Significance
Grain weight is an important crop yield component; however, its underlying regulatory mechanisms are largely unknown. Here, we identify a grain-weight quantitative trait locus (QTL) in rice encoding a new-type GNAT-like protein that harbors intrinsic histone acetyltransferase activity (OsglHAT1). Elevated OsglHAT1 expression enhances grain weight and yield by enlarging spikelet hulls via increasing cell number and accelerating grain filling, and increases global acetylation levels of histone H4. Our findings reveal the first example, to our knowledge, of a QTL for a yield component trait being due to a chromatin modifier that has the potential to improve crop high-yield breeding.
Keywords: grain size, weight, yield, plant biomass, rice
Abstract
Grain weight is an important crop yield component; however, its underlying regulatory mechanisms are largely unknown. Here, we identify a grain-weight quantitative trait locus (QTL) encoding a new-type GNAT-like protein that harbors intrinsic histone acetyltransferase activity (OsglHAT1). Our genetic and molecular evidences pinpointed the QTL-OsglHAT1’s allelic variations to a 1.2-kb region upstream of the gene body, which is consistent with its function as a positive regulator of the traits. Elevated OsglHAT1 expression enhances grain weight and yield by enlarging spikelet hulls via increasing cell number and accelerating grain filling, and increases global acetylation levels of histone H4. OsglHAT1 localizes to the nucleus, where it likely functions through the regulation of transcription. Despite its positive agronomical effects on grain weight, yield, and plant biomass, the rare allele elevating OsglHAT1 expression has so far escaped human selection. Our findings reveal the first example, to our knowledge, of a QTL for a yield component trait being due to a chromatin modifier that has the potential to improve crop high-yield breeding.
Rice (Oryza sativa L.) is the staple food for one-half of the world population (1). To meet the ever-growing demand for this crop, it is essential to develop rice varieties with higher yield potential. Grain weight is an important yield-related trait in rice; however, because it is regulated by multiple naturally occurring quantitative trait loci (QTLs), attempts to maximize it have proved difficult. Additionally, the potential size of the rice grain is physically restricted by the size of the hull, which is determined 1 wk before flowering (2, 3). Therefore, even with ideal grain filling, the size of the spikelet hull (i.e., grain length, width, and thickness) determines the final grain weight.
Recent cloning studies have identified some of the underlying QTLs for grain weight, such as the transmembrane protein GS3 (4, 5) and its homolog DEP1 (6), the Kelch-like domain Ser/Thr phosphatase GL3.1 (also called OsPPKL1) (7, 8), the RING-type E3 ubiquitin ligase GW2 (grain width and weight 2) (9), the arginine-rich domain nuclear protein qSW5/GW5 (10, 11), the putative serine carboxypetidase GS5 (12), the SBP domain transcription factor GW8 (OsSPL16) (13), and the newly reported IAA-glucose hydrolase protein TGW6 (14). However, the current understanding of the mechanisms of grain weight regulation remains fragmentary, and the precise mechanism by which any of the proteins is unknown.
Here, we present the identification and functional analysis of a QTL regulating grain weight, hull size, yield, and plant biomass. We reveal a previously unidentified member of histone acetyltransferases (HATs) that function as positive regulators of these traits. These findings provide the first mechanistic demonstration, to our knowledge, of HAT modulation of important agronomic traits.
Results and Discussion
QTL Cloning for Grain Weight at GW6a.
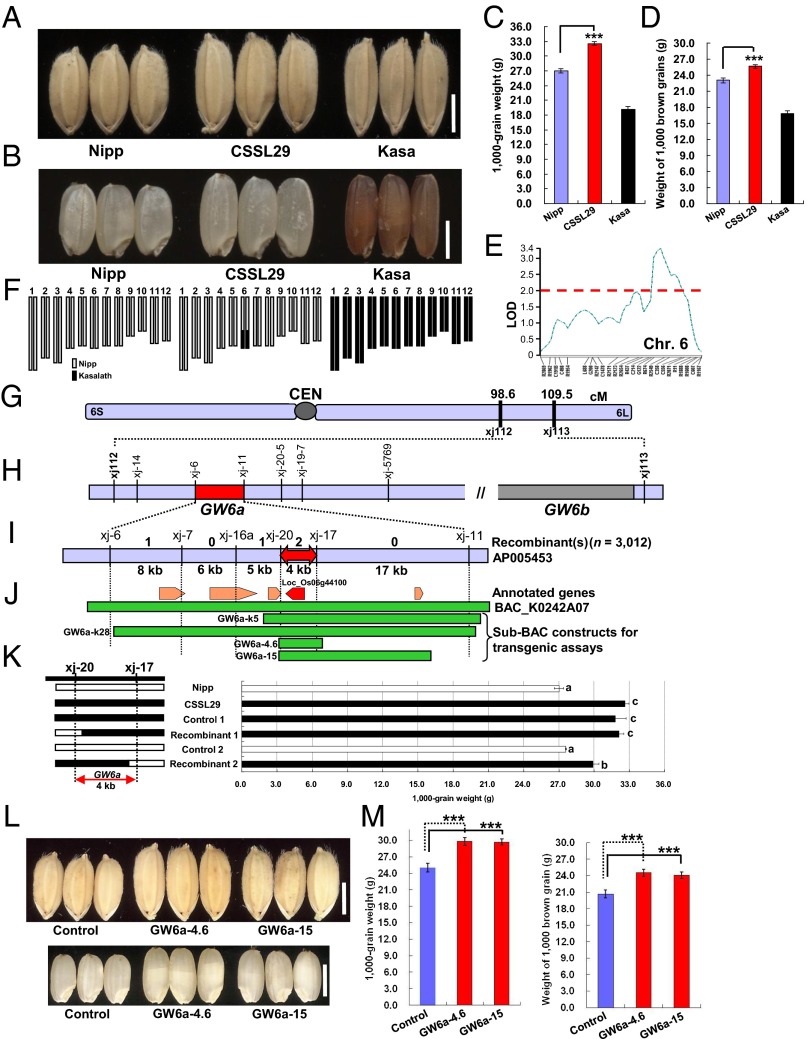
To clone QTLs for grain weight, we applied a QTL detection approach on a set of backcrossed inbred lines derived from a cross of Kasalath (Kasa, a rice indica variety) with the heavier Nipponbare (Nipp, a japonica variety) (15) (Fig. 1 A–D and SI Appendix, Fig. S1). Our QTL analysis identified a QTL, Grain weight on chromosome 6 (GW6) (Fig. 1E), enhanced by the Kasa allele. We then selected CSSL29, a chromosome segment substitution line that harbors an introgression of this Kasa region in the Nipp genetic background (Fig. 1F). As expected, CSSL29 had a significant increase in grain weight and brown grain weight (20.6% and 11.2%, respectively) compared with Nipp (P < 0.001) (Fig. 1 A–D).
Fig. 1.
QTL cloning at GW6a. (A–D) Grain and brown grain phenotypes. (E) QTL GW6 detection. A threshold of 2.0 as LOD (log likelihood) was used to declare the presence of significant QTL in a genomic region. (F) Graphical genotypes. (G) The candidate region of GW6 defined by markers xj112 and xj113. (H) GW6 consisted of two loci: GW6a, mapped between markers xj-6 and xj-11, and GW6b. (I) Fine-mapping of GW6a to a portion of Nipp PAC clone AP005453, where four recombinants were identified by using 3,012 plants. (J) Five annotated genes exist within the mapped region of ∼40 kb, and the Kasa genomic BAC clone K0242A07 and four sub-BACs for the transgenic assays are shown. (K) Progeny testing shows that the QTL GW6a effect is placed within a 4-kb interval. (L) Grain and brown grain phenotypes of indicated plants. (M) Comparisons of grain weight between plants shown in L. ***P < 0.001; Student’s t test was used to generate the P values in C, D, and M and a pairwise test was used to determine significance in K. (Scale bars: 3 mm.)
Next, we obtained an F2 population of CSSL29 crossed with Nipp and initially mapped GW6 to a region between markers xj112 and xj113 (Fig. 1G). Unexpectedly, however, this region consisted of two loci (GW6a and GW6b) that impacted grain weight equally (Fig. 1H). Analysis of both loci demonstrated more frequent recombination events at GW6a; we therefore focused on this locus and mapped it to a region between markers xj-6 and xj-11 (Fig. 1H). Upon analyses of an additional 3,012 F2 plants, we identified four recombinants that we used for a subsequent high-resolution linkage analysis (Fig. 1I). We identified an interval of ∼40 kb containing five predicted genes (Fig. 1J). To verify this result, we screened a bacterial artificial chromosome (BAC) genomic library of Kasa, and obtained a positive clone (K0242A07), from which two sub-BACs (GW6a-k5 and GW6a-k28) were derived. These BACs were cloned into a binary vector (Fig. 1J) and used for Agrobacterium tumefaciens-mediated transgenic assays. We identified two key recombinants, xj-20 and xj-17, resolved GW6a to a 4-kb region through progeny testing of fixed recombinant plants (Fig. 1K), and then constructed additional sub-BACs (GW6a-4.6 and GW6a-15; Fig. 1J) for transgenic assays. We observed significantly heavier grains in the transgenic lines containing these clones (Fig. 1 L and M and SI Appendix, Fig. S2). Thus, we conclude that the mapped 4-kb interval contains GW6a.
GW6a Encodes a Functional GNAT-like Protein: OsglHAT1.
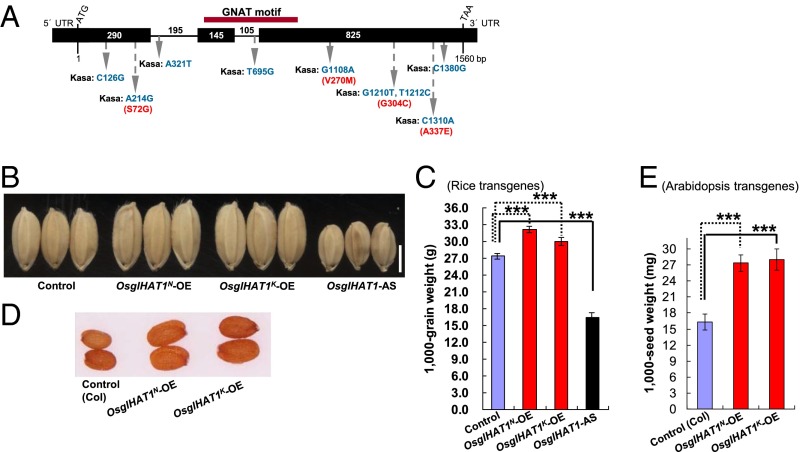
We found that the candidate GW6a region contained one ORF (Loc_Os06g44100) (Fig. 1 I–J). On comparing its cDNA sequence of the Nipp allele with the corresponding genomic DNA (gDNA), we found three exons and two introns (Fig. 2A). The Rice Genome Automated Annotation System (http://riceGAAS.dna.affrc.go.jp) annotated this gene as GCN5-related N-acetyltransferase-like (GNAT-like) (OsglHAT1), containing a conserved GNAT motif (Fig. 2A). Comparisons of gDNAs of the parental ORFs identified nine single-nucleotide polymorphisms (SNPs), of which five caused changes in four amino acids; however, none of the changed amino acids were localized within the conserved GNAT domain (Fig. 2A).
Fig. 2.
GW6a encodes a functional GNAT-like protein: OsglHAT1. (A) OsglHAT1 structure and mutation sites, including SNPs (blue) and changed amino acid residues (red). (B) Grain phenotypes of plants overexpressing the OsglHAT1 Nipp allele (OsglHAT1N-OE) and the Kasa allele (OsglHAT1K-OE), and OsglHAT1 antisense transgene (OsglHAT1-AS). (C) Comparison of grain weight. (D) Seed phenotypes of Arabidopsis plants overexpressing the OsglHAT1N-OE and OsglHAT1K-OE. (E) Comparison of seed weight of Arabidopsis transgenes. ***P < 0.001. Student’s t test was used to generate the P values in C and E.
To evaluate the functional consequences of the OsglHAT1 alleles in plants, we overexpressed the Nipp allele OsglHAT1N (OsglHAT1N-OE) and Kasa allele OsglHAT1K cDNA ORFs (OsglHAT1K-OE), and a series of alleles with SNP combinations from the parental alleles driven by the 35S promoter. These transgenic plants all displayed enhanced grain weights and elevated OsglHAT1 transcript expressions (Fig. 2 B and C and SI Appendix, Figs. S3 and S4 A and B). In contrast, transgenic plants overexpressing OsglHAT1 (the entire cDNA ORF) in the antisense direction (OsglHAT1-AS) showed markedly decreased grain weights and reduced endogenous OsglHAT1 transcripts (Fig. 2 B and C and SI Appendix, Fig. S4 A, C, and D). In addition, transgenic plants overexpressing the OsglHAT1 alleles in Arabidopsis produced larger, significantly heavier seeds than the wild type (Fig. 2 D and E and SI Appendix, Fig. S4E). Together, these observations support the notions that both parental OsglHAT1 alleles can functionally influence grain weight and that OsglHAT1 has a crucially conserved role in modulating seed size and weight in both monocots and dicots. The results also suggested that none of the amino acid differences between the Kasa and Nipp alleles are the cause of the phenotypic difference and that altered expression of the alleles alone may be responsible.
Altered OsglHAT1 Promoter Activity Underlies the QTL Effect on Grain Weight Regulation.
To examine the expression profile of OsglHAT1, we carried out reverse transcription-PCR (RT-PCR) and quantitative real-time PCR (qPCR) analyses to compare Nipp with its nearly isogenic line, NIL(OsglHAT1). Whereas OsglHAT1 transcripts were present in all organs and tissues examined, with preferential expression in young panicles (consistent with its function in grain weight regulation), higher OsglHAT1 transcripts were consistently observed in the NIL(OsglHAT1) genotypes (SI Appendix, Fig. S6). These results were confirmed by qPCR analysis (Fig. 3A).
Fig. 3.
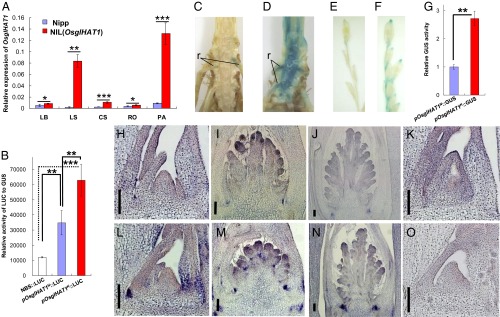
Altered OsglHAT1 promoter activity underlies the QTL effect on grain weight regulation. (A) qPCR analysis of OsglHAT1 expression pattern. RNA was isolated and quantitated by qPCR, normalized to ubiquitin. CS, culm tissue containing shoot apical meristem; LB, leaf blade; LS, leaf sheath; PA, young panicle; RO, root. (B) Transient assay using maize leaf protoplasts to test OsglHAT1 promoter activity. GUS staining of transgenic samples containing pOsglHAT1N::GUS (C and E) and pOsglHAT1K::GUS construct (D and F). r, root hair. (G) Quantification of the GUS signal that harbors the construct as indicated. In situ RNA hybridization of OsglHAT1 shows expression in the vegetative stage (H and L) and during the reproductive stage (I, J, M, and N); (K and O) Negative controls of OsglHAT1 in situ RNA hybridization that uses a sense probe. (Scale bars: 100 μm.) The length of the promoters pOsglHAT1N and pOsglHAT1K used in B–G was 1,681 and 1,652 bp, respectively, upstream of the ORF of OsglHAT1 alleles. Sample sections in H–K are Nipp genotypes, and in L–O are Kasa genotypes. *P < 0.1; **P < 0.05; ***P < 0.001. Student’s t test was used to generate the P values.
To identify the causes of the observed differences in OsglHAT1 allelic expressions we focused on the gene promoter region, which we had previously linked to the QTL effect (Fig. 1K and SI Appendix, Fig. S7). We used a transient assay with maize leaf protoplasts to test the effects of individual segments of the promoter region on gene expression. The promoter segments of the Nipp (pOsglHAT1N) and Kasa (pOsglHAT1K) alleles were cloned into reporter constructs, and relative luciferase (LUC) expression was measured. Both OsglHAT1 promoter constructs led to significant increases of LUC expression relative to vector control alone, with an approximately twofold greater increase for pOsglHAT1K than pOsglHAT1N (Fig. 3B). Furthermore, we analyzed transgenic rice plants expressing the OsglHAT1 promoter segments fused with β-glucuronidase (GUS) reporter clones. Signals were much stronger in transgenic plants carrying the pOsglHAT1K::GUS clone than in those with the pOsglHAT1N::GUS clone (Fig. 3 C and D versus E and F). Quantification of these signals revealed that the Kasa construct signal was two to threefold higher than that of the Nipp construct (Fig. 3G). Thus, the promoter activity of the OsglHAT1 Kasa allele was relatively stronger than its counterpart in Nipp.
We further analyzed the specific expression patterns of OsglHAT1 through in situ hybridization. The OsglHAT1 mRNAs were expressed at the basal part of the abaxial side of leaves (Fig. 3 H and L) in the vegetative phase. A similar expression pattern was observed throughout the reproductive phase, whereas during the primary and secondary branch differentiation stages, OsglHAT1 mRNA accumulated in the bracts of initiating branches (Fig. 3 I, J, M, and N and SI Appendix, Fig. S7). In accordance with the GUS staining results, Kasa OsglHAT1 mRNA expression was markedly stronger than that of Nipp OsglHAT1 (Fig. 3 H–J versus L–N). In addition, we checked our data from progeny testing of NILs of the QTL; OsglHAT1 gene had a d/a (dominance deviation/additivity) of 0.14, which indicated that the large-grain allele for OsglHAT1 is semidominant to the small-grain allele. Together, these data suggest that changes at the transcription level cause the OsglHAT1 allelic phenotypic variation in grain weight, and confirm OsglHAT1 as a positive regulator of this trait.
OsglHAT1 Regulates Grain Weight, Yield, and Plant Biomass.
Quantitative analysis of grain shape components demonstrated that, relative to Nipp, NIL(OsglHAT1) has increased grain length (7.4%) and width (by 4.3%), with no change in grain thickness (SI Appendix, Fig. S8). Thus, OsglHAT1 regulates grain weight principally via regulation of grain length. Similarly, the spikelet hulls of NIL(OsglHAT1) were significantly longer at prefertilization than those of Nipp (4.2%, P = 1.39 × 10−5; Fig. 4 A and B). We next analyzed the longitudinal inner epidermal cell of lemmas by scanning electron microscopy (SEM) (Fig. 4C). The average cell length of NIL(OsglHAT1) (125.6 μm) did not differ significantly from that of Nipp (126.3 μm) (Fig. 4D). These data indicate that OsglHAT1 regulates grain weight through alteration of cell numbers.
Fig. 4.
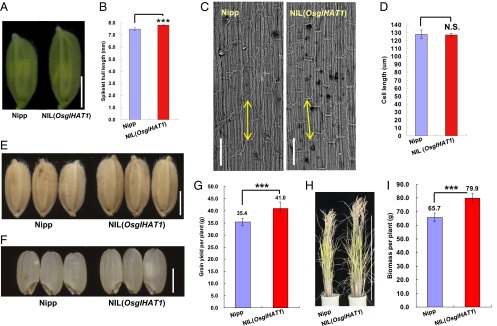
OsglHAT1 affects the number of cells in spikelet hulls and modulates grain yield and plant biomass. (A) Spikelet hull phenotypes used for SEM inspection. (B) Comparison of spikelet hull length between Nipp and NIL(OsglHAT1) at the same stage as A. (C) Histological examination in the central portion of inner epidermal cells of lemma by SEM. (Scale bars: 100 μm.) Double-headed arrows indicate cell lengths. (D) Comparison of inner epidermal cell length of Nipp (counted cells, n = 499) and NIL(OsglHAT1) (n = 496) lemmas. (E) Grain phenotypes. (F) Brown grain phenotypes. (G) Quantification and comparison of grain yield per plant. (H) Plant phenotypes of indicated plants at harvest. (I) Quantification and comparison of plant biomass per plant. ***P < 0.001; N.S., not significant. Data are given as the means ± SD, n > 20 plants in B, G, and I. Student’s t test was used to generate the P values.
We postulated that the larger spikelet hulls possibly facilitated grain milk filling, as seen with the GW2 gene (9). To test this postulation, we measured the fresh weight of brown grain at several time points after fertilization. No differences were observed at day 3 after fertilization (dpf); however, starting at 6 dpf, the fresh weight of brown grain of NIL(OsglHAT1) was significantly higher, and at 17 dpf was 16.7% greater, than that of Nipp (SI Appendix, Fig. S9). Thus, OsglHAT1 might play a role in dry matter accumulation during grain milk filling, thereby regulating grain weight.
We next assessed the effects of OsglHAT1 on grain production. In comparison with Nipp, NIL(OsglHAT1) had a significantly increased grain weight (+8.3%, P = 1.55 × 10−4; Fig. 4E and SI Appendix, Fig. S10A) and brown grain weight (+6.2%, P = 2.12 × 10−3; Fig. 4F and SI Appendix, Fig. S10B); other components of grain yield, such as grain number per main panicle (SI Appendix, Fig. S10C), and panicle number per plant (SI Appendix, Fig. S10D) showed no difference. As anticipated, the grain yields per NIL(OsglHAT1) plant increased by 15.8% (P < 0.05; SI Appendix, Fig. S4G). Moreover, the plants with pyramiding GW6a and GW6b loci (i.e., CSSL29) had a much greater grain yield potential (SI Appendix, Fig. S11A). Although the grain yield per plant determined in plants grown in paddy field under standard agronomic procedures (30 plants with three replicates) showed that NIL(OsglHAT1) could boost grain yield, we still need to carefully test this result in plots with randomized blocks in paddy field. Nevertheless, OsglHAT1 could be of value for grain yield improvement.
The finding that the plants of both NIL(OsglHAT1) and CSSL29 lines are taller than Nipp plants (Fig. 4H and SI Appendix, Figs. S10E and S11 B and C) prompted us to investigate whether OsglHAT1 also regulates plant height. Phenotypic analyses of the key recombinants suggested that this situation indeed is the case (SI Appendix, Fig. S12A). During early seeding stages, rice plants harboring the OsglHAT1-OE construct outgrew Nipp plants, whereas OsglHAT1-AS plants showed a markedly stunted growth (SI Appendix, Fig. S12 B and C). Thus, OsglHAT1 appears to control plant vegetative growth; importantly, when growing in paddies under standard cultivation conditions, NIL(OsglHAT1) and CSSL29 lines exhibited superior plant biomass compared with Nipp plants (Fig. 4I and SI Appendix, Fig. S11D). Collectively, these results suggest that OsglHAT1, in line with its ubiquitous expression pattern, has multiple effects on at least two beneficial agronomic traits—grain yield and plant biomass.
OsglHAT1 Is a Nuclear-Localized Histone H4 Acetyltransferase and Functions Presumably via Regulation of Gene Expression.
The OsglHAT1 protein contains a conserved segment Arg146-X-X-Gly149-X-Gly151 (i.e., R146-X-X-G149-X-G151, where X denotes variation) in its GNAT motif, which corresponds to the highly conserved acetyl-CoA (CoA) binding site of acetyltransferases (16). To test whether OsglHAT1 is an active histone acetyltransferase, we expressed a six-histidine (HIS) OsglHAT1 fusion protein in Escherichia coli (SI Appendix, Fig. S13), and subjected the purified OsglHAT1 protein to an in vitro HAT assay. We failed to detect any change of acetylation levels when free core histones were used as a substrate. However, when we used Xenopus chromatin as an alternative, the fusion OsglHAT1 protein showed the ability to enhance acetylation levels of Xenopus chromatin on histone H4, as did a typical HAT protein p300 (17) (SI Appendix, Fig. S14A). In addition, a smaller fragment (residues 1–165, HIS-OsglHAT1-N; SI Appendix, Fig. S13) that contains the conserved R146-X-X-G149-X-G151 segment could also acetylate chromatin histone H4, whereas a mutated version of OsglHAT1 protein (OsglHAT1-m (R146W); SI Appendix, Fig. S13) abolished its activity (SI Appendix, Fig. S14B). We also determined the substrate specificity of OsglHAT1 activity by Western blot using antibodies against specific acetylation sites in the histone H4 N-terminal tail (SI Appendix, Fig. S14A). In vitro acetylation by OsglHAT1 occurred preferentially at lysines 5, 12, and 16 of histone H4 (K5, K12, and K16; SI Appendix, Fig. S14A). By contrast, using nuclear protein extracts from plants at the reproductive stage, OsglHAT1 overexpression caused increased acetylation activity toward all four histone H4 lysine residues tested (SI Appendix, Fig. S14C). This discrepancy between the in vivo and in vitro assays suggests that OsglHAT1 may have associated partner proteins in vivo that increase its lysine acetylation spectrum, as has been demonstrated for Gcn5 (18). Collectively, these results suggest that OsglHAT1 is a histone H4 acetyltransferase.
Subcellular localization analysis using a green fluorescent protein (GFP)-OsglHAT1 fusion construct transiently expressed in onion epidermal cells revealed that GFP-OsglHAT1 localized to the nucleus (SI Appendix, Fig. S15), suggesting that it most likely catalyzes transcription-related acetylation events as proposed (19, 20). Thus, we compared the transcriptome of wild-type, GW6a-4.6, and OsglHAT1-OE samples by messenger RNA sequencing (RNA-seq). Hierarchical clustering, global correlation, and principal component analysis indicated that the samples were clearly separated by their genotypes, with Spearman correlation coefficients of 0.99 within biological replicates (SI Appendix, Fig. S16). Enhanced OsglHAT1 expression resulted in differential expression of 3,970 genes (false discovery rate < 0.05), of which 53.3% (2,117 genes) were up-regulated and 46.7% (1,853 genes) down-regulated (SI Appendix, Fig. S17 A and B and Dataset S1). Gene Ontology (GO) analysis showed significant enrichment in pathways related to transcription, stress, transport, protein metabolism, hormone response, and development (SI Appendix, Fig. S17 C and D), and enriched molecular functions including hydrolase, DNA binding, ATP binding, and transcription regulation (SI Appendix, Fig. S17 E and F and Dataset S2). As expected, there was up-regulation of genes involved in the cell cycle (P < 1.2 × 10−19), including G2- and S-phase genes (SI Appendix, Table database S1 and Dataset S1); this finding is consistent with OsglHAT1’s function in cell division (Fig. 4). Interestingly, we found that the expression of PGL2, a basic helix–loop–helix (bHLH) protein that positively regulates grain length (21), was activated by the OsglHAT1 transgenes. TH1/BSG1, a DUF640 domain-containing gene, was also clearly up-regulated, consistent with prior studies correlating deficiency of this gene with reduced grain size/weight (22–24) (SI Appendix, Table database S1 and Fig. S18A, and Dataset S1). Furthermore, we compared the relative expressions of another 12 previously identified grain-size genes among the samples (wild-type, GW6a-4.6, and OsglHAT1-OE) in our RNA-seq analyses, and the results revealed that 3 of these genes (i.e., GS5, SG1, and XIAO) were significantly up-regulated in the GW6a-4.6 genotype, whereas 5 genes (i.e., GS5, SG1, XIAO, GW8, and qSW5/GW5) were significantly up-regulated in the OsglHAT1-OE genotype in contrast to the wild type (10–13, 25, 26) (SI Appendix, Fig. S18B). Collectively, these results support the notion that OsglHAT1 functions as a transcription regulator.
The Rare Allele Elevating OsglHAT1 Expression Has So Far Escaped Human Selection.
Previous studies have shown that transcriptional regulators are central players in domestication (27). We therefore examined whether OsglHAT1 had been the target of human selection during rice domestication and modern breeding, by analyzing genetic variations at three sites: the OsglHAT1 promoter in a representative set of O. sativa and O. rufipogon (28) (SI Appendix, Table S3), as well as the regions ∼50 kb upstream and ∼60 kb downstream of this gene. Analyses of nucleotide diversity and coalescent simulation revealed no signature of selection (SI Appendix, Table S2), indicating that the advantages conferred by the OsglHAT1 alleles have not been actively exploited. The Kasa allele was not found in any of the japonica cultivars tested, whereas it was present in 26 of 50 indica cultivars; additionally, geographical distributions showed no biases for the locations in which the indica alleles of OsglHAT1 were found. Thus, we propose that the OsglHAT1 allele could be used to improve agronomic traits in crops, especially in japonica cultivars.
Sequence blast analysis against public databases identified 59 putative OsglHAT1 homologs, including one known gene, HOOKLESS1 (AtHLS1, At4G37580), that functions in differential cell elongation in the Arabidopsis hypocotyls (29), although biochemical features and functional analyses of AtHLS1 have not yet been reported. We found that OsglHAT1 homologs were restricted to the plant kingdom and are found within several important crop species including maize (Zea mays), soybean (Glycine max), sorghum (Sorghum bicolor), and rapeseed (Brassica napus). Phylogenetic analysis of these homologs suggests that, unlike AtHLS1, OsglHAT1 appears to function as a representative member of an undefined subclass of GNAT-like proteins (SI Appendix, Fig. S19). Given that our studies showed effects in both rice and Arabidopsis, it is plausible that OsglHAT1 homologs could be tailored to improve agronomic traits in other crop species.
Materials and Methods
We roughly mapped the GW QTL by using a BIL set derived from Nipponbare and Kasalath, and then chose CSSL29 that possessed the introgressed segment of chromosome 6 from Kasalath and crossed with Nipponbare to produce a F2 population and derived F3 or F4 population for QTL genetic mapping. Gene expression analyses were conducted by semiquantitative RT-PCR and qPCR by using gene specific primers, and in situ RNA hybridization experiments. The intrinsic HAT activities of OsglHAT1 were confirmed by using in vitro and in vivo HAT assays. Microscopic inspections of inner epidermal cell of lemmas of spikelet hulls were observed by SEM. A transient assay with maize leaf protoplasts was performed to assess the effects of individual control segment. An RNA-Seq experiment that compared the transcriptomes of the OsglHAT1 transgenes with that of Nipponbare was performed to support that OsglHAT1 functions as a transcription regulator and to investigate its possible downstream genes. Genetic diverity and coalescent simulation analyses were conducted by using a diverse set of rice accessions to examine whether OsglHAT1 was the target of human selection. Details of all of the experiments performed in this paper and any associated references are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank N. Ueda for making the transgenic Arabidopsis plants, K. Yano for providing the protocol for the qPCR analysis, J. Kyozuka for suggestions on in situ hybridization, H. Tagami for providing proten materials, and S. Mizuno for maintaining the paddy field. This work was supported mainly by the Program for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, in part by Grants in Aid for Scientific Research 22119007 (to M. Ashikari) from the Ministry of Education, Culture, Sports, Science, by the Japan Science and Technology (JST) Agency-Japan International Cooperation Agency within the framework of the Science and Technology Research Partnership for Sustainable Development (SATREPS) (M. Ashikari) and by the Core Research for Evolutional Science and Technology, JST. This work was supported in part by the Funding Program for Next Generation World-Leading Researchers NEXT Program GS-024 (to K.M.). S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: OsglHAT1 sequence data have been deposited in DNA Data Bank of Japan nucleotide core database (accession nos. LC003015–LC003018) and the GEO database (accession no. GSE62554).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421127112/-/DCSupplemental.
References
- 1.Khush GS. Green revolution: The way forward. Nat Rev Genet. 2001;2(10):815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- 2.Li J, et al. 2012. The rice HGW gene encodes a ubiquitin-associated (UBA) domain protein that regulates heading date and grain weight. PLoS One 7(3):e34231.
- 3.Yoshida S. Physiological aspect of grain yield. Annu Rev Plant Physiol. 1972;23:437–464. [Google Scholar]
- 4.Fan C, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112(6):1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 5.Mao H, et al. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA. 2010;107(45):19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41(4):494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 7.Qi P, et al. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22(12):1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA. 2012;109(52):21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39(5):623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 10.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40(8):1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 11.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18(12):1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43(12):1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 14.Ishimaru K, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45(6):707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 15.Lin S, Sasaki T, Yano M. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet. 1998;96:997–1003. [Google Scholar]
- 16.Wolf E, et al. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94(4):439–449. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 17.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87(5):953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 18.Grant PA, et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274(9):5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 19.Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92(14):6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 21.Heang D, Sassa H. An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed Sci. 2012;62(2):133–141. doi: 10.1270/jsbbs.62.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol Biol. 2012;78(4-5):351–359. doi: 10.1007/s11103-011-9868-8. [DOI] [PubMed] [Google Scholar]
- 24.Yan D, et al. Beak-shaped grain 1/TRIANGULAR HULL 1, a DUF640 gene, is associated with grain shape, size and weight in rice. Sci China Life Sci. 2013;56(3):275–283. doi: 10.1007/s11427-013-4449-5. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa H, et al. Short grain1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 2012;158(3):1208–1219. doi: 10.1104/pp.111.187567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, et al. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 2012;70(3):398–408. doi: 10.1111/j.1365-313X.2011.04877.x. [DOI] [PubMed] [Google Scholar]
- 27.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127(7):1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Asano K, et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci USA. 2011;108(27):11034–11039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85(2):183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.