Significance
Adaptation to hypoxia promotes cancer progression, resulting in enhanced patient mortality. Activation of hypoxia-inducible factor 1 (HIF-1) leads to a transcriptional switch, which, regulating angiogenesis, metabolism, and survival, results in hypoxia adaptation. In cancer, increased HIF-1 levels can be a result of either intratumoral hypoxia or the altered function of tumor suppressors. Our study demonstrates that the tumor suppressor TAp73, a member of the p53 family of genes, opposes HIF-1 activation in cancer cells, resulting in reduced angiogenesis and tumor progression. TAp73-depleted mice show increased tumorigenicity, associated with increased HIF-1 signaling and angiogenesis. Expression of TAp73 in human cancers predicts good survival outcome and retrocorrelates with HIF-1 expression and activation. The TAp73/HIF-1 axis plays a critical role in cancer pathogenesis.
Keywords: p73, p53 family, tumor progression, VEGF, tumor vascularization
Abstract
Tumor hypoxia and hypoxia-inducible factor 1 (HIF-1) activation are associated with cancer progression. Here, we demonstrate that the transcription factor TAp73 opposes HIF-1 activity through a nontranscriptional mechanism, thus affecting tumor angiogenesis. TAp73-deficient mice have an increased incidence of spontaneous and chemically induced tumors that also display enhanced vascularization. Mechanistically, TAp73 interacts with the regulatory subunit (α) of HIF-1 and recruits mouse double minute 2 homolog into the protein complex, thus promoting HIF-1α polyubiquitination and consequent proteasomal degradation in an oxygen-independent manner. In human lung cancer datasets, TAp73 strongly predicts good patient prognosis, and its expression is associated with low HIF-1 activation and angiogenesis. Our findings, supported by in vivo and clinical evidence, demonstrate a mechanism for oxygen-independent HIF-1 regulation, which has important implications for individualizing therapies in patients with cancer.
Adaptation to hypoxia allows for cancer progression in the hypoxic environment within tumors, resulting in enhanced patient mortality (1). A major mechanism mediating this adaptive response to reduced O2 is the activation of the hypoxia-inducible factors (HIF-1 and HIF-2). HIF gain of function results in increased tumor growth, vascularization, and metastasis, and high HIF-1α and HIF-2α protein levels are associated with a poor prognosis in many human cancers (2). HIF-1 is a heterodimeric transcription factor comprising a constitutive HIF-1β subunit and an oxygen-labile HIF-1α subunit (3). Prolyl-hydroxylases promote HIF-1α subunit degradation in response to high oxygen levels, whereas prolyl-hydroxylase enzymatic activity is reduced under low-oxygen tension, allowing for HIF-1α stabilization (4). HIF-1 activation results in a wide transcriptional reprogramming of cancer cells. HIF-1 directly controls the expression of crucial genes involved in proliferation/survival (IGF2, PDGFB), metabolism (GLUTs, HKs), angiogenesis (VEGF-A, PDGFB, ANGPTs), and metastasis (ZEB-1, MMP9). This transcriptional switch is an important determinant of tumor progression (5). Increased HIF-1α levels in cancer cells can be a result of either intratumoral hypoxia or the altered function of tumor suppressors and oncogenes. The most frequent mutation is the loss of function of the tumor suppressor Von Hippel-Lindau (VHL) (6), which increases HIF-1α half-life, nuclear accumulation, and transactivation under nonhypoxic conditions. The relationship between HIF-1 and tumor suppressors/oncogenes can be crucial in determining the fate of preneoplastic lesions. Among the different biological processes controlled by HIF-1, angiogenesis represents a limiting step for cancer progression. In addition to the enhanced blood supply, this angiogenic switch contributes to the tumor microenvironment, which enables the transition from dysplastic lesions to overt carcinoma (7).
Although TAp73 shares transcriptional control of proapoptotic genes in the response to genotoxic stress with p53, the precise role of p73 in tumorigenesis has been debated for some time. Initially, the observation that human TP73 gene mutations are rare in human cancer, together with the lack of spontaneous tumors in Trp73−/− mice, suggested that the contribution of p73 to tumorigenesis was marginal. However, the severity of the Trp73−/− mouse phenotype did not allow a full analysis of the tumor spectrum in aged mice (8). Studies on the small (25%) cohort of Trp73−/− mice surviving at 10 mo of age revealed a 60% lung adenocarcinoma incidence (9). Isoform-specific deletion of TAp73 (TAp73−/− mice) also resulted in both spontaneous and carcinogen-induced tumors with a particularly high incidence of lung adenocarcinomas (10), indicating TAp73 as a genuine tumor suppressor. Conversely, the p73 N-terminal truncated isoform, ΔNp73, acts as an oncogene. Thus, ΔNp73−/−-derived MEFs transformed with Kirsten rat sarcoma failed to form tumors when injected in BALB/c nu/nu mice (11). Moreover, increasing evidence suggests that an altered ratio between TAp73 and ΔNp73 isoforms affects tumor progression (10). Contradictory and preliminary evidence has linked p73 to VEGF expression, but not to angiogenesis (12, 13).
Here, we report the ability of TAp73 to oppose tumor progression and angiogenesis by directly promoting ubiquitin-dependent degradation of HIF-1α. TAp73 might act through the recruitment of mouse double minute 2 homolog (MDM2) to HIF-1α, facilitating its ubiquitin-dependent proteasomal degradation in an oxygen-independent manner. In human lung adenocarcinomas, TAp73 is a good prognostic factor, and its expression is inversely correlated with HIF-1α activation and the angiogenesis pathway. These data indicate that TAp73 loss contributes to tumor progression and activation of the angiogenic switch by stabilizing HIF-1 and the HIF-1-dependent angiogenic response.
Results
TAp73 Ablation Promotes Tumor Progression and Angiogenesis.
Three in four TAp73−/− mice and one in three TAp73+/− mice spontaneously develop malignancies (10). Hence, TAp73 is a genuine tumor suppressor gene. To investigate whether TAp73 could also control the later stages of tumorigenesis and directly affect tumor progression, we used a chemical-induced skin carcinogenesis protocol. The two-stage chemical carcinogenesis process (7,12-dimethylbenz[a]anthracene [DMBA]/12-O-tetradecanoyl-phorbol 13-acetate) in mouse skin offers the possibility of monitoring early and late events in cancer development and progression (14). The induced benign skin papillomas can either regress or progress to squamous cell carcinoma. TAp73−/− mice displayed increased tumor susceptibility and accelerated tumor progression. At 14 wk after the initial treatment, 77% of TAp73−/− mice had developed at least one lesion, whereas only 23% of TAp73+/+ mice showed any carcinogenic effect at the same point. The highest incidence (38%) in the TAp73+/+ group was reached at the 24th week, 9 wk after the TAp73−/− group (Fig. S1 A–C). The average tumor number per mouse was significantly greater in the TAp73−/− group (1.4 tumors/mouse compared with 0.5 in wild-type mice), and this result was associated with a larger tumor size. The lesions in TAp73+/+ mice never exceeded a 2-mm size, whereas TAp73 ablation led to a larger number of lesions greater than 2 mm in size, together with significant numbers of tumors 2–4 mm and greater than 4 mm in size (Fig. S1C). In addition, the progression from premalignant hyperplasia (papillomas) to invasive carcinoma (squamous cell carcinoma) was substantially more evident in the TAp73−/− group, leading to skin ulceration (Fig. S1 A and D). Overall, the accelerated initial lesion development, larger tumor sizes, and increased progression to squamous cell carcinoma suggested that TAp73 ablation promoted tumor progression. Notably, deficiency of TAp73 in topically carcinogen-treated mice also determined the onset of lung malignant lesions (Fig. S2A), confirmed by histological analysis (Fig. S2B). This supported the particular sensitivity of TAp73−/− mice to lung tumorigenicity. Lung adenocarcinomas indeed represent the most frequent cancer subtype spontaneously developed by Trp73−/− and TAp73−/− mice, being observed in 60% and 32% of these animals, respectively (9, 10).
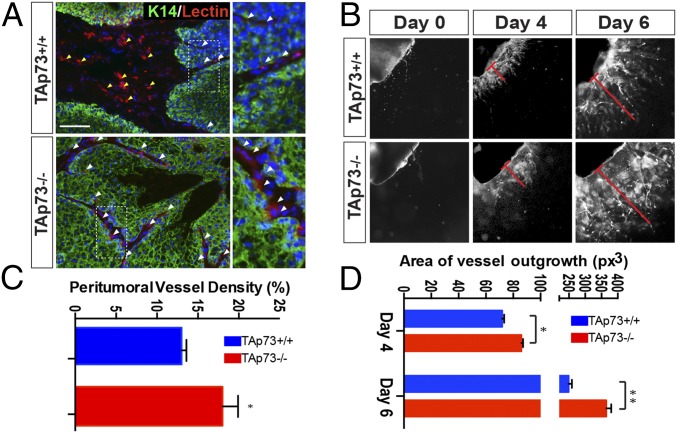
Tumor microenvironment remodeling is the rate-limiting step for cancer progression, and neoangiogenesis is particularly important for cancer cell niche maturation in a tumor microenvironment. Hyperplasia does not necessarily require neoangiogenesis; however, neovascularization induction correlates with the transition to neoplasia (7, 15). Thus, we analyzed peritumoral vascularization in TAp73−/− mice. The skin lesions that developed in TAp73−/− mice had increased numbers of small vessels in the peritumoral region compared with the tumors from TAp73+/+ mice (Fig. 1A, white arrows). Vasculature covered 18% of the peritumoral area in the TAp73−/− mice, but only 13% in the TAp73+/+ mice (a 40% increase; Fig. 1C). To confirm the propensity of the TAp73−/− genotype toward increased angiogenesis, we performed an aortic ring assay by culturing aorta ring samples from knockout and wild-type mice ex vivo and measuring new vessel formation. Strikingly, the aorta rings from TAp73−/− mice displayed a significant increase (20% at day 4 and 50% at day 6) in neocapillary formation compared with TAp73+/+ samples (Fig. 1 B and D). Thus, TAp73 loss had a stimulatory effect on neoangiogenesis in vitro, supporting the in vivo data that TAp73 affects an important biological process for tumor progression.
Fig. 1.
TAp73 depletion drives tumor progression. (A) Immunofluorescence staining for keratin-14 (green), lectin (red), and DAPI (blue) shows dermic (yellow arrows) and peritumoral (white arrow) vascularization in TAp73+/+ and TAp73−/− mice. (Scale bar, 50 μm.) (B) Peritumoral vessel density was assessed through the digital measurement of the area covered by tumor-associated vessels (relative mean ± SD, n = 3 per genotype, two-tailed unpaired t test, P = 0.018). (C and D) The aortic ring assay of TAp73−/− mice shows an increased propensity to form new vessels (relative mean ± SD, n = 3 per genotype, two-tailed unpaired t test, *P < 0.05, **P < 0.01).
TAp73 Interacts with HIF-1α, Facilitating Its MDM2-Dependent Ubiquitination and Consequent Proteasomal Degradation.
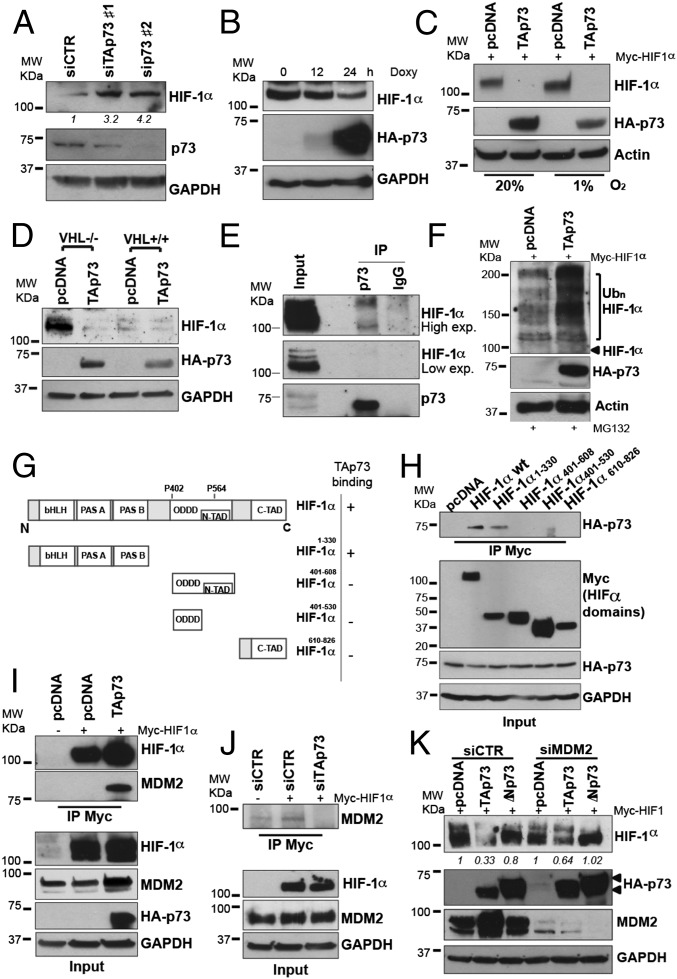
HIF-1 is a well-known master regulator of tumor progression. To investigate the potential mechanism by which TAp73 affects tumor growth and angiogenesis, we hypothesized that p73 could control HIF-1α protein levels. To test this hypothesis, we used two different p53-null cancer cell lines: H1299 cells, which express detectable levels of endogenous TAp73α protein (Fig. S3E), and SaOs-2 cells, which are negative for any p73 isoforms. Knockdown of endogenous TAp73 in H1299 promoted a marked increase in endogenous HIF-1α protein (Fig. 2A), whereas restoration of TAp73 expression in SaOs-2 cells was able to reduce endogenous HIF-1α protein (Fig. 2B). HIF-1α steady-state levels are regulated by oxygen-dependent, ubiquitin-mediated proteasomal degradation, which normally employs the tumor suppressor protein VHL (6). Hence, because HIF-1α protein levels are tightly controlled by intracellular oxygen concentrations, we investigated whether the TAp73-dependent HIF-1α reduction was sensitive to oxygen concentrations. However, the effect of TAp73 was independent of oxygen concentrations, and down-regulation of Myc-tagged or endogenous HIF-1α protein occurred efficiently in cells cultured in either normoxic (20% O2) or hypoxic (1% O2) conditions (Fig. 2C and Fig. S3A). In addition, TAp73 promoted HIF-1α degradation in VHL-mutant renal cell carcinoma (RCC4) cells, suggesting that TAp73-mediated degradation is independent of VHL (Fig. 2D). Finally, to further prove the independence of TAp73 degradation from the VHL pathway, we tested a hydroxylation-mutant HIF1α construct (HIF1α PA: P402A/P564A). Substitution of prolines by alanines in positions 402 and 564 impairs the ability of proline hydroxylase enzymes to promote VHL binding and oxygen-dependent degradation of HIF-1α (16). Concurrent overexpression of HIF1α PA with TAp73 showed that TAp73 can down-regulate HIF1α independent of the HIF1α hydroxylation status. (Fig. S3B). This result confirms that TAp73 can degrade HIF-1α by an oxygen-independent mechanism.
Fig. 2.
p73 interacts with HIF-1α and promotes its ubiquitin-dependent degradation. (A) p73 knockdown increased endogenous HIF-1α levels in the H1299 cells. (B) TAp73β overexpression reduced endogenous HIF-1α protein levels in SaOs Tet-On cells under basal conditions. (C) TAp73β reduced HIF-1α protein levels in both normoxic (20% O2) and hypoxic (1% O2) conditions. (D) TAp73β overexpression reduces endogenous HIF-1α protein levels in the VHL-mutant RCC4 parental cell line and the VHL-overexpressing RCC4 cell line. (E) Immunoprecipitation in H1299 in the presence of proteasomal inhibitor MG132 and cobalt chloride of endogenous p73 demonstrates the protein–protein interaction between TAp73 and HIF-1α. (F) TAp73β overexpression increased HIF-1α polyubiquitination in H1299 cells in the presence of CoCl. (G) Schematic representation of HIF-1α fragments generated to map the binding region of HIF-1α with TAp73. (H) Immunoprecipitation in H1299 cells of Myc-tagged HIF-1α fragments demonstrates the involvement of N-terminal bHLH/PAS domain of HIF-1α in the interaction with TAp73. (I) TAp73 drives the E3 Ub ligase MDM2 to HIF-1α. Immunoprecipitation shows that TAp73α facilitated the MDM2 interaction with HIF-1α. (J) Myc-HIF-1α immunoprecipitation in siTAp73-transfected H1299 cells demonstrated that TAp73 depletion impaired HIF1α-MDM2 interaction. (K) MDM2 knockdown in H1299 cells partially affects the TAp73-dependent HIF-1α degradation. Actin or GAPDH were used as loading controls.
HIF-1α accumulation can also be induced by exposure to cobalt chloride, which mimics hypoxia and inhibits VHL-dependent ubiquitination (3). To examine whether TAp73 influences HIF-1α protein stability in this model, the HIF-1α half-life was analyzed in H1299 cells treated with cobalt. In control-transfected cells, the HIF-1α protein decayed with a half-life of 20–40 min, whereas the HIF-1α half-life was extended beyond 40 min in sip73-transfected cells (Fig. S3 C and D). These data indicate that the oxygen-independent regulation of HIF-1α expression by TAp73 is exerted by controlling its protein stability.
To investigate the molecular mechanism by which TAp73 regulates HIF-1α protein stability, we next determined whether, similar to p53 (17, 18), a direct interaction with TAp73 was involved. Indeed, an immunoprecipitation assay in H1299 revealed the coexistence of endogenous TAp73 and HIF-1α in the same molecular complex (Fig. 2E). Consistently, protein interaction was confirmed by coimmunoprecipitation in either direction, overexpressing HA-TAp73 and Myc-HIF-1α in H1299 (Fig. S4A). To further investigate the detailed molecular mechanism of the TAp73/HIF-1α complex, we mapped the region of HIF-1α involved in the interaction. We generated different fragments of Myc-tagged HIF-1α: the N-terminal region HIF-1α1–330, the oxygen-dependent degradation domain HIF-1α401–608, the oxygen-dependent degradation domain lacking the N-terminal transactivation domain HIF-1α401–530, and the C-terminal region HIF-1α610-826 (Fig. 2G and Fig. S3B). Coimmunoprecipitation experiments demonstrate that only the N-terminal region of HIF-1α retained the ability to bind TAp73 (Fig. 2H). The N-terminal region of HIF-1α contains the basic helix-look-helix/PAS (bHLH/PAS) domain, which is involved in the dimerization of HIF-1α with HIF-1β and in DNA binding of the transcription factor. TAp73 might, therefore, potentially affect dimerization and transcriptional activity of HIF-1 before driving the degradation of the HIF-1α subunit.
To determine whether increased HIF-1α ubiquitination was a consequence of the TAp73–HIF-1α interaction, we analyzed ubiquitin-HIF-1α levels after TAp73 overexpression. To discriminate the TAp73 effects from oxygen-induced HIF-1α ubiquitination, we mimicked the hypoxia blockage of the HIF-1α–VHL interaction, using cobalt. In the presence of the 20S proteasome inhibitor, MG132, TAp73 transfection strongly increased the expression of the polyubiquitinated forms of HIF-1α 24 h after transfection (Fig. 2F). We next tested whether the proteasome was required for TAp73-induced HIF-1α degradation. Treatment of H1299 cells with MG132 inhibited TAp73-mediated HIF-1α degradation (Fig. S4C). Finally, to confirm the requirement of HIF-1α-polyubiquitination for TAp73-promoted proteasomal degradation, we used the ts20R cell line, which harbors a thermolabile E1 Ub-activating enzyme (19). As expected, TAp73 overexpression at the permissive temperature (34 °C) resulted in reduced HIF-1α in ts20R. However, when the cells were switched to the restrictive temperature (39 °C), the impairment of the thermolabile E1 enzyme strongly abrogated TAp73-dependent HIF-1α degradation (Fig. S4D, Left). Correction of the temperature-sensitive E1 enzyme through the introduction of human E1 cDNA in the ts20R-derived H38 cells (19) confirmed the ability of TAp73 to promote HIF-1α reduction at both temperatures (34 °C and 39 °C; Fig. S4D, Right). Thus, TAp73 promoted HIF-1α polyubiquitination and consequent proteasomal degradation through a protein–protein interaction with HIF-1α.
Ubiquitin conjugation to proteins destined for proteasomal degradation involves complex machinery that includes an E1 Ub-activating enzyme, E2 Ub-conjugating enzyme, and E3 Ub-protein ligase (20). Multiple E3 Ub-ligase enzymes are responsible for the selection of proteins destined for the proteasome. The E3 Ub-protein ligase MDM2 binds TAp73 at the N-terminal region but does not promote its ubiquitination or proteasomal degradation. Paradoxically, this interaction results in increased p73 stability (21), but inhibition of its transcriptional activity (22). Thus, the biological significance of this MDM2-TAp73 interaction is not completely clear. We hypothesized that the TAp73-HIF-1α interaction functioned to facilitate MDM2-dependent HIF-1α ubiquitination, with TAp73 playing a role as a scaffold protein. We indeed analyzed the TAp73 and HIF-1α protein complex for the presence of MDM2. Anti-Myc-HIF-1α immunoprecipitation from HA-TAp73-transfected H1299 cells displayed significantly higher MDM2 levels in the complex compared with empty vector-transfected cells (Fig. 2I). The opposite experiment, performed by selective knockdown of endogenous TAp73 in H1299 cells, displayed an impaired HIF-1α-MDM2 interaction when TAp73 was depleted, confirming that TAp73 might play a role in presenting HIF-1α to MDM2 for ubiquitination (Fig. 2J).
To weight the importance of MDM2 in TAp73-dependent HIF-1α degradation, we evaluated the effect of MDM2 depletion. MDM2 knockdown in H1299 cells partially reduced TAp73-induced HIF-1α degradation (Fig. 2K). Taken together, the data in Fig. 2 demonstrate that TAp73 interacts with HIF-1α, promoting its ubiquitin-dependent degradation and possibly acting as a scaffold protein to facilitate MDM2 recruitment in the molecular complex.
TAp73 Predicts Lung Adenocarcinoma Patient Survival and Correlates with HIF-1α Activity and Angiogenesis Repression.
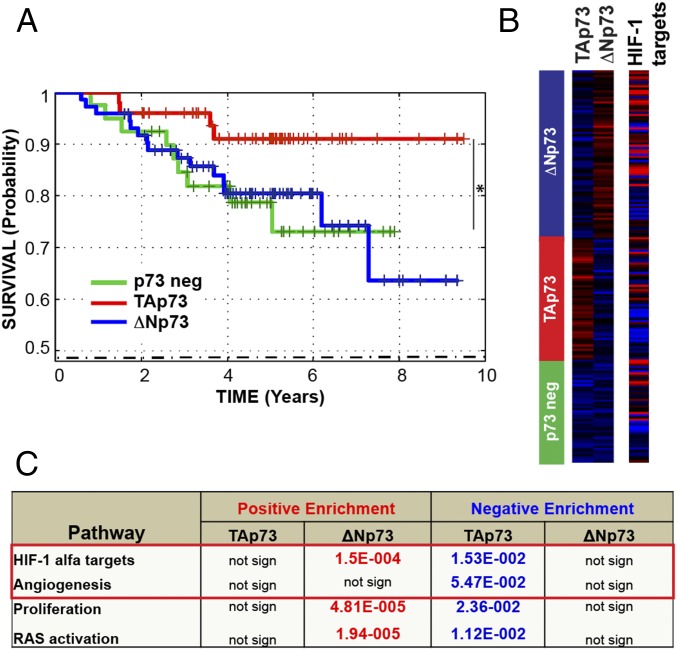
We next assessed whether this TAp73/HIF-1α complex was also relevant in human cancer and performed transcriptional signature analysis on a large group of biopsies from human cancers. Lung adenocarcinoma is the most frequent cancer subtype spontaneously developed by TAp73−/− mice and is also highly frequent in different chemical-induced carcinogenesis (10). Thus, we focused our attention on this particular tumor in humans, analyzing 226 biopsies from a human lung adenocarcinoma dataset (GSE31210) (23). Unsupervised sample clustering by TAp73 or ΔNp73 expression revealed three groups: TAp73-expressing (red group), ΔNp73-expressing (blue group), and no-p73-expressing (p73 neg; green group) (Fig. 3B). Using univariate Kaplan-Meier analyses, the individuals in the lung adenocarcinoma group with high TAp73 expression were shown to have a significantly better prognosis compared with the ΔNp73 and p73 neg groups (Fig. 3A). To gain insights into the mechanisms by which TAp73 was linked to improved survival in this cancer dataset, we investigated whether its expression was linked to other known tumorigenic pathways by gene set enrichment analyses. The “HIF-1α activity” and “angiogenesis” signatures were inversely associated with TAp73 expression, suggesting that TAp73 expression counteracts their activation in cancer (Fig. 3 B and C). Strikingly, this finding validated the functional connection between TAp73 and HIF-1α, confirmed the implication on tumor angiogenesis, and underlined the high relevance of our findings for human cancers. These results clearly demonstrate that TAp73 significantly inhibits HIF-1α activation and angiogenesis in human cancer.
Fig. 3.
TAp73 predicts patient survival and correlates with HIF-1 activity in the human cancer. (A) A Kaplan-Meier graph representing the probability of survival in patients with lung adenocarcinoma (dataset, GSE31210), stratified according to p73 expression levels. TAp73 strongly predicts a good patient prognosis (TAp73 group, n = 45; no-p73 group, n = 64; ΔNp73 group, n = 86; *P = 0.018). (B and C) Geneset enrichment analysis indicated negative enrichment of “HIF-1α target genes” and “angiogenesis pathway” in tumors expressing high TAp73 levels. Numbers in the table indicate P values.
TAp73 Affects HIF-1α Signaling in Vitro and in Vivo.
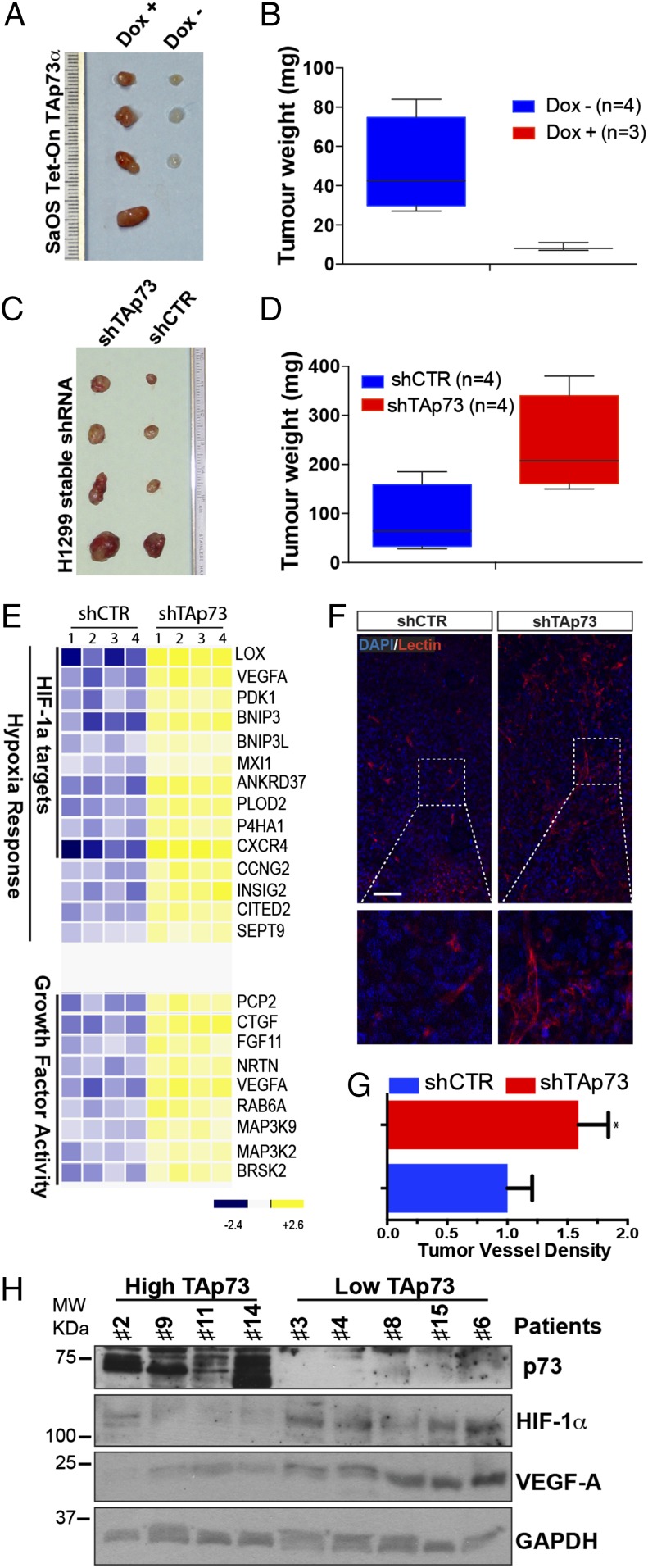
To confirm that TAp73 is also functionally affecting HIF-1α activity, we evaluated the expression of the HIF-1α downstream targets. TAp73 overexpression in H1299 cells under both normoxic and hypoxic conditions reduced VEGF-A and VEGF-R2 mRNA levels (Fig. S5 A and B). VEGF-R2 represents the transmembrane receptor for VEGF-A, which is indirectly induced by HIF-1α, hypoxia, and the proangiogenic (24). To confirm that TAp73 is able to affect HIF-1 signaling in a valid tumor environment, we used mouse tumor xenograft models s.c., implanting human cancer cell lines in athymic BALB/c mice (nu/nu). Similar to Fig. 2, we reverted the expression of TAp73 in two human cancer cell lines, which express (H1299) or do not express (SaOs-2) endogenous TAp73. Induction of TAp73α by doxycycline oral administration in nu/nu mice, after inoculation of SaOs-2 Tet cells, harshly abolished tumor growth (Fig. 4 A and B and Fig. S5 C and D). Tumor weight was reduced by ∼80% in three mice, and one inoculation completely failed to show any tumor. This experiment highlighted the powerful extent of TAp73’s tumor-suppression properties. Next, we stable knocked-down TAp73 in H1299 cells (Fig. S6 A and B), and we again inoculated athymic BALB/c mice (nu/nu). ShTAp73 H1299 cells displayed a significantly higher tumor weight compared with control cells, confirming that TAp73 depletion favors tumorigenesis (Fig. 4 C and D and Figs. S5 C and D and S6C). To determine the signaling pathways affected in TAp73-depleted tumors, we analyzed the expression profiles of the tumor xenografts, using gene microarray analysis. We identified 200 genes modulated in TAp73-depleted tumors, 123 of which were up-regulated (Fig. S7A and Dataset S1). Moreover, Gene Ontology term analysis identified “Response to Hypoxia” as the most significantly enriched pathway among the up-regulated genes (Fig. 4E and Fig. S7 B and C). Specifically, 14 up-regulated genes were associated with hypoxia; and 10 of these, including VEGF-A, were direct transcriptional targets of HIF-1 (Fig. 4E). A selected gene group was analyzed using quantitative (q)PCR to validate the microarray analysis (Fig. S8). The expression profile analysis strikingly confirmed that TAp73 depletion enables HIF-1 signaling in the tumor environment. Indeed, vessel density in tumor xenograft tissues indicated increased vascularization in tumors lacking TAp73 (Fig. 4 F and G).
Fig. 4.
TAp73 affects HIF-1 signaling in tumors. (A and B) Xenograft tumors derived from transplantation of SaOs-2 Tet on TAp73α in nu/nu mice. TAp73 expression was induced by oral doxycycline administration. Tumor weight was assessed 5 wk after inoculation (mean ± SD, n = 4 per group, two-tailed unpaired t test, P < 0.05). (C and D) Xenograft tumors derived from transplantation of H1299 cells stably transfected with shCTR or shTAp73. TAp73 depletion increased the tumor weight of H1299-derived tumors (n = 4 per group, two-tailed unpaired t test, P < 0.05). (E) A heat map of “HIF-1 Response” and “Growth Factor Activity” genes identified by GO term analysis, as differentially expressed between TAp73-intact and TAp73-depleted tumors (n = 4 per group). (F) Immunofluorescence staining for lectin (red) and DAPI (blue) indicated intratumoral vascularization in shCTR-H1299-derived or shTAp73-H1299-derived tumor xenograft tissues. (Scale bar, 100 μm.) (G) Intratumoral vessel density was assessed by digital measurement of the area covered by lectin-positive vessels (mean ± SD of n = 4 animals per group, two-tailed unpaired t test, *P < 0.01). (H) “High TAp73” fresh specimens from patients with lung cancer exhibited lower HIF-1α and VEGF-A protein levels compared with “Low TAp73” samples; GAPDH was used as a loading control.
To confirm that TAp73 can disrupt the HIF-1α/VEGF axis in human cancers, we performed an additional analysis on different human lung cancer datasets. By splitting the samples into tumors expressing high or low TAp73 levels, we found that HIF-1α expression only correlates with its target, VEGF-A, in the low-TAp73-expression group, whereas the correlation was absent when TAp73 was highly expressed (Table 1). These data indicate that altered TAp73 expression affects HIF-1α activity in clinical settings. Finally, we analyzed protein content in fresh lung cancer specimens. We again split the samples into two groups (“High TAp73” and “Low TAp73”), depending on TAp73 Western blot data. Validating our data, the human lung cancers expressing high TAp73 displayed reduced HIF-1α and VEGF-A protein levels; conversely, low TAp73 expression was associated with higher HIF-1α and VEGF-A levels (Fig. 4H).
Table 1.
Correlation between HIF-1 and VEGF-A in conditions of p73 high expression and low expression
| Transcriptional factor | TF targets | p73 Low | p73 High | Dataset | P value |
| HIF1 | VEGF | 0.269818674 | −0.111102663 | GSE12771 | 0.01 |
| HIF1 | VEGF | 0.138108818 | −0.446627573 | GSE20189 | 0.04 |
| HIF1 | VEGF | 0.335418822 | 0.089737037 | GSE31210 | 0.05 |
The table shows a correlation between HIF1a and its targets in conditions of p73 high expression and low expression in different cancer patient datasets.
Discussion
HIF-1α stabilization in cancer cells enables tumor progression and results in poor survival. We have identified a novel TAp73-dependent tumor suppression mechanism that controls HIF-1α signaling. TAp73 physically interacts with HIF-1α and affects its protein stability, promoting ubiquitin-dependent proteasomal degradation in an oxygen-independent manner. TAp73-induced HIF-1α degradation is reduced in the absence of MDM2, although MDM2 knockdown does not completely rescue the TAp73 effect, suggesting that additional mechanisms may be involved in TAp73-dependent HIF-1α regulation. MDM2 has previously been implicated in HIF-1α regulation, via both a direct (25) and a p53-dependent mechanism (18). However, in our model, we have investigated the TAp73/HIF-1α/MDM2 axis in a p53 null background (H1299 and SaOs-2 cell lines), which might contribute to the different interpretations previously reported, as no interference by MDM2/p53 and p53/HIF-1α signaling are present in our system. MDM2 interacts with the transcriptional activation (TA) domains of p53, p63, and p73; however, these interactions with the p53 family members have different consequences. MDM2 is the major negative regulator of p53 (26). MDM2 protein masks the TA domain of p53 and promotes its proteasomal degradation through polyubiquitination. MDM2 also inhibits TAp73 transcriptional activity but, importantly, does not induce its ubiquitination and degradation (21). Here, we propose that the biological meaning of the physical interaction between TAp73 and MDM2 might lie in the role of TAp73 as a scaffold protein to serve HIF-1α to MDM-2. Our model seems to suggest that a TAp73/MDM2 molecular complex may act in TAp73-expressing cancer cells to target HIF-1α signaling and counteract tumor progression and angiogenesis. We provide a possible mechanism to explain the molecular determinants of HIF-1α interaction with the ubiquitination machinery in an oxygen-independent manner. In addition, considering the high degree of similarity between HIF-1α and HIF-2α, particularly in the bHLH/PAS domain, where TAp73 has been shown to bind, it is a reasonable hypothesis that TAp73 exerts a possible similar regulatory effect on HIF-2α.
TAp73 deficiency results in increased tumorigenesis in both TAp73−/− mice and human lung carcinoma cells (shTAp73 H1299) transplanted into nu/nu mice. Notably, multistep skin carcinogenesis in TAp73−/− mice clearly displayed a worse tumor progression, with larger skin papillomas and earlier squamous cell carcinoma onset. TAp73 deficiency also resulted in a higher neo-angiogenic propensity, which was associated with higher VEGF-A expression levels, suggesting that TAp73 depletion releases HIF-1α signaling. Thus, TAp73 deficiency enabled the HIF-1α-dependent angiogenic switch and promoted tumor progression. Further supporting this idea is the observation that TAp73 represents a powerful predictor of survival in patients with lung cancer. Strikingly, in large-scale transcriptome analysis, TAp73 expression correlated with low HIF-1α activity and the angiogenic pathway; we also validated this result by examining protein expression levels in fresh human lung carcinoma specimens. These latter findings provide strong clinical relevance to our in vitro and in vivo data. In addition, given that HIF-1α overexpression is widespread in human cancers through tumor hypoxia or oncogenic mechanisms, the effect of the TAp73-HIF-1α pathway may extend beyond lung cancer. Thus, our study may provide molecular insights into the biological consequences of TAp73 loss in diverse cancer types.
TAp73 has also been proven to control metastasis onset. Interacting with nuclear factor Y (NF-Y), p73 represses the expression of platelet-derived growth factor receptor beta (PDGFRb) in pancreatic noninvasive cells. Expression of mutant p53 disrupts the p73/NF-Y interaction, determining a PDGFRb-dependent metastatic potential of pancreatic cancer cells (27). Our data suggest that the TAp73–HIF-1 axis might also have a strong effect on the metastatic potential of cancer cells. Similarly, mp53 might affect p73/HIF-1 interaction, promoting HIF-1 proinvasion and prometastasis programming.
In conclusion, our findings demonstrate that TAp73 is a controller of tumor progression and angiogenesis. We provide evidence that TAp73 is a regulator of HIF-1 signaling through a nontranscriptional mechanism mediated through protein–protein interactions. TAp73 loss enables HIF-1 signaling, promoting tumor growth, angiogenesis, and progression. Thus, TAp73 functions as a tumor suppressor, whose inactivation contributes to tumorigenesis in part through the induction of the HIF-1 pathway.
Materials and Methods
Mice and Xenograft Tumor Model.
The two-step DMBA/PMA skin carcinogenesis has been performed on TAp73+/+ and TAp73−/− mice of the C57B6/129Ola (F7) genetic background by dorsal skin administration once of DMBA (25 µg in 0.1 mL), followed by biweekly exposure to PMA (6.25 µg in 0.1 mL of acetone), starting at 2 wk after DMBA exposure. For the xenograft tumor model, nu/nu mice were purchased from Charles River. A total of 1.5 × 106 SaOs Tet on TAp73alfa or 2.5 × 106 shCTR/shTAp73 H1299 cells were injected s.c. into 6–8-wk-old nude (nu/nu) mice in a one-to-one mix of RPMI:Matrigel (Gibco).
Aortic Ring Assay.
Thoracic aortas were removed from 8–12-wk-old TAp73+/+ and TAP73−/− mice and sectioned in 1-mm-long aortic rings, sealed in place with an overlay of 70 μL of basement matrix extract (BME). Tubular structures were examined using a phase contrast microscope (Axiovert 200M, Zeiss) and photographed.
Immunoprecipitation and Western Blot.
Immunoprecipitation of endogenous or overexpressed proteins have been performed by incubating overnight (ON) at 4 °C with anti-p73 (Imgenex) and then 1 h at 4 °C with Protein G-Agarose (Roche) or ON at 4 °C with Ez view Red anti-HA affinity gel (E6779, Sigma) or Myc-Tag Sepharose beads (3400S, Cell Signaling). Immunoblotting was performed using a standard protocols list of antibodies, available in SI Materials and Methods. Full materials and methods information is available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank H. Ozer and C. Borner for kindly providing the Ts20 and H38 cell lines, D. J. Read and J. M. Edwards for their expert assistance in microscopy and histology studies, and K. Dudek for assistance in gene microarray analysis. We also thank V. Petrova, M. Mancini, and S. Singh for their technical support and M. Agostini for scientific discussion. This study was supported by the Medical Research Council, United Kingdom, by Ministero dell'Università e Ricerca, MinSan/Istituto Dermatopatico dell'Immacolata–Istituto di Ricovero e Cura a Carattere Scientifico (RF73, RF57), and by Italian Association for Cancer Research investigator grants (to G.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410609111/-/DCSupplemental.
References
- 1.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. HIF-1, O(2), and the 3 PHDs: How animal cells signal hypoxia to the nucleus. Cell. 2001;107(1):1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 5.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 7.Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13(7):511–518. doi: 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
- 8.Yang A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404(6773):99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 9.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22(19):2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilhelm MT, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24(6):549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salimath B, Marmé D, Finkenzeller G. Expression of the vascular endothelial growth factor gene is inhibited by p73. Oncogene. 2000;19(31):3470–3476. doi: 10.1038/sj.onc.1203672. [DOI] [PubMed] [Google Scholar]
- 13.Vikhanskaya F, et al. p73 Overexpression increases VEGF and reduces thrombospondin-1 production: Implications for tumor angiogenesis. Oncogene. 2001;20(50):7293–7300. doi: 10.1038/sj.onc.1204896. [DOI] [PubMed] [Google Scholar]
- 14.Filler RB, Roberts SJ, Girardi M. Cutaneous two-stage chemical carcinogenesis. CSH Protoc. 2007;2007:pdb prot4837. doi: 10.1101/pdb.prot4837. [DOI] [PubMed] [Google Scholar]
- 15.Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339(6219):58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- 16.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 17.Hansson LO, Friedler A, Freund S, Rudiger S, Fersht AR. Two sequence motifs from HIF-1alpha bind to the DNA-binding site of p53. Proc Natl Acad Sci USA. 2002;99(16):10305–10309. doi: 10.1073/pnas.122347199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravi R, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14(1):34–44. [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14(3):1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 21.Bálint E, Bates S, Vousden KH. Mdm2 binds p73 alpha without targeting degradation. Oncogene. 1999;18(27):3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 22.Zeng X, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19(5):3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okayama H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 24.Elvert G, et al. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J Biol Chem. 2003;278(9):7520–7530. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- 25.Joshi S, Singh AR, Durden DL. MDM2 regulates hypoxic hypoxia-inducible factor 1α stability in an E3 ligase, proteasome, and PTEN-phosphatidylinositol 3-kinase-AKT-dependent manner. J Biol Chem. 2014;289(33):22785–22797. doi: 10.1074/jbc.M114.587493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prives C. Signaling to p53: Breaking the MDM2-p53 circuit. Cell. 1998;95(1):5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 27.Weissmueller S, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell. 2014;157(2):382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.