Significance
The number of driver events required for human tumorigenesis has remained one of the fundamental issues in cancer research since the seminal studies of Armitage and Doll. This question has become even more important with the recent genome-wide sequencing studies of cancer, whose major goal is the identification of the driver genes responsible for tumor initiation and progression. By using a novel approach that combines conventional epidemiologic studies with genome-wide sequencing data, we show that only three sequential mutations are required to develop lung and colon adenocarcinomas, a number that is lower than what is typically thought to be required for the formation of cancers of these and other organs. This finding has important implications for the design of future cancer genome-sequencing efforts.
Keywords: cancer, driver mutations, somatic mutation rate, cancer incidence, cancer evolution
Abstract
Cancer arises through the sequential accumulation of mutations in oncogenes and tumor suppressor genes. However, how many such mutations are required for a normal human cell to progress to an advanced cancer? The best estimates for this number have been provided by mathematical models based on the relation between age and incidence. For example, the classic studies of Nordling [Nordling CO (1953) Br J Cancer 7(1):68–72] and Armitage and Doll [Armitage P, Doll R (1954) Br J Cancer 8(1):1–12] suggest that six or seven sequential mutations are required. Here, we describe a different approach to derive this estimate that combines conventional epidemiologic studies with genome-wide sequencing data: incidence data for different groups of patients with the same cancer type were compared with respect to their somatic mutation rates. In two well-documented cancer types (lung and colon adenocarcinomas), we find that only three sequential mutations are required to develop cancer. This conclusion deepens our understanding of the process of carcinogenesis and has important implications for the design of future cancer genome-sequencing efforts.
Somatic mutation theories of cancer have been unequivocally substantiated by the identification of the genes responsible for neoplasia over the last 40 y. The conceptual foundation for this field of research was established by the seminal work of Nordling (1) and Armitage and Doll (2). These investigators realized that the relationship between age and cancer incidence was a power function, suggesting that the process was driven by sequential (rather than single) mutations. Moreover, by examining the slope of the curve depicting incidence against age (or incidence curve), they predicted that six or more mutations were required for most common cancer types. These insights have guided the field for the last half-century.
The research stimulated by these studies has led to several conceptual challenges (3–8). For example, there are relatively large fluctuations in the slopes of incidence curves, leading to great uncertainty in the estimation of the number of rate-limiting events required for cancer. Moreover, clonal expansions during the neoplastic process complicate the analysis and interpretation of incidence curves. Inclusion of such clonal expansions into conventional models can substantially reduce the estimated number of required events. As a result of these uncertainties, estimates of the number of rate-limiting events required for cancer range from two to seven (3–9) and are still the subject of active debate.
With the advent of genome-wide sequencing, one might envision that issues such as these could be conclusively addressed. However, instead of providing definitive answers, the sequencing studies have actually raised new questions related to these issues. Typical solid tumors each contain hundreds or thousands of genetic alterations, the vast majority of which are point mutations or small insertions or deletions. Only a few of these are “drivers,” conferring selective growth advantages to the cancer cell in which they occur (9–11). The remaining thousands of mutations are “passengers” that coincidentally occurred during the large number of cell divisions associated with the neoplastic process (12). Driver genes are defined as genes containing driver mutations. Although genes can be confidently identified as drivers because mutations in them are observed in many tumors, the identification of driver genes that are infrequently mutated is more difficult. Several criteria for identifying driver mutations have been proposed (9–11), but none has been validated in an objective fashion. In addition to point mutations, alterations such as gene fusions, chromosomal translocations, and copy number changes further complicate our understanding of tumors’ genomic landscapes (13).
Recent reviews have emphasized that, in most patients with solid tumors, it is challenging to identify the six or seven driver gene mutations predicted by the original incidence curve analyses (9, 10). This could result from imperfect sequencing or limitations in sequence analysis, even when genomes are sequenced to high coverage.
Alternatively, the paucity of mutations could indicate that there is “dark matter” in the cancer genome, i.e., epigenetic changes and genomic alterations that cannot be easily identified by massively parallel sequencing or other commonly used methods.
We are therefore confronted with a frustrating situation: we do not always know how to identify a driver mutation when we see one, and we do not even know how many we are looking for in an individual cancer. The current study addresses the latter issue. If we knew the number of driver mutations we should expect in a cancer, it would both simplify the interpretation of individual cancer genomes and contribute to our understanding of cancer progression. As a corollary, the number of driver mutations that can be targeted by therapeutic agents would become clearer. In this work, we describe a method to infer the number of driver mutations required for cancer development. It combines, in a novel fashion, genome-wide sequence data with epidemiologic data. Using this approach in colorectal and lung cancers, we show that the number of required driver mutations, even for advanced cancers, is likely to be three.
Results
Quantifying Mutation Rates in Individual Cancers.
Cells with higher mutation rates develop into cancer more rapidly than those with lower mutation rates. This principle is widely accepted, long recognized, and supported by numerous independent lines of investigation (14–16). It explains why patients whose cells are exposed to exogenous mutagens or have endogenous defects in DNA repair enzymes are at greater risk for cancer than other individuals (14, 17, 18). The novel aspect of the analysis presented here is the quantitative comparison of cancer incidence in groups of patients whose cells have different mutation rates. This simple comparison allows us to infer the number of rate-limiting mutations required for cancer development in a time-independent fashion.
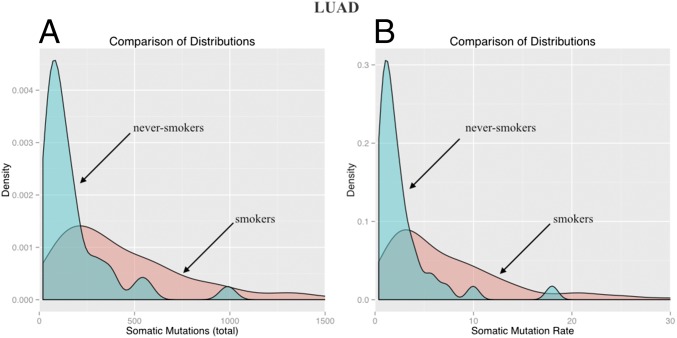
To reach this objective, we first need to estimate the mutation rates in individual cancers. Cancers with higher mutation rates are expected to have higher numbers of somatic mutations in their tumors, as documented in many cancer types (9). We chose two tumor types, lung adenocarcinomas (LUADs) and colorectal cancers (CRCs), for our analyses because these tumors are common, each can be easily divided into subgroups expected to have different mutation rates, and exome-wide sequencing data on large numbers of patients have been made available by the The Cancer Genome Atlas (TCGA) (cancergenome.nih.gov). In LUAD, the two subgroups we compare are those whose members have smoked at some point in their lives (“smokers”) and those whose members never smoked (“never-smokers”). Fig. 1A presents the distributions of mutation counts in these cohorts. The median number of somatic mutations per tumor is 357.5 (57–2,487, 2.5–97.5% quantiles) in smokers, 3.15 times higher [median ratio; 2.48–3.90, 95% confidence interval (CI); P = 2.5 × 10−9] than in never-smokers (median, 111; 24–610, 95% CI) (Materials and Methods). The number of somatic mutations per tumor can be converted to a somatic mutation rate, R, by dividing the number of mutations by the age of the patient at diagnosis. As shown in Fig. 1B and Table 1, this somatic mutation rate is 3.23 higher (median ratio; 2.58–4.05, 95% CI) in lung cancer patients who smoked vs. those who did not; this difference is highly statistically significant (P = 1.5 × 10−9). The definition and units of this rate (somatic mutations per year) are different from those usually used to describe mutation rates (somatic mutations per base pair per cell generation). Assuming a constant rate of cell division implies that R is a constant multiple of the conventional mutation rate, in which case, the ratio of the R values between two groups is equivalent to the ratio of the conventional mutation rates.
Fig. 1.
Distributions of the total number of somatic mutations and of the somatic mutation rates (somatic mutations per year) in smokers (blue) and never-smokers (red) in LUAD. The rightmost parts of the red distributions have been excluded to facilitate comparison. Distributions of the number of somatic mutations (A) and of the somatic mutation rates (B).
Table 1.
Comparison of the somatic mutation rate R (somatic mutations per year) between smokers and never-smokers with LUAD, and between MMR-deficient and -proficient CRCs
| Tissue type | Min | First quartile | Median | Mean | Third quartile | Max |
| LUAD smoker | 0.55 | 2.77 | 5.72 | 9.10 | 10.56 | 114.30 |
| LUAD never-smoker | 0.39 | 0.97 | 1.54 | 2.74 | 2.83 | 17.98 |
| MSI | 2.82 | 7.13 | 11.76 | 13.97 | 16.64 | 38.97 |
| MSS | 0.36 | 0.96 | 1.33 | 5.95 | 1.82 | 564 |
| MLH1 silent | 2.27 | 7.07 | 10.46 | 11.8 | 13.36 | 27.7 |
| MLH1 normal | 0.36 | 0.97 | 1.36 | 6.86 | 1.83 | 564 |
MMR deficiency can be assessed either by showing that a tumor has MSI or by showing that a tumor has silenced MLH1. Tumors that are MMR-proficient are MSS and have normal MLH1 expression.
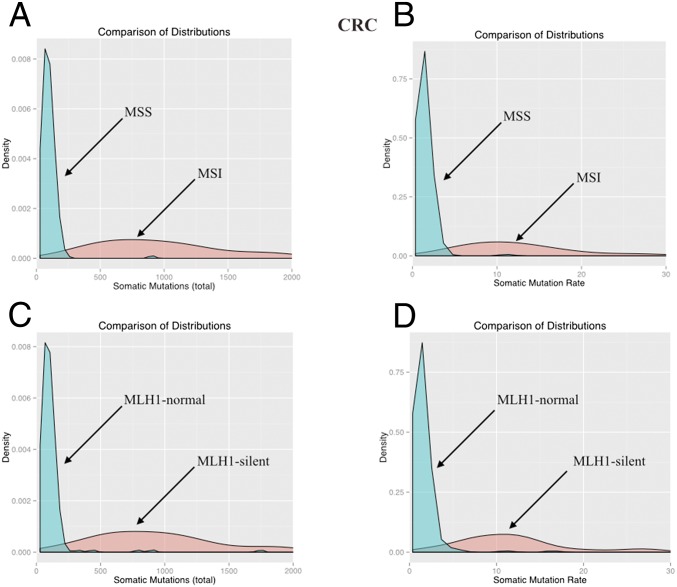
In CRC, we compare patients with and without a mismatch repair (MMR) deficiency. Cells with defects in MMR are mutation-prone, particularly at microsatellite sequences but also throughout the genome (19, 20). To determine whether individual cancers have defective MMR, two markers are commonly used: microsatellite instability (MSI) and MLH1 (MutL homolog 1) silencing (19, 20). MLH1 silencing is used because it can be easily assayed with conventional immunohistochemical staining, and this silencing is the mechanism underlying most cancers with defective MMR. The number of mutations in CRCs with MMR deficiency is about 10 times higher than in MMR-proficient CRC. Specifically, based on MSI, the ratio is 10.05 (median ratio; 8.79–11.52, 95% CI; P = 1.8 × 10−15), whereas based on MLH1 silencing it is 9.13 (median ratio; 8.05–10.43, 95% CI; P = 1.4 × 10−12) (Fig. 2 A and C). The rate of somatic mutations, as defined above for LUAD, is also higher in CRCs with MMR deficiency (P < 2.5 × 10−12). When categorized by MSI, the somatic mutation rate in MMR-deficient tumors was 8.85 times larger (median ratio; 7.77–10.11, 95% CI) than the non–MMR-deficient tumors (Fig. 2B and Table 1). When categorized by MLH1 silencing, the somatic mutation rate was 7.81 times larger (median ratio; 6.89–8.78, 95% CI) in the MMR-deficient tumors (Fig. 2D and Table 1).
Fig. 2.
Distributions of the total number of somatic mutations (A and C) and of the somatic mutation rates (somatic mutations per year) (B and D) in MMR-proficient (MSS or MLH1-normal) and MMR-deficient (MSI or MLH1-silent) CRCs. The rightmost parts of each red distribution have been excluded to facilitate comparison.
Comparing Mutation Rates with Rates of Cancer Development.
In the classic results of Nordling and Armitage and Doll, it is known that n, the number of rate-limiting mutational events in cancer development, is exponentially related to cancer incidence. The basic insight that inspired the current analysis is the following. If the average mutation rate in cancer subgroup A is twice that in cancer subgroup B, then the cancer incidence at any chosen age should be 2n times higher in subgroup A than in subgroup B (an adjustment to the power n is needed when n is larger than 2; see Materials and Methods for details). Similarly, a threefold increase in mutation rate should result in approximately a 3n-fold increase in cancer incidence. We use this insight to infer the number of driver gene mutations required to develop lethal malignancies. A critical point is that, even though cancer incidence is a function of age, the fold increase in incidence resulting from a higher mutation rate is the same for all ages. Interestingly, this is supported by epidemiological data on the cumulative risk of lung cancer with respect to age, showing approximately exponential incidence curves with the same zero intercept and different bases across different smoking subgroups [see figure 3 in Peto et al. (18)].
Doll and Hill (21) were the first to show that smoking significantly increases the incidence of lung cancer. Their 50-y study of British physicians found that smoking increased LUAD incidence by fourfold for former smokers and by 25-fold in those who smoke 25 or more cigarettes a day (22). More recent analyses show that the incidence of LUAD is 16.2-fold (10.25–25.6, 99% CI) in smokers compared with never-smokers (23).
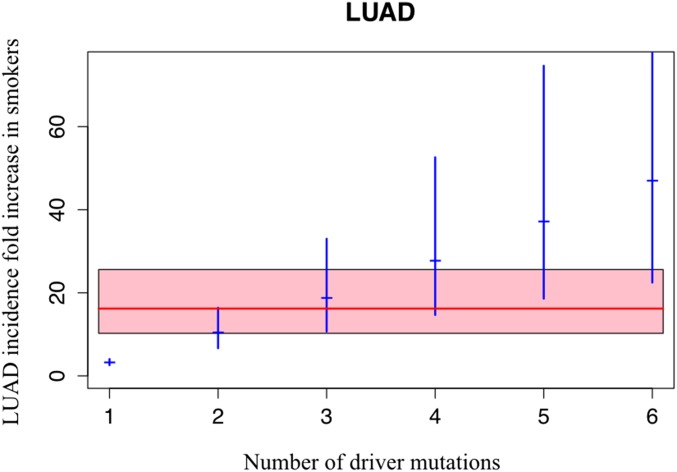
Now compare this increased LUAD incidence in smokers to the 3.23-fold increase in somatic mutations in LUADs from smokers noted above. If LUADs occurred through the sequential acquisition of two driver mutations, then the expected fold increase in incidence in smokers would be (3.23)2 = 10.4 (6.66–16.4, 95% CI), which is smaller than the actual increase. Alternatively, if LUAD formation required three mutations, the incidence in smokers would be 18.75 (10.69–33, 95% CI)-fold higher than in never-smokers, whereas with four mutations it would be 33.7 (17.17–66.43, 95% CI) higher, which is larger than the actual increase (see Materials and Methods for an explanation of why an adjustment to the power of 3 or 4 is needed). By performing a goodness-of-fit test on these data (Materials and Methods), we can infer the number of mutations required. For LUAD, three mutations provide the best fit to the actual increase observed, as graphically depicted in Fig. 3.
Fig. 3.
Relation between the number of rate-limiting driver mutations and the increase in LUAD incidence observed in smokers. The observed average increase in LUAD incidence associated with smoking is 16.2-fold (red line); the pink area is its 99% CI (10.25–25.6) (23). The expected increase in LUAD incidence associated with the indicated number of rate-limiting mutations is represented by the blue marks, with the corresponding 95% CI indicated by blue vertical segments.
A similar analysis can be performed in the CRC subgroups. Patients with an inherited MMR deficiency are cancer-prone (17, 19). Recent studies have shown that the age-adjusted CRC incidence in such patients is 114.2-fold increased (60.7–217, 95% CI) (14). Compare this increase in CRC incidence to the 7.7- to 8.8-fold increase in somatic mutations in MMR-deficient CRCs noted above.
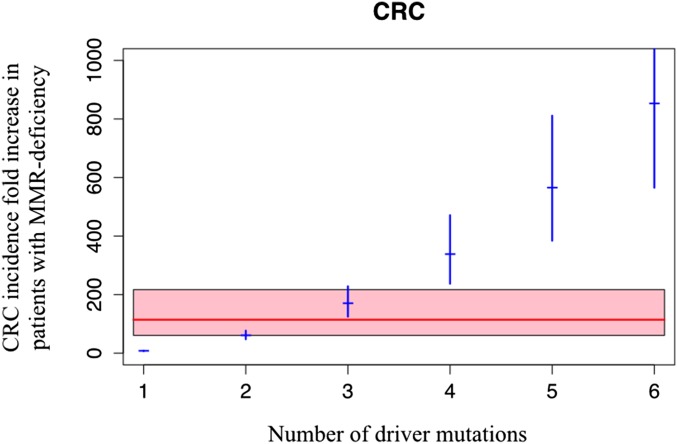
If CRCs occurred through the sequential acquisition of an average of two driver gene mutations, then the expected increase in incidence in MMR-deficient cancers would be too small. If it occurred through four or more sequential driver gene mutations, the expected increase would be too large (Fig. 4). A goodness-of-fit test on these data shows that the most likely number of mutations required for CRC in the general population is three, just as it was for LUAD.
Fig. 4.
Relation between the number of rate-limiting driver mutations and the increase in CRC incidence observed in patients with MMR deficiency. The observed average increase in incidence associated with MMR deficiency is 114.2-fold (red line); the pink area is its 95% CI (60.7–217) (14). The expected increase in CRC incidence associated with the indicated number of rate-limiting mutations is represented by the blue marks, with the corresponding 95% CI indicated by blue vertical segments.
Discussion
One of the major goals of cancer genomics is the identification of the driver genes responsible for tumor initiation and progression. A key question in this quest is a simple one: how many driver genes are needed? In certain neoplastic types, the number of drivers is small. For example, in chronic myeloid leukemia, a single mutation (a chromosome translocation juxtaposing the BCR and ABL genes) may be all that is required to convert a normal bone marrow stem cell into a tumor cell (24), and the transformation of myelodysplastic syndrome to acute myeloid leukemia appears to be the result of a single event (25). For most solid tumors, however, it is generally thought that a larger number of driver gene mutations is required. This number has been debated for decades, and the only way of estimating it, before genome-wide sequencing, has been through modeling of incidence vs. age curves. Using an approach that is independent of the steep dependence of cancer incidence on age, we show that, in two common solid tumors, the number of driver mutations required for cancer is likely to be three. This number represents a good fit for the total number of driver gene mutations typically found in CRC, as well as in a variety of other solid tumors (figure 5 in ref. 9).
Does this mean that three mutations are sufficient for lethal cancers, i.e., those that are metastatic, or just for the initial stages of invasion that distinguish cancer from benign tumors? This question can be addressed through the analysis of LUAD, which is the predominant form of lung cancer. The great majority (89%) of LUAD patients already have a nonlocalized tumor, either regionally spread to lymph nodes (22%) or metastatic (57%) at diagnosis, and their 5-y survival rate is only 26.5% and 4%, respectively (26). Over one-half of LUAD patients die from their disease within 1 y of diagnosis (26). Thus, our analysis of LUAD indicates that only three driver gene mutations are required for the appearance of late-stage cancers.
A related point is that our analysis should not be taken to imply that there are a maximum of three driver genes in lung or colorectal cancers. Cancer continually evolves, responds to changing microenvironments (including those associated with chemotherapy and radiation), and can develop new mutations that confer a selective growth advantage at any time. Our analysis suggests that three mutations are sufficient for a lethal cancer to develop, but we would expect that additional mutations could develop thereafter, often in a heterogeneous manner within a single tumor. This expectation is consistent with the numerous studies documenting genetic heterogeneity within tumors (9–11, 27, 28).
What are the limitations of our approach? One is that we have only studied two cancer types—LUAD and CRC. Other cancers may require more or fewer driver gene mutations. We have used the approach described here to evaluate head and neck cancers and pancreatic ductal adenocarcinomas, two other cancer types associated with smoking. The results on these tumors are consistent with the trend suggesting that three mutations are sufficient, but the number of cases that we could evaluate (i.e., those complete with clinical information and somatic mutation data) was too small to allow confident interpretation.
We have ignored driver gene mutations that are not rate-limiting steps, because, by definition, their occurrence is not a bottleneck for the appearance of cancer. Another potential limitation of our analysis is that we have assumed that driver gene mutations are the only rate-limiting steps. In reality, it is likely that epigenetic changes represent rate-limiting steps in some cancers (29). However, the existence of such epigenetic drivers could only lower, not raise, our estimate of the average number of driver gene mutations required for cancer development (Materials and Methods). Thus, in certain cancers, two conventional driver gene mutations plus one epigenetic change might be sufficient for cancer development. In mathematically formal terms, the approach used here provides an upper bound for the required average number of rate-limiting driver gene mutations to develop cancer (explained in detail in Materials and Methods).
In conclusion, our analysis shows that only a relatively small number of driver gene mutations appears to be required for the development of advanced cancers of the lung and colon. In addition to the understanding of human tumor genetic data it provides, this number should prove useful for implementing experimental model systems of cancer in vitro and in vivo. As more sequencing and epidemiologic data are gathered on cancers of various types, it will be of interest to assess what our approach reveals about other types of cancers and various types of exposures and hereditary risk factors.
Materials and Methods
Statistical Analysis.
We analyzed two whole-exome sequencing datasets publicly available on the TCGA website: LUAD (281 patients) and CRC (276 patients) (30). We considered all patients for whom the required smoking or MMR information was available. Sequencing for all datasets was performed using Illumina GA DNA sequencing as described elsewhere (cancergenome.nih.gov).
We defined a “smoker” as an individual that reports to be (or to have been) a smoker (244 patients), independently of smoking duration. We considered all other individuals to be “never-smokers” (37 patients). In the CRC dataset, the number of patients in each group was as follows: MSI (28 patients), microsatellite stable (MSS) (160 patients), MLH1 silent (23 patients), and MLH1 normal (196 patients).
We defined the somatic mutation rate R as the ratio of the total number of somatic mutations found in a patient and the patient’s age. To test the null hypothesis of no difference between the distributions of R or of the total number of somatic mutations, or more specifically, against the alternative hypothesis that one distribution is stochastically greater than the other (and therefore that one distribution has a larger median value than the other), we used the two-sided Wilcoxon–Mann–Whitney test (Wilcoxon rank sum test). We computed medians and CIs for the ratio of the R values between two groups via bootstrap (10,000 values). We performed the goodness-of-fit analysis using the Kolmogorov–Smirnov test statistics for comparing the distribution for the increase in R due to smoking or silent MLH1 (raised to the appropriate powers), as estimated from the TCGA data via bootstrap, with the distribution of the fold increase in cancer incidence determined by epidemiological studies. We used Jha et al. (23) for LUAD and Dowty et al. (14) for CRC. We averaged CIs for men and women. Because only a CI rather than the full distribution for the fold increase in cancer incidence was available from these studies, we assumed a normal distribution consistent with that CI. The Kolmogorov–Smirnov statistic was D = 0.80, 0.21, and 0.69 for two, three, and four drivers, respectively, in LUAD; and D = 1, 0.91, 0.42, and 0.98, for one, two, three, and four drivers, respectively, in CRC. For robustness, other distributions (uniform, gamma) were considered, and these yielded equivalent results. All statistical analyses were performed using R software, version 3.0.3 (31).
Mathematical Modeling.
There are various ways to derive the equation for cancer incidence found in Armitage and Doll (2). Here, we briefly review the basic assumptions behind this equation and show its derivation. We also discuss our new modeling approach and the methodology used in this paper.
Assume that in a healthy tissue the mutation rate of a given driver gene is approximately constant in time. Then the time, X, until the occurrence of a driver mutation in this gene can be modeled by an exponential random variable with (constant) rate u, where u represent the probability for that gene to mutate in a unit interval of time. This implies that the probability of that gene to be mutated by a given age t is 1 – e−ut, which is approximately equal to ut, given that u is many orders of magnitude smaller than t−1 (12). Now assume that, to evolve a given tumor at a given stage, a sequence of n driver mutations (rate-limiting steps) are required. It follows that the time of cancer occurrence is given by X1 + X2 + … + Xn, a sum of exponentially distributed independent random variables, where Xj+1 is the time it takes for driver j + 1 to occur once driver j has occurred, with probability density approximately (for ut 1):
where ui is the rate for Xi, i = 1, 2,…n. This probability density then represents the incidence for that cancer type at age t, if we disregard competing risks (in our analysis, epidemiological cancer incidence estimates at old ages will be avoided for this reason).
By taking the logarithm in the above equation, we obtain the well-known result that the slope of the cancer incidence curve in a log-log plot of incidence vs. age is given by the number of required rate-limiting steps minus 1, i.e., (n − 1), because
This assumes that all tumors in the individuals used to estimate the incidence require the same number n of rate-limiting steps, as n is a constant (see below for letting n have a distribution).
All of the above is widely known, and it originated with the work of Armitage and Doll (2). Estimating the number of rate-limiting steps using the slope of the log-log plot can be used to estimate the total number of rate-limiting steps, irrespective of whether they arise from point mutations or other genomic alterations and/or epigenetic events. A new element is introduced by considering dependence of gene mutation rates on gene length. Assume for now that all rate-limiting steps are driver gene mutations and that n of them are required for cancer. Even assuming a constant (average) mutation probability u per base pair per unit of time, different genes have different base pair lengths l1, l2, …, ln. It is also possible to include the effects of differing nucleotide compositions by multiplying the various l values by a factor depending on the specific nucleotide composition of each gene (32). The formula above may be written as follows:
Another relevant element can be introduced into the model using the following argument. Evidence indicates that a given cancer type may result from different combinations of driver gene mutations. It is also possible that n, the total number of driver gene mutations required by that cancer type, may not be a constant among patients. It is reasonable to assume that the range for n is relatively small, as we will explain below, and given the large body of evidence from age vs. incidence curves estimating n to be between 2 and 7 in various tissues. Thus, instead of assuming a fixed n for all patients of a given cancer type (at a given stage), we can consider a distribution for n. To be precise, let n be a variable taking values of positive integers, and m and M be defined, respectively, as the minimum and maximum of n required to evolve a given tumor at a given stage. Let jn be an index for all (mutually exclusive) combinations of exactly n drivers yielding a given detectable cancer type and let li(jn) be the length of the ith gene in the n-gene combination jn. Then the incidence at age t in the population will be given by the following:
where In is defined as the incidence for that cancer type at age t through the occurrence of n driver gene mutations.
A key element of the model is that an increase by a factor x (where x > 1) of the average mutation rate (per base pair across all nucleotides) u will cause an increase of that cancer incidence by a factor xn, independent of t,
In other words, the ratio between the incidence of two subgroups, where the first has a mutation rate x time larger than the second, will yield a ratio of their incidences equal to xn, independently of age (if we disregard competing risks, which represent a problem for incidence curves at older ages, and affect all models over that age range). Even for nonlinear transformations of time [for example, if the cell division rate is not constant with age, but rather any function of it, f(t)], where the standard approach would strongly suffer the age dependency due to the need of estimating the power of age t, our approach is independent of such transformations, and therefore more robust to time dependencies.
Our model can be naturally extended to also allow for the somatic mutation rate to have a distribution (even time dependent), making it robust against variation in the mutation rate.
It is possible to include in the model also other genetic alterations, like gene fusions, chromosomal translocation, and copy number changes (amplifications, large deletions, gains or losses of whole chromosomes or chromosome arms) as well as methylation changes, if the rate of their occurrence v were known. Thus, let ip be an index for all (mutually exclusive) possible combinations of exactly p of these other genetic and epigenetic alterations, which, together with n point mutations, yield a given detectable cancer type, and let vi(ip) be the mutation rate of the ith gene in the p-gene combination ip. Then, by considering these p more rate-limiting steps, we have the following:
Again, an increase by a factor x of the average somatic mutation rate u will cause an increase of that cancer incidence by a factor xn, independently of the other p rate-limiting steps to cancer. Note how environmental or inherited factors causing an increase also in the mutation rate of drivers that are not gene mutations would then lower, not raise, the estimate of the average number of driver gene mutations required for cancer development.
For x > 1 (always satisfied in our study), where m, M 1,
Therefore, for a given cancer with incidence , an increase by a factor x of the average mutation rate per base pair will cause an increase in incidence that will be bounded below by and above by . Let be defined as the constant for which the following equality holds:
that is, is the weighted sum for the increase in incidence, where the weights are given by ,
It is precisely the mathematical average of the fold increase xn (across all possible values for n, and where the weights are given by the frequencies of the various ns in the population). This is exactly what is identifiable in the data, whereas the standard weighted average of n would not. However, what is
From the fact that ut 1, and if we assume that the driver gene length distribution is not different across the various values of n, it follows that and
that is, the overall incidence of a given cancer type should be approximately determined by the combinations of minimum length (i.e., those for which n = m) of different driver gene mutations required to get to cancer status. Thus, may approximate well its lower bound m, the minimum number of rate-limiting driver mutations required to get to cancer.
More importantly, let n* be the standard weighted average of n,
By Jensen’s inequality,
thus, n* , and therefore is an upper bound for the average number of driver mutation hits, n*, required to evolve to a given tumor type. With close to m, should also be a good approximation of n*.
Clearly, implies that , which provides a way to estimate an upper bound.
The model presented up to this point has the important limitation of assuming that the sequential mutations resulting in cancer occur independently of each other. This represents a clear violation of what is known about cancer progression. For example, driver gene mutations, by definition, confer growth fitness advantages, some of which have been measured (33). Thus, the acquisition of a driver gene mutation induces a subclonal exponential growth that will affect the rate at which the next mutations are acquired. Each driver gene mutation increases the pool of cells already possessing mutation j and therefore at risk for the next mutation. Although a thorough analysis of all possible dependencies among driver gene mutations is beyond the scope of this work, we will consider two fundamental types of known dependencies and show that, even in these cases, an increase by a factor x of the rate u will still cause an overall increase in cancer incidence by a factor xn, with a potential adjustment to n. If the added fitness advantage conferred by subsequent drivers is equivalent to or higher than that conferred by a previous driver gene mutation, this adjustment to n is needed, as described below. If the added fitness advantage conferred by subsequent drivers is exponentially decreasing, no adjustment is needed. The main point is that our approach appears to be therefore sound, even when more complicated dependencies are considered.
First, we consider the case where the acquisition of a driver gene mutation induces a subclonal exponential growth caused by the conferred fitness advantage. For simplicity of exposition, we start by considering the case of n = 2, where the two mutations have a given order of occurrence, letting be the rate of the continuous exponential growth induced by the first driver, and where the following discrete deterministic approximation is used. The healthy cell is assumed to divide deterministically in time, according to a constant division rate, and time is counted by the (discrete) number of times the healthy cell has divided, i.e., the time to division is used as the time unit. We also assume, for simplicity of exposition, that cells divide asymmetrically, and that the possibly required mutation in the second allele of the first driver is not rate-limiting. The probability to get cancer by time t is as follows:
As
and , for all , it is apparent that So the two mutation rates factor out, and the result will not be affected by this dependency. Note that here the mutation rate probabilities ui are per cell, and not for the whole tissue's cell population.
When there are more than two driver gene mutations (n >2), it is possible to show, by combining results from Tomasetti et al. (12) for the self-renewal phase of a tissue and from Durrett et al. (34) for tumoral clonal expansion, that the mutation rates factor out approximately as follows:
where the are the growth rates induced by driver i. In our analysis, as in Figs. 3 and 4, we assume that the fitness advantage added by each successive driver is constant, a typical assumption given the lack of information on the actual values. This assumption yields the following values: = 1/2, = 1/3, and so on. This implies that if the mutation rate is x times higher in a subpopulation, then the incidence for that subpopulation should be
higher than in the control group. Note that for small n values this adjustment is not large. If instead the fitness advantages added by each successive driver were to be decreasing significantly, i.e., exponentially fast, then the situation reverts to the standard case with the incidence increasing by approximately a factor xn and our estimate of three driver gene mutations would represent an upper bound.
Another type of dependency occurs where a driver mutation will not appear (i.e., confer a fitness advantage) unless it occurs before the tumor has reached a certain critical size K. Such a driver gene mutation might, for example, only be observed once cells outgrow their blood supply. In such cases, equation 5 of Tomasetti et al. (35) applies, where it is shown that the probability for that mutation to occur before reaching size K is as follows:
where C is a constant. Because the above expression is , the mutation rate again factors out and our conclusions are not affected.
We want to note that, with the classical approach of estimating the number of rate-limiting steps, the results would be much more strongly dependent on the specific form of the assumed model for clonal expansions, timing of driver gene mutations, etc. Overall, then, we believe our approach allows a more robust inference of the number of rate-limiting steps required for cancer with respect to both time dependencies and model misspecifications.
Acknowledgments
We thank Donald Geman for useful comments on the manuscript. C.T. and L.M. were supported by NIH Core Grant P30CA006973 (to Johns Hopkins Kimmel Cancer Center). B.V. was supported by The Virginia and D. K. Ludwig Fund for Cancer Research and by NIH Grant R37CA43460.
Footnotes
The authors declare no conflict of interest.
References
- 1.Nordling CO. A new theory on cancer-inducing mechanism. Br J Cancer. 1953;7(1):68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8(1):1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1(2):157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 4.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little MP. Are two mutations sufficient to cause cancer? Some generalizations of the two-mutation model of carcinogenesis of Moolgavkar, Venzon, and Knudson, and of the multistage model of Armitage and Doll. Biometrics. 1995;51(4):1278–1291. [PubMed] [Google Scholar]
- 6.Luebeck EG, Moolgavkar SH. Multistage carcinogenesis and the incidence of colorectal cancer. Proc Natl Acad Sci USA. 2002;99(23):15095–15100. doi: 10.1073/pnas.222118199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moolgavkar SH, Dewanji A, Venzon DJ. A stochastic two-stage model for cancer risk assessment. I. The hazard function and the probability of tumor. Risk Anal. 1988;8(3):383–392. doi: 10.1111/j.1539-6924.1988.tb00502.x. [DOI] [PubMed] [Google Scholar]
- 8.Moolgavkar SH, Luebeck EG. Multistage carcinogenesis: Population-based model for colon cancer. J Natl Cancer Inst. 1992;84(8):610–618. doi: 10.1093/jnci/84.8.610. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci USA. 2013;110(6):1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowty JG, et al. Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat. 2013;34(3):490–497. doi: 10.1002/humu.22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank SA. Dynamics of Cancer: Incidence, Inheritance, and Evolution. Princeton Univ Press; Princeton: 2007. [PubMed] [Google Scholar]
- 16.Jackson AL, Loeb LA. The mutation rate and cancer. Genetics. 1998;148(4):1483–1490. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 18.Peto R, et al. Smoking, smoking cessation, and lung cancer in the UK since 1950: Combination of national statistics with two case-control studies. BMJ. 2000;321(7257):323–329. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 21.Doll R, Hill AB. Smoking and carcinoma of the lung; preliminary report. BMJ. 1950;2(4682):739–748. doi: 10.1136/bmj.2.4682.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doll R, Peto R, Boreham J, Sutherland I. Mortality from cancer in relation to smoking: 50 years observations on British doctors. Br J Cancer. 2005;92(3):426–429. doi: 10.1038/sj.bjc.6602359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jha P, et al. 21st-century hazards of smoking and benefits of cessation in the United States. N Engl J Med. 2013;368(4):341–350. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- 24.Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340(17):1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 25.Shukron O, Vainstein V, Kündgen A, Germing U, Agur Z. Analyzing transformation of myelodysplastic syndrome to secondary acute myeloid leukemia using a large patient database. Am J Hematol. 2012;87(9):853–860. doi: 10.1002/ajh.23257. [DOI] [PubMed] [Google Scholar]
- 26.Howlader N, et al. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute; Bethesda, MD: 2013. [Google Scholar]
- 27.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 28.Owens AH, Coffey DS, Baylin SB. Tumor Cell Heterogeneity: Origins and Implications. Academic; New York: 1982. [Google Scholar]
- 29.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. R Development Core Team (2013) R software, version 3.0.3. Available at www.r-project.org. Accessed July 1, 2014.
- 32.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 33.Vermeulen L, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342(6161):995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 34.Durrett R, Moseley S. Evolution of resistance and progression to disease during clonal expansion of cancer. Theor Popul Biol. 2010;77(1):42–48. doi: 10.1016/j.tpb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Tomasetti C, Levy D. Role of symmetric and asymmetric division of stem cells in developing drug resistance. Proc Natl Acad Sci USA. 2010;107(39):16766–16771. doi: 10.1073/pnas.1007726107. [DOI] [PMC free article] [PubMed] [Google Scholar]