful establishment of a symbiotic relationship, which would allow for later targeted genetic and molecule manipulations. Our transcriptomic data showed that roughly 340 genes in XH001 were differentially regulated (greater than threefold; false discovery rate, P < 0.01) under coculture conditions (Table S4). A total of 70 XH001 genes were up-regulated more than threefold when XH001 was physically associated with TM7x. Of the 35 most up-regulated genes, eight (23%) encode functions related to general stress responses. Besides genes encoding proteins homologous to general stress proteins (accession nos. ActOdo67396_0867, ActOdo67396_0743, ActOdo67396_0069, and ActOdo67396_1406) and a few stress-related transcriptional regulators (ActOdo67396_0857 and ActOdo67396_1614), a gene (ActOdo67396_0259) that encodes a ribosomal subunit interface protein, which binds to ribosomal machinery and inhibits protein biosynthesis, was increased almost 13-fold. Additionally, a fourfold increase was observed for YbaK (ActOdo67396_1917), which encodes a Cys-tRNAPro deacylase that prevents the addition of amino acids to the tRNA molecule, thus inhibiting protein translation.
Some of the most highly up-regulated genes corresponded to four TA-encoding systems, including the following: prevent-host-death family protein (ActOdo67396_0959), toxin component GNAT family (ActOdo67396_0525), addiction module toxin-RelE family (ActOdo67396_0988), and YefM TA system (ActOdo67396_1874). Increasing evidence indicates that these chromosomal TAs could potentially function as stress regulators involved in different stress responses (42, 43). Furthermore, the association with TM7x also induced turgor stress-related responses, which were manifested by an eightfold increase in the gene-encoding potassium efflux system KefA homolog and a more than threefold down-regulation of potassium uptake protein-encoding gene. These changes could be a response to the increased turgor pressure when XH001 is associated with TM7x, as indicated by its often-enlarged cell shape compared with XH001 alone. Considering the up-regulation of the many stress-related genes in XH001 when physically associated with TM7x, the transcriptomic data, together with phenotypic observation that long cocultivation could lead to the lysis of XH001 cells under starvation conditions, supports our hypothesis that TM7x forms a parasitic rather than commensal epibiont relationship with XH001. Furthermore, we also found that the presence of TM7x induced up-regulation of many genes necessary for biosynthesis of essential amino acids, as well as genes encoding transporters in XH001 (Table S4), whereas a strong repression of ompA expression, known to encode an immunogenic protein, was monitored, which could contribute to the observed reduction in TNF-α gene expression. Interestingly, two genes (ActOdo67396_0146 and ActOdo67396_1612) encoding putative membrane proteins were up-regulated in the presence of TM7x, and their potential roles in TM7x/XH001 interaction warrant further investigation.
A focused metabolomic study revealed many putative molecules that were uniquely produced in the coculture (Fig. S6). By comparing all obtained MS2 spectra (i.e., ionization spectra of precursor ions) from LC/MS with the extensive Global Natural Products Social Molecular Networking database (gnps.ucsd.edu), it was clear that most of these putative molecules have not yet been identified. However, the cyclic peptide cyclo(l-Pro-l-Val) was uniquely identified in the coculture (Fig. S6). In a previous study of the role of cyclic dipeptides in quorum sensing in Gram-negative bacteria, cyclo(l-Pro-l-Val) inhibited the 3-oxo-C6-homoserine lactone molecule that is the natural ligand for activation of the lux operon via the LuxR protein in E. coli JM109 (pSB401) (44). Cyclo(l-Pro-l-Val) also activated violacein pigment production in Chromobacterium violaceum, which causes acute toxicity response in nanoflagellates (44). The roles of these potential signaling molecules may be of critical importance for the interaction between TM7x and XH001, as well as for the human immune response, and needs to be addressed further to gain a more complete understanding of key mechanisms in oral health and disease.
Future Perspective
In summary, we are reporting a major breakthrough in TM7 research. Our unique culturing approach led to the in vitro domestication of a human-associated TM7 phylotype, which enabled us to explore its physiological and pathological nature. TM7x’s small cell size, reduced genome, lack of biosynthetic capacity for amino acids, and possible killing of XH001 under extensive starvation represent features consistent with its lifestyle as a parasitic bacterium. The fact that it appears to “mask” XH001-induced immune responses makes it even more interesting. Questions arise, such as why TM7x associates with XH001 (an A. odontolyticus strain) as its partner (host) and how these two species may interact with each other at the molecular level during metabolism and pathogenesis. We are also highly interested in the ecology and evolution of the TM7 phylum, especially its unique relationships with Actinomyces spp. Further detailed genomic, transcriptomic, and metabolomic investigations of TM7x will help to reveal answers to these intriguing questions for this fascinating bacterium.
Materials and Methods
Detailed experimental procedures are provided in SI Materials and Methods. TM7x was cultivated in SHI medium (22) as part of a human saliva-derived microbial community and enriched via streptomycin selection. Saliva sample collection was performed under University of California, Los Angeles, Institutional Review Board no. 09-08-068-02A as previously described (22). Briefly, saliva samples were collected from two healthy individuals (one male and one female, 30 and 32 y of age, respectively) and informed consent was obtained from these subjects. The TM7x phylotype was coisolated with XH001 as singles colonies by plating TM7x-enriched culture on SHI solid agar plates. The physical association between TM7x and XH001 was observed by FISH using confocal laser scanning microscopy and TEM. XH001 alone and XH001/TM7x coculture were used to infect J2 BMM cells, and quantitative PCR was conducted to monitor their TNF-α induction. Furthermore, XH001/TM7x cocultures grown in the SHI medium were shotgun-sequenced and assembled using SPAdes, followed by contig binning and annotation as described by McLean et al. (31) to assemble the genomes independently. Genome completeness for all TM7x and published TM7 assemblies was estimated based on 111 universal, single-copy genes.
Supplementary Material
Acknowledgments
We thank Youngik Yang and Jonathan Badger for additional assistance with TM7 genome and transcriptome analyses, and Son Pham for whole-genome alignment and synteny analyses. We thank Venkat Macherla and Sirenas Marine Discovery for providing LC/MS analysis, and Mark Adams and Ken Nealson for manuscript review. This work was supported by NIH National Institute of Dental and Craniofacial Research (NIDCR) Grants 1R01DE023810-01 (to W.S., X.H., and J.S.M.), 1R01DE020102 (to W.S., R.L., and J.S.M.), and 1-R01-DE021108 (to R.L. and W.S.); National Institute of General Medical Sciences Grant 1R01GM095373 (to J.S.M.); and NIDCR K99/R00 Pathway to Independence Award K99DE024543 and NIDCR T90 Training Award DE022734 (to A.E.).
Footnotes
Conflict of interest statement: W.S. is a part-time chief science officer of C3 Jian, Inc., which has licensed technologies from the University of California Regents that could be indirectly related to this research project.
This article is a PNAS Direct Submission.
Data deposition: Assembled TM7x genomes have been deposited in the National Center for Biotechnology Information BioProject database, www.ncbi.nlm.nih.gov/bioproject (BioProject PRJNA241438), and the National Center for Biotechnology Information Genome database, www.ncbi.nlm.nih.gov/genome (accession no. CP007496). MS data are accessible though the Center for Computational Mass Spectrometry repository at the University of California, San Diego, ftp://MSV000078570:a@massive.ucsd.edu, and molecule identification results are available at gnps.ucsd.edu/ProteoSAFe/result.jsp?task=3f3cacc08e4244c48ff29706c16e67c4&view=view_all_annotations_DB. Additional datasets are available at depts.washington.edu/jsmlab/downloads/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419038112/-/DCSupplemental.
References
- 1.Chen T, et al. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson J, et al. NIH HMP Working Group The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasken RS, McLean JS. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet. 2014;15(9):577–584. doi: 10.1038/nrg3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rheims H, Rainey FA, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17(3-4):159–169. [Google Scholar]
- 7.Abrams M, et al. Genomic characteristics of an environmental microbial community harboring a novel human uncultivated TM7 bacterium associated with oral diseases. Open Access Scientific Reports. 2012;1:276. [Google Scholar]
- 8.Bik EM, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4(8):962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 2003;69(3):1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353(18):1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 11.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67(1):411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE. 2012;7(6):e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paster BJ, et al. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Ann Periodontol. 2002;7(1):8–16. doi: 10.1902/annals.2002.7.1.8. [DOI] [PubMed] [Google Scholar]
- 15.Rylev M, Bek-Thomsen M, Reinholdt J, Ennibi OK, Kilian M. Microbiological and immunological characteristics of young Moroccan patients with aggressive periodontitis with and without detectable Aggregatibacter actinomycetemcomitans JP2 infection. Mol Oral Microbiol. 2011;26(1):35–51. doi: 10.1111/j.2041-1014.2010.00593.x. [DOI] [PubMed] [Google Scholar]
- 16.Adler CJ, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45(4):450–455, e1. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehbacher T, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57(Pt 12):1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 18.Podar M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73(10):3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104(29):11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soro V, et al. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl Environ Microbiol. 2014;80(20):6480–6489. doi: 10.1128/AEM.01827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edlund A, et al. An in vitro biofilm model maintaining a high species and metabolic diversity similar to the human oral microbiome. Microbiome. 2013;1(1):25. doi: 10.1186/2049-2618-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Y, et al. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol. 2010;25(5):357–367. doi: 10.1111/j.2041-1014.2010.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber H, et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417(6884):63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 25.Waters E, et al. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA. 2003;100(22):12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo AV, Silva CM, Haffajee A, Colombo AP. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55(Pt 5):609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- 27.Sato T, et al. Peptidoglycan of Actinomyces naeslundii induces inflammatory cytokine production and stimulates osteoclastogenesis in alveolar bone resorption. Arch Oral Biol. 2012;57(11):1522–1528. doi: 10.1016/j.archoralbio.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Flannagan RS, Cosío G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7(5):355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 29.Kantor RS, et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 2013;4(5):e00708–e00713. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albertsen M, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31(6):533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 31.McLean JS, et al. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA. 2013;110(26):E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis JJ, Xia F, Overbeek RA, Olsen GJ. Genomes of the class Erysipelotrichia clarify the firmicute origin of the class Mollicutes. Int J Syst Evol Microbiol. 2013;63(Pt 7):2727–2741. doi: 10.1099/ijs.0.048983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gil R, Sabater-Muñoz B, Latorre A, Silva FJ, Moya A. Extreme genome reduction in Buchnera spp.: Toward the minimal genome needed for symbiotic life. Proc Natl Acad Sci USA. 2002;99(7):4454–4458. doi: 10.1073/pnas.062067299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Kadouri DE, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rendulic S, et al. A predator unmasked: Life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303(5658):689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 36.Baldridge GD, Burkhardt NY, Simser JA, Kurtti TJ, Munderloh UG. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni. Appl Environ Microbiol. 2004;70(11):6628–6636. doi: 10.1128/AEM.70.11.6628-6636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hrabak EM, Willis DK. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174(9):3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun. 2000;68(10):5690–5695. doi: 10.1128/iai.68.10.5690-5695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sevin EW, Barloy-Hubler F. RASTA-Bacteria: A web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007;8(8):R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fineran PC, et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA. 2009;106(3):894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen DD, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci USA. 2013;110(28):E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 43.Hayes CS, Sauer RT. Toxin-antitoxin pairs in bacteria: killers or stress regulators? Cell. 2003;112(1):2–4. doi: 10.1016/s0092-8674(02)01282-5. [DOI] [PubMed] [Google Scholar]
- 44. Holden MT, et al. (1999) Quorum sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Molecular Microbiology 33(6):1254–1266. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.